Abstract
Staphylococcus saprophyticus is a uropathogenic Staphylococcus frequently isolated from young female outpatients presenting with uncomplicated urinary tract infections. We sequenced the whole genome of S. saprophyticus type strain ATCC 15305, which harbors a circular chromosome of 2,516,575 bp with 2,446 ORFs and two plasmids. Comparative genomic analyses with the strains of two other species, Staphylococcus aureus and Staphylococcus epidermidis, as well as experimental data, revealed the following characteristics of the S. saprophyticus genome. S. saprophyticus does not possess any virulence factors found in S. aureus, such as coagulase, enterotoxins, exoenzymes, and extracellular matrix-binding proteins, although it does have a remarkable paralog expansion of transport systems related to highly variable ion contents in the urinary environment. A further unique feature is that only a single ORF is predictable as a cell wall-anchored protein, and it shows positive hemagglutination and adherence to human bladder cell associated with initial colonization in the urinary tract. It also shows significantly high urease activity in S. saprophyticus. The uropathogenicity of S. saprophyticus can be attributed to its genome that is needed for its survival in the human urinary tract by means of novel cell wall-anchored adhesin and redundant uro-adaptive transport systems, together with urease.
Keywords: hemagglutination, paralog expansion, divalent-cation transporter, capsule, staphylococcal cassette chromosome
Staphylococcus belongs to the Gram-positive low GC content group of the Firmicutes division of bacteria. Three species, Staphylococcus aureus, Staphylococcus epidermidis, and Staphylococcus saprophyticus, are recognized as major human pathogens. S. saprophyticus is a coagulase-negative Staphylococcus and frequently causes uncomplicated urinary tract infection (UTI) in young and middle-aged female outpatients (1-4). S. saprophyticus is notable uropathogen without the involvement of indwelling catheters, whereas two other staphylococci are often clinically isolated from hospitalized patients who have indwelling catheters rather than from outpatients (5). This finding indicates that S. saprophyticus has a potential ability to adhere to and grow persistently in the urinary tract.
Besides S. saprophyticus is a leading Gram-positive uropathogen of uncomplicated UTI, a restricted group of Gram-negative bacteria, including Escherichia coli, Proteus mirabilis, and Klebsiella spp. is often observed to cause uncomplicated UTI (4). These bacteria possess surface structures known as fimbriae or pili responsible for mediating the adherence to uroepithelial cells (6). A recent genomic analysis of uropathogenic E. coli CFT073 revealed that additional fimbriae loci seemed to be acquired during evolution to facilitate the colonization in urinary tracts (7). Thus, such specific adhesin should play a crucial role in the ascension of uropathogenic bacteria through the urinary tract. In this regard, most strains of S. saprophyticus show strong adherence to uroepithelial cells and sheep erythrocytes causing hemagglutination (8). In addition to adherence, urease has been also reported to play a role in the persistent growth and invasiveness of the bacteria in the bladder (9). Urease of P. mirabilis and Klebsiella spp. has been also implicated as a uropathogenic factor that prompts the persistent growth and renal stone formation (10, 11). Because uroepithelial adhesion and urease have been implicated as major virulence factors for UTI, it has been suspected that they are more effective in S. saprophyticus than in other staphylococci and Gram-positive bacteria (9, 12).
Although the whole genome sequence of uropathogenic E. coli CFT073 has been completed (7), as yet, little genome information for uropathogenic bacteria such as P. mirabilis and Klebsiella spp. is available. Because the whole genome sequences of S. aureus and S. epidermidis strains have been made available (13-16), comparative genomic analyses of S. saprophyticus with these species as well as with other uropathogenic bacteria are expected to elucidate the specific characteristics of the factors in this leading Gram-positive bacterium, which causes uncomplicated UTI. Here we present the whole genome sequence of S. saprophyticus type strain ATCC 15305 and elucidate the uropathogenesis of this strain with some experimental demonstrations.
Methods
Supporting Information. For further details, see Figs. 6-8 and Tables 2-6, which are published as supporting information on the PNAS web site.
Genome Sequencing and Assembly. A S. saprophyticus GTC 265 strain that is equivalent to type strain ATCC 15305 was obtained from the Japanese Society for Bacteriology (Tokyo). Type strain ATCC 15305 was isolated from a human urine specimen (17). The genome sequence was determined by a whole-genome shotgun strategy with 44,160 reads of two libraries comprising 1- to 2-kb or 10-kb inserts. All of the data were assembled with phred/phrap/consed programs (18), giving 9.4-fold redundancy. Remaining gaps were closed by PCR direct sequencing with specific primers at the ends of each contig. Accuracy of the finished sequence was estimated to have an error of <1 per 10,000 bases (phrap score ≥40). The complete sequence agreed with the digestion profiles using AscI, RsrII, SgrA, and SmaI on the pulsed-field gel electrophoresis.
