Abstract
Inhibitory receptors for MHC class I molecules increase the threshold of lymphocyte activation. Natural Killer (NK) cells express a large number of such inhibitory receptors, including the human killer Ig-like receptors (KIR). However, activating members of the KIR family have poorly defined ligands and functions. Here we describe the use of activating KIR tetramer reagents as probes to detect their ligands. Infection of cells with Epstein–Barr virus leads to expression of a detectable ligand for the activating receptor KIR2DS1. In this case, KIR2DS1 interacts with up-regulated peptide–MHC class I complexes on Epstein–Barr virus-infected cells in a transporter associated with antigen processing (TAP)-dependent manner. In tetramer-based cellular assays and direct affinity measurements, this interaction with MHC class I is facilitated by a broad spectrum of peptides. KIR2DS1 and its inhibitory homologue, KIR2DL1, share sensitivity to peptide sequence alterations at positions 7 and 8. These results fit a model in which activating and inhibitory receptors recognize the same sets of self-MHC class I molecules, differing only in their binding affinities. Importantly, KIR2DS1 is not always sufficient to trigger NK effector responses when faced with cognate ligand, consistent with fine control during NK cell activation. We discuss how our results for KIR2DS1 and parallel studies on KIR2DS2 relate to the association between activating KIR genes and susceptibility to autoimmune disorders.
Keywords: natural killer cell
Natural killer (NK) cells recognize normal or aberrant cells through multiple receptors that detect normal host molecules, stress-induced ligands, and pathogen-expressed motifs (1, 2). Their role as innate immune effectors is apparent through their rapid recruitment to sites of inflammation, production of proinflammatory cytokines, and cytolytic responses without prior sensitization. Recently, attention has been drawn to their role in shaping adaptive immune responses where they modulate the response and function of B cells, T cells, and dendritic cells (3–8). One long-known interplay with the adaptive immune system is the ability of both T cells and NK cells to recognize MHC class I molecules. The “missing self hypothesis” was proposed to explain observations that NK cells are responsive to cells lacking MHC class I, stating that NK cells would thus complement cytotoxic T cell responses, which require the presence of self-MHC class I on the target cell surface (9). A mechanistic understanding was provided in the identification of inhibitory MHC class I receptors that contain immuno-tyrosine-based inhibitory motifs (ITIM) in their cytoplasmic tails, conferring association with the tyrosine phosphatases SHP-1 and SHP-2 (10–12).
Inhibitory HLA class I NK receptors fall into three groups (11, 13, 14). The CD94/NKG2 dimers recognize HLA-E, which presents leader peptides of classical HLA class I molecules. Some Ig-like transcript (or leukocyte Ig-like receptor) family members recognize moieties present on many HLA class I molecules. Inhibitory members of the killer Ig-like receptor (KIR) family (KIR-L) are more selective in their recognition of certain allotypes of HLA class I. KIR3DL2 recognizes HLA-A3 and HLA-A11, KIR3DL1 recognizes certain HLA-B allotypes, and certain KIR2DL molecules recognize HLA-C allotypes (11). The HLA-C specificity of KIR2DL members is largely determined by the amino acid at position 80. Group 1 HLA-C (HLA-C1) allotypes have an asparagine residue at position 80, conferring recognition by KIR2DL2 and KIR2DL3, whereas group 2 HLA-C (HLA-C2) allotypes, with position 80 lysine, are recognized by KIR2DL1 (14). Variegated expression of these receptors leads to a repertoire of HLA specificities within any individual's NK cell population (15).
The KIR family also has activating members (KIR-S) that have a short cytoplasmic tail lacking an immuno-tyrosine-based inhibitory motif, and a charged lysine residue in the transmembrane domain conferring association with the immunoreceptor tyrosine-based activation motif-containing DAP12/KARAP signaling adaptor (1, 2, 11, 12). Despite their homology to inhibitory HLA class I-binding KIR, very few studies have been able to document binding of KIR-S to HLA class I molecules. KIR2DS1, a close homologue of KIR2DL1, can promote NK cytotoxicity in an HLA class I-dependent manner (16). Direct binding of KIR2DS1 to HLA-C group 2 allotypes has been demonstrated (17), but the reduced level of this binding compared with that of KIR2DL1 has brought into question the physiological relevance of this interaction.
Because of very weak, or sometimes possibly nonexistent, binding of activating NK “class I” receptors to MHC class I molecules, a number of roles have been suggested involving MHC or non-MHC ligands (15). As such, KIR-S could recognize alternative forms of classical HLA class I, differing from their normal counterparts by alternative folding or in the peptides presented (18). A high level of HLA class I expression may be necessary for KIR-S function, occurring under conditions of IFN-γ production associated with infections. It is also possible that KIR-S bind non-HLA class I molecules either expressed by stressed self cells or encoded by pathogens. Roles for KIR-S involving alternative-MHC or non-MHC ligands are supported by some examples. A non-MHC ligand for one KIR-S molecule has been reported on melanoma cells (19). In addition, the murine Ly49 receptor family, which appears functionally analogous to human KIR, includes activating members that recognize viral homologues of MHC class I or altered forms of host MHC class I. Both of these are found on cells after infection with murine CMV, and the genetic presence of Ly49H in combination with its murine CMV m157 ligand, or Ly49P in combination with H-2Dk, confers resistance to murine CMV infection (20–26).
To our knowledge, we here describe the first detailed characterization of the interaction between the activating KIR2DS1 and its ligands, group 2 allotypes of HLA-C molecules.
Methods
Cells and HLA. 221/Cw3, 221/Cw4, and 221/Cw6 are transfectants of the L721.221 cell line expressing the HLA-C alleles HLA-Cw*0302, HLA-Cw*040, and HLA-Cw*0602, respectively [kindly provided by R. Biassoni, Istituto Giannina Gaslini