Annotation and Functional Assignment. Putative ORFs were predicted by the combination of genome gambler 1.51 (19) and glimmer 2.0 programs (20). The 807 orthologs of S. aureus N315 strain showing high similarity to the proteins in the cluster of orthologous groups of proteins (COGs) (21) were used as a training data set for initial ORF prediction by glimmer 2.0. Remaining ORFs ≥30 aa were extracted by genome gambler 1.51 between ORFs, and then possible ORFs, having potential initiation codons (ATG, TTG, and GTG) and a ribosome-binding site, were included in the former prediction.
Functional assignments of the predicted ORFs were based on a blastp homology search against the nonredundant (nr) protein database and COGs (21). Putative tRNAs were identified by trnascan-se (22). Putative localization of predicted proteins was evaluated by the combination of signalp and psort programs. Predicted cell wall-anchored protein was identified by the existence of the N-terminal signal peptide and C-terminal cell wall sorting signal (23). The complete sequences and annotation of the S. saprophyticus ATCC 15305 genome and the two plasmids have been deposited in DDBJ (accession nos.: AP008934 for the chromosome, AP008935 and AP008936 for the plasmids pSSP1 and pSSP2, respectively).
Comparative Genomics. The genome sequences of S. aureus N315 (13) and of S. epidermidis ATCC 12228 (15) were obtained through the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov). Homologous genes were identified by homology searches in an amino acid sequence by using the blastp filtering expectation value of e < 10-10 (24). When the best hit was identical with the query, the ORFs were regarded as being orthologous genes. Paralogous genes were defined by using blastclust with ≥30% identity in the amino acid sequence and ≥70% in their length (24). Multiple sequence alignment was performed by using clustalw with 1,000 times bootstrapping (25). mega3 software was used for displaying the phylogenetic tree of aligned sequences (26).
Bacterial Strains for the Adherence Assays. A DNA fragment containing the whole SSP0135 ORF (uafA) was amplified by PCR using Elongase (Invitrogen). Primers contained BamHI restriction sites at both of the 5′ ends (5′-AAAAAggatccAAGAAATGTATTGTGCTAAAAGGAA-3′ and 5′-AAAAAggatccTCAGTCATTTGCTACTTAGTAGGT-3′; italics indicate the BamHI site). The PCR product was cloned into the BamHI site of shuttle vector pRIT5H. The resultant plasmid pRLC(+) was electroporated into S. aureus RN4220 (27), and selected on brain heart infusion agar with chloramphenicol.
The SCC15305cap excised mutant C1 was obtained by the screening of primary adherence to a polystyrene surface from the serially passaged culture of ATCC 15305. The excised junction was determined by PCR using primers (5′-AAAGCATTAGCACATCCAAA-3′ and 5′-CAATTCCTTCTGTTACCTTT-3′). The uafA gene (SSP0135) deleted mutant G5 was obtained as nonhemagglutination derivatives from the serially passaged culture of ATCC 15305. The results of pulsed-field gel electrophoresis analysis and Southern hybridization on both strains are shown in Fig. 6.
Hemagglutination Assay. Bacteria harboring the plasmid were grown in Muller-Hinton broth supplemented with chloramphenicol for 16 h at 37°C. The cell was washed twice with 0.9% sodium chloride and resuspended with it at an OD600 of 1.0. Hemagglutination assay was performed by mixing an equal volume of washed cells and 2% sheep erythrocyte suspension on a 96-well U-bottom titer plate. The mixture was kept at a static condition at room temperature for 2 h.
Bacterial Adherence Assay to Tissue Culture Cells. Human T24 bladder carcinoma cells were maintained in culture by using DMEM (high glucose) supplemented with 10% heat-inactivated FCS at 37°C in 5% carbon dioxide. Bacteria cells were grown in Muller-Hinton broth at 37°C overnight, followed by washing and suspending with PBS. The cultivated T24 cells (90% confluent growth) on 12-well tissue culture plate was washed with PBS once and incubated with 500 μl of the bacterial suspension containing 1 × 106 colony-forming units for 15 min at 25°C with gentle agitation. The supernatant was removed, and the wells were washed with 1 ml PBS three times. The T24 cells were lysed vigorously in 1 ml of prechilled sterilized water. Adherent bacteria were counted by plating on tryptic soy agar with serial dilutions. Statistical difference of the adherence was calculated by Student's t test using the quadruplicate experiments.
Results and Discussion
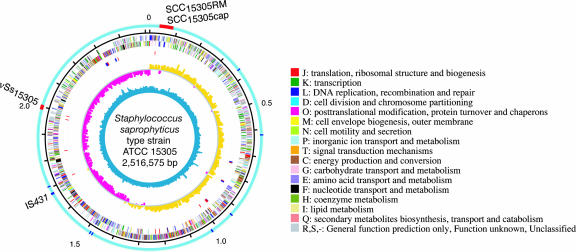
General and Specific Features of the S. saprophyticus Genome. The length of the whole genome sequence of S. saprophyticus is 2,516,575 bp with G+C content of 33.2% similar to those of other staphylococci (Fig. 1). The genome contains six ribosomal RNA operons and 60 tRNAs for all amino acids. Its chromosomal origin and terminus determined by GC-deviation (G-C/G+C) locate to the position similar to those of S. aureus and S. epidermidis. The genome contains 2,446 ORFs and a variety of mobile genetic elements including one 39.3-kb prophage remnant, two IS431 elements, nine putative transposases, two staphylococcal cassette chromosomes (SCCs, designated SCC15305RM and SCC15305cap), and one genomic island (designated νSs15305). The strain has two plasmids (designated pSSP1 and pSSP2) that are 38.4 and 22.9 kb in size, respectively. Comparison of the general features of S. saprophyticus with those of other staphylococci genomes sequenced to date was shown in Table 2.
Fig. 1.
Schematic circular diagrams of the S. saprophyticus ATCC 15305 chromosome. From the outside, the seven circles display: (i) possible horizontally acquired genomic islands (red) and IS elements (blue) on the light blue outer ring; (ii) scale in Mbp; predicted coding regions transcribed in the clockwise (iii) or counterclockwise (iv) direction, and colored according to the category of COGs shown beside the circular diagrams; (v) tRNA (red) and rRNA (blue); (vi) GC deviation (G-C/G+C); (vii) G+C content.
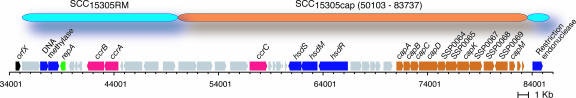
Most methicillin-resistant S. aureus horizontally acquired some types of SCCmec involved in multiple antibiotic resistance (28, 29). Instead of antibiotic resistance, S. saprophyticus SCC15305cap carries a capsular polysaccharide synthesis locus consisting of ORFs with similarity to several distinct lineages as in a mosaic organization, including some of orthologs to S. aureus-type I Cap enzymes, three additional putative glycosyl transferase, and a putative polysaccharide polymerase (Fig. 2). SCC15305RM carries neither a virulence factor nor an antibiotic resistance determinant. These SCCs seem to be individually integrated into the genome in a two-step insertion manner (Fig. 2) and commonly carry two remarkable features: the restriction-modification system and the cassette chromosome recombinase (Ccr). The SCCs might be stabilized after Ccr-mediating integration by the selfish restriction-modification system (30), and disseminate such accessory pathogenicity as well as antibiotic resistance among staphylococci.
Fig. 2.
Mobile genomic islands of S. saprophyticus ATCC 15305. SCC15305cap was integrated into the potential attachment site on SCC15305RM as a nested structure.
Besides SCC, the genomic island νSs15305 associated with antibiotic resistance against streptomycin (SSP1924) and fosfomycin (SSP1926) was identified. Although cognate genomic islands are widely found to carry enterotoxins and toxic shock syndrome toxin-1 genes on the staphylococcal genome (13, 14, 28, 31), this island carries antibiotic resistance determinants instead of virulence factors.
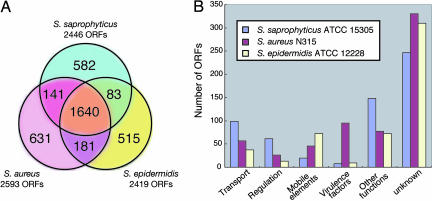
Comparative genomic analyses of S. saprophyticus with S. aureus and S. epidermidis identified the 582 (23.8%) S. saprophyticus-specific ORFs (Fig. 3A). Those species-specific ORFs are classified according to the COGs category (see detailed category description shown in Fig. 1). The categories of transcription (K), energy production/conversion (C), and carbohydrate and amino acids transport/metabolism (G and E) are significantly abundant in S. saprophyticus-specific ORFs (see Fig. 7). Because the COGs category is classified as physiological functions, those species-specific ORFs are separately classified as protein functions by using the pfam databases (Fig. 3B). The ORFs involved in transport and regulation systems are relatively abundant, whereas the ORFs involved in mobile elements and virulence factors are much less when compared with those in S. aureus and S. epidermidis. All of the virulence factors of S. aureus, such as coagulase, hemolysins, enterotoxins, extracellular matrix-binding proteins, and exoenzymes, are absent in the S. saprophyticus genome. In fact, the actual activities of exoenzymes described above were not detected (data not shown). Instead, the S. saprophyticus genome information reveals the presence of a cell wall-anchored protein and unique paralog expansion of transport systems related to ion contents of urine. The specific features of uropathogenicity are described in detail below.
Fig. 3.
Orthologous classification of predicted ORFs compared with those of S. aureus N315 and S. epidermidis ATCC 12228. (A) Orthologs were defined by BLAST reciprocal best hit with an e-value cutoff of e-10. (B) Classification of nonorthologous ORFs based on a motif search using the COGs and pfam databases. See Tables 3-5.
Adherent Phenotype by the Cell Wall-Anchored Hemagglutinin. S. saprophyticus ATCC 15305 shows hemagglutination that seems to be mediated by its cell wall-anchored or associated surface proteins (12). Most staphylococcal extracellular matrix-binding proteins are anchored to peptidoglycan mediated by sortase A that recognizes the LPXTG amino acids motif (32). Intriguingly, a single unique ORF (SSP0135) can be predicted as a sortase A-mediating cell wall-anchored protein in S. saprophyticus, even though at least 20 and 11 cell wall-anchored proteins can be predicted in S. aureus and S. epidermidis, respectively (23). This finding may account for the distinct colonization manner and virulence of S. saprophyticus, except for UTI, from those of other staphylococci.
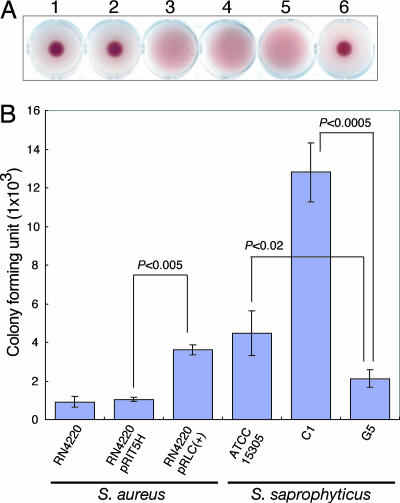
SSP0135 is the largest of all predicted ORFs on the genome, composed of 2,316 amino acids, with a unique 6-aa imperfect tandem repeat (SESESL) from the position of 812 to 2,263 aa that shows no significant similarity to any other known proteins. Overexpression of SSP0135 protein in S. aureus RN4220, a restriction-deficient mutant of strain 8325-4 with nonhemagglutination, shows hemagglutination with sheep erythrocytes, as does S. saprophyticus ATCC 15305 (Fig. 4A). S. saprophyticus hemagglutination has been reported to correlate well with the level of adherence to human ureteral epithelium (12, 33). Indeed, SSP0135 overexpressed S. aureus RN4220 adheres well to human T24 bladder carcinoma cells, and S. saprophyticus G5 strain (ΔSSP0135) causes no hemagglutination and the reduced adherent phenotype compared with other hemagglutination-positive strains (Fig. 4B). Interestingly, the SCC15305cap rescued C1 strain shows significantly higher hemagglutination titer (data not shown) and adherence to T24 cell than ATCC 15305 (Fig. 4B), implying that the polysaccharide may cover the cell surface of S. saprophyticus, resulting in the inhibition of the SSP0135-mediated adhesion to T24 cell. These results revealed that S. saprophyticus has a specific adhesin UafA (SSP0135, standing for uro-adherence factor A) associated with adherence to the eukaryotic cell in the urinary tract.
Fig. 4.
Adherence to eukaryotic cell by UafA (SSP0135). (A) Cell wall-anchored protein encoded by SSP0135 is involved in hemagglutination. The positive control S. saprophyticus ATCC 15305 (well 4) and SSP0135 introduced S. aureus RN4220 (well 3) showed hemagglutination, whereas parental strain RN4220 (well 1) and RN4220 harboring control vector pRIT5H (well 2) did not. S. saprophyticus C1 (ΔSCC15305cap) shows hemagglutination (well 5), but the uafA gene (SSP0135) deleted mutant G5 (well 6) did not. (B) Adherence assay to human T24 bladder carcinoma cell. The detailed procedure was shown in Methods. The number of adherent bacteria was expressed as the means ± SD for quadruplicate experiments. Significant difference (P < 0.02) in the adherent phenotype was measured by Student's t test.
Paralog Expansion of Transporter Systems for the Adaptation to Urine. Comparative genomic analysis revealed that S. saprophyticus has abundant transporter systems to facilitate adaptation to human urine by paralog expansion, in which the pH, osmolarity, concentration of urea, inorganic ions, and other organic substances are unstable and variable. The noteworthy transporter paralog expansion is listed in Table 1.
Table 1. Paralog expansion of transport systems related to urine environment.
| Protein function | Common ortholog* | Additional paralog† |
|---|---|---|
| Osmotolerance | ||
| Proline/betaine transporter ProP | SSP2142 | SSP0211 |
| High-affinity proline permease PutP | SSP0889 | SSP0399 |
| Glycine betaine/choline transporter | SSP1409 | SSP1344 |
| SSP0701 | SSP0323 | |
| SSP0193 | ||
| Proline/glycine betaine ABC transporter | ||
| ATPase component OpuCA/CB/CD | SSP1997 | SSP0451 |
| SSP0452 | SSP0454 | |
| Periprasmic component OpuCC | SSP0453 | SSP0993 |
| Water channel AqpZ | SSPP142‡ | |
| SSPP212‡ | ||
| Inorganic ion transport | ||
| Divalent cation transporter NRAMP§ | SSP1158 | SSP2176 |
| SSP1685 | ||
| Na+/H+ antiporter NhaC | SSP0612 | SSP0329 |
| SSP0578 | SSP0560 | |
| Sodium/sulfate symporter | SSP0875 | SSP0304 |
| SSP0395 | ||
| Sulfate symporter | SSP0298 | |
| SSP0574 |
Osmotolerance is one of the characteristic features of staphylococci. The three compared staphylococci basically share orthologs that function for importing osomoprotectants such as proline, choline, and glycine betaine. In addition to such orthologs, S. saprophyticus remarkably possesses additional sets of osomoprotectant transport systems generated by paralog expanding on the genome (Table 1). Such paralog expansion may contribute to the prompt balancing of intracellular osmotic pressure against highly variable osmotic change (≈50-1,400 mOsm per kg of H2O) in urine by using multiple driven-motive forces with those coordinated actions. Furthermore, two plasmids pSSP1 and pSSP2 carry the putative water channel aquaporin Z gene (aqpZ) that might contribute to osmotic balancing. Multicopy number of plasmids may give an advantage to S. saprophyticus to express more water channels, and has the similar dose-dependent effect to paralog expanding.
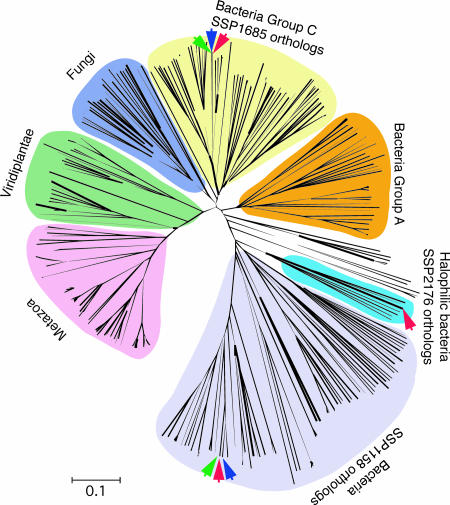
Iron is an absolute requirement for bacterial survival. Most uropathogenic bacteria including E. coli and Klebsiella spp. use iron-acquiring systems such as siderophores and iron transporters. Proteus spp. do not produce common siderophores but can acquire iron with α-keto acids, phenylpyruvate, and indolpyruvate (34). S. aureus produces the unique siderophore staphylobactin, but S. saprophyticus has neither common siderophores nor α-keto acid siderophore-producing systems. One of the probable candidates for the iron acquisition system in S. saprophyticus is the NRAMP (natural-resistance-associated macrophage protein) family involved in the transport of divalent metals (35). Three homologues of the NRAMP family were found in S. saprophyticus (Table 1). These seem to show distinct pylogenetical origins in the phylogenetic tree inferred from the ortholog comparison, including their mammalian counterparts (Fig. 5). SSP1158 and SSP1685 are similar to other staphylococcal homologues, whereas SSP2176 belong to the clearly distinct group that includes orthologs of halophilic and archaeal bacteria isolated from marine. A functional motif search also suggests that SSP2176 is a sodium-dependent symporter, whereas SSP1685 counterparts are pH-dependent symporters. This observation suggests that sodium ion is a more favorable solute for cotransporting divalent cations by SSP2176. Basically, the mammalian NRAMP family acts in a pH-dependent manner in the bacterial phagolysosome of a macrophage to deplete free available divalent cations, and the bacterial counterparts manage to compete for the uptake of divalent cations in the same pH-dependent manner under acidic pH conditions. In contrast, in sodium-rich urine, the sodium-driven motive force should be a more effective source than other ions. Thus, the sodium-dependent symporter SSP2176 seems to contribute to the sufficient acquisition of divalent cations, including iron, to grow in hyperosmotic urine without siderophores.
Fig. 5.
The phylogenetic tree shows polyphyletic origins for bacterial orthologs of the NRAMP family. S. saprophyticus ATCC 15305 carries three paralogs shown as a red arrow. SSP2176 is classified as a notable group including halophilic bacteria isolated from marine, deep-sea, or Dead Sea. Orthologs of S. aureus and S. epidermidis are indicated as green and blue arrows, respectively. The analyzed 336 orthologs showing significant similarity (identity ≥30% and alignment length ≥60%) were obtained from the nonredundant database. The scale of 0.1 amino acid replacement per site is indicated. Individual references of orthologs are provided in Table 6.
Sodium and sulfate ions are rich component of urine, and a unique paralog expansion of these ion transporters was also found (Table 1). Two additional Na+/H+ antiporters of the NhaC family involved in alkali tolerance may contribute to intracellular pH homeostasis by uptake of proton. Three additional sulfate transporters were identified and likely maintain the adequate utilization of sulfate ion.
Intriguingly, S. saprophyticus lacks the potassium-importing ATPase KdpABC and the responsible two-component system KdpDE, suggesting that a large amount of potassium ion in urine might be able to support the bacterial growth without the active uptake system. In the case of S. aureus N315 and Mu50, they have extra paralogs of KdpABC and KdpDE on mobile element SCCmec (13) in addition to chromosomal genes. This finding suggests that acquisition of potassium ion could be an important pathogenesis for S. aureus causing diseases in a potassium-limited environment. It may also explain one reason why S. saprophyticus very rarely causes severe infectious diseases.
Because all of the above-described paralogs of transporters specifically act on the ion contents in urine, our findings suggest that S. saprophyticus exclusively requires osomoregulation and ion transport systems in terms of the redundancy and diversity to grow rapidly in the urinary tract before being washed out by micturition.
Nitrogen Metabolism Is Involved in Persistent Growth in Urinary Tracts. S. saprophyticus can grow on a medium containing ammonium sulfate as the sole nitrogen source, but cannot obtain ammonium by nitrate/nitrite reduction because of the lack of respiratory nitrate reductase NarGHI and assimilatory nitrite reductase NasDE on the genome. The organism can use ammonia that is derived from urea hydrolyzed by urease and transported by the ammonium transporter. The urease of S. saprophyticus is known to be a virulent factor for the persistent infection in the urinary tract (9), as in Proteus mirabilis (36) and Klebsiella pneumoniae (37) but not in E. coli. The urease activity of S. saprophyticus is significantly higher and was detected 2 h earlier on Christensen urea agar and by quantitative measurement of ammonium than the other two Staphylococcus species, the activities of which were detected only after 24 h of incubation (see Fig. 8). The activity was inhibited by the potent urease inhibitor acetohydroxamate. Although the three staphylococci species possess the urease operon composed of the ureAB-CEFGD gene cluster in the same orientation of the chromosome, the expression regulation system or the activation mechanism seems to be different in S. saprophyticus than the other two. Urease production at the early proliferation stage could be an important event for the organism to cause infection. The detailed regulatory mechanism of urease remains to be elucidated.
Staphylococcal Virulence Regulatory Systems. The three staphylococci share 12 two-component systems (TCS), which are responsible for cell morphology, cell wall biosynthesis, autolysis, exoprotease expression, respiration, and uptake of phosphate. The agr locus is a quorum-sensing TCS modulating the multiple regulation of virulence factors in S. aureus (38). Interestingly, the system is also well conserved in S. saprophyticus, whereas it does not have the typical virulence factors carried in S. aureus described above. Unlike S. aureus, S. saprophyticus lacks the five TCSs including the above described potassium homeostasis, the regulation of coagulase and hemolysins, and uncharacterized others. Instead, S. saprophyticus has more regulatory genes as specific ORFs (Fig. 3B). It also has more putative metabolic enzymes, as well as a transport system, as seen in the S. saprophyticus-specific ORFs (Fig. 3B). Thus, the abundant regulation systems may act individually for the modulation of those transport systems and metabolism, and may give an advantage for a prompt and individual response in metabolite-rich urine.
Conclusions
We sequenced the whole genome sequence of S. saprophyticus type strain ATCC 15305, and the genome analysis suggests why S. saprophyticus causes uncomplicated UTI but no severe bacterial infectious disease as caused by S. aureus. S. saprophyticus does not have the extracellular matrix-binding proteins possessed by S. aureus or S. epidermidis, but does have a single and unique cell wall-anchored protein involved in the adherence to uroepithelial cells. Further intriguing findings are the high population of transport systems due to expansion of the paralogs involved in osomotolerance and inorganic ions transport related to the highly variable ion contents of urine. Such paralog expansion might result in facilitating rapid growth in the urinary tract by means of abundant and overall transport systems and persistent growth after the establishment of initial attachment with cell wall-anchored adhesin. The high urease activity of S. saprophyticus may also contribute to persistent growth, despite our finding that the gene organization is very similar to those of other staphylococci, implying that the specific regulation system may be involved in urease expression. Our observations suggest that S. saprophyticus may have evolved to adapt specifically to the urinary environment by exhibiting a notable ability for adherence and rapid growth in the urinary tract. The analysis of the genome sequence of S. saprophyticus reveals it to be a leading species among Gram-positive bacteria in the uropathogenesis of uncomplicated UTI.
Supplementary Material
Acknowledgments
We are grateful to Drs. Naotake Ogasawara, Hideo Hayashi, Tetsuya Hayashi, and Takeshi Shimizu for their suggestions in the project. We also thank K. Furuya, C. Yoshino, A. Nakazawa, Y. Yamashita, and N. Ito for technical assistance. Human T24 bladder carcinoma cell was kindly given by Dr. Kaoru Takeuchi (University of Tsukuba). This study was supported by Research for the Future Program of the Japan Society for the Promotion of Science Grants JSPS-RFTF 00L01411 and JSPS-RFTF 00L01412. This work was also supported in part by Grant of the 21st Century Centers of Excellence Program, Ministry of Education, Culture, Sports, Science and Technology, Japan (to Kitasato University).
Author contributions: M. Kuroda, S.K., M. Hattori, and T.O. designed research; M. Kuroda, A.Y., M. Kumano, K. Morikawa, M. Higashide, A.M., Y.I., K. Matoba, and H.T. performed research; M. Kuroda, A.Y., H.H., H.T., and S.K. analyzed data; M. Kuroda and T.O. wrote the paper; and A.Y., H.T., and M. Hattori performed whole genome sequencing and assembly.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: UTI, urinary tract infection; COG, cluster of orthologous groups; SCC, staphylococcal cassette chromosome.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AP008934-AP008936).
References
- 1.Torres Pereira, A. (1962) J. Clin. Pathol. 15, 252-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meers, P. D., Whyte, W. & Sandys, G. (1975) J. Clin. Pathol. 28, 270-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallmark, G., Arremark, I. & Telander, B. (1978) J. Infect. Dis. 138, 791-797. [DOI] [PubMed] [Google Scholar]
- 4.Kahlmeter, G. (2003) J. Antimicrob. Chemother. 51, 69-76. [DOI] [PubMed] [Google Scholar]
- 5.von Eiff, C., Peters, G. & Heilmann, C. (2002) Lancet Infect. Dis. 2, 677-685. [DOI] [PubMed] [Google Scholar]
- 6.Anderson, G. G., Dodson, K. W., Hooton, T. M. & Hultgren, S. J. (2004) Trends Microbiol. 12, 424-430. [DOI] [PubMed] [Google Scholar]
- 7.Welch, R. A., Burland, V., Plunkett, G., III, Redford, P., Roesch, P., Rasko, D., Buckles, E. L., Liou, S. R., Boutin, A., Hackett, J., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hell, W., Meyer, H. G. & Gatermann, S. G. (1998) Mol. Microbiol. 29, 871-881. [DOI] [PubMed] [Google Scholar]
- 9.Gatermann, S., John, J. & Marre, R. (1989) Infect. Immun. 57, 110-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones, B. D., Lockatell, C. V., Johnson, D. E., Warren, J. W. & Mobley, H. L. (1990) Infect. Immun. 58, 1120-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Podschun, R., Sievers, D., Fischer, A. & Ullmann, U. (1993) J. Infect. Dis. 168, 1415-1421. [DOI] [PubMed] [Google Scholar]
- 12.Gatermann, S., Marre, R., Heesemann, J. & Henkel, W. (1988) FEMS Microbiol. Immunol. 1, 179-185. [DOI] [PubMed] [Google Scholar]
- 13.Kuroda, M., Ohta, T., Uchiyama, I., Baba, T., Yuzawa, H., Kobayashi, I., Cui, L., Oguchi, A., Aoki, K., Nagai, Y., et al. (2001) Lancet 357, 1225-1240. [DOI] [PubMed] [Google Scholar]
- 14.Baba, T., Takeuchi, F., Kuroda, M., Yuzawa, H., Aoki, K., Oguchi, A., Nagai, Y., Iwama, N., Asano, K., Naimi, T., et al. (2002) Lancet 359, 1819-1827. [DOI] [PubMed] [Google Scholar]
- 15.Zhang, Y. Q., Ren, S. X., Li, H. L., Wang, Y. X., Fu, G., Yang, J., Qin, Z. Q., Miao, Y. G., Wang, W. Y., Chen, R. S., et al. (2003) Mol. Microbiol. 49, 1577-1593. [DOI] [PubMed] [Google Scholar]
- 16.Holden, M. T., Feil, E. J., Lindsay, J. A., Peacock, S. J., Day, N. P., Enright, M. C., Foster, T. J., Moore, C. E., Hurst, L., Atkin, R., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 9786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaw, C., Stitt, J. M. & Cowan, S. T. (1951) J. Gen. Microbiol. 5, 1010-1023. [DOI] [PubMed] [Google Scholar]
- 18.Gordon, D., Desmarais, C. & Green, P. (2001) Genome Res. 11, 614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakiyama, T., Takami, H., Ogasawara, N., Kuhara, S., Kozuki, T., Doga, K., Ohyama, A. & Horikoshi, K. (2000) Biosci. Biotechnol. Biochem. 64, 670-673. [DOI] [PubMed] [Google Scholar]
- 20.Delcher, A. L., Harmon, D., Kasif, S., White, O. & Salzberg, S. L. (1999) Nucleic Acids Res. 27, 4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tatusov, R. L., Natale, D. A., Garkavtsev, I. V., Tatusova, T. A., Shankavaram, U. T., Rao, B. S., Kiryutin, B., Galperin, M. Y., Fedorova, N. D. & Koonin, E. V. (2001) Nucleic Acids Res. 29, 22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowe, T. M. & Eddy, S. R. (1997) Nucleic Acids Res. 25, 955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Comfort, D. & Clubb, R. T. (2004) Infect. Immun. 72, 2710-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higgins, D. G., Thompson, J. D. & Gibson, T. J. (1996) Methods Enzymol. 266, 383-402. [DOI] [PubMed] [Google Scholar]
- 26.Kumar, S., Tamura, K. & Nei, M. (2004) Brief Bioinform. 5, 150-163. [DOI] [PubMed] [Google Scholar]
- 27.Kreiswirth, B. N., Lofdahl, S., Betley, M. J., O'Reilly, M., Schlievert, P. M., Bergdoll, M. S. & Novick, R. P. (1983) Nature 305, 709-712. [DOI] [PubMed] [Google Scholar]
- 28.Hiramatsu, K., Watanabe, S., Takeuchi, F., Ito, T. & Baba, T. (2004) Vaccine 22, Suppl. 1, S5-S8. [DOI] [PubMed] [Google Scholar]
- 29.Ito, T., Ma, X. X., Takeuchi, F., Okuma, K., Yuzawa, H. & Hiramatsu, K. (2004) Antimicrob. Agents Chemother. 48, 2637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naito, T., Kusano, K. & Kobayashi, I. (1995) Science 267, 897-899. [DOI] [PubMed] [Google Scholar]
- 31.Novick, R. P. (2003) Plasmid 49, 93-105. [DOI] [PubMed] [Google Scholar]
- 32.Mazmanian, S. K., Liu, G., Ton-That, H. & Schneewind, O. (1999) Science 285, 760-763. [DOI] [PubMed] [Google Scholar]
- 33.Fujita, K., Yokota, T., Oguri, T., Fujime, M. & Kitagawa, R. (1992) Urol. Res. 20, 399-402. [DOI] [PubMed] [Google Scholar]
- 34.Drechsel, H., Thieken, A., Reissbrodt, R., Jung, G. & Winkelmann, G. (1993) J. Bacteriol. 175, 2727-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forbes, J. R. & Gros, P. (2001) Trends Microbiol. 9, 397-403. [DOI] [PubMed] [Google Scholar]
- 36.Burall, L. S., Harro, J. M., Li, X., Lockatell, C. V., Himpsl, S. D., Hebel, J. R., Johnson, D. E. & Mobley, H. L. (2004) Infect. Immun. 72, 2922-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Podschun, R. (1990) Zentralbl. Hyg. Umweltmed. 189, 527-535. [PubMed] [Google Scholar]
- 38.Novick, R. P. (2003) Mol. Microbiol. 48, 1429-1449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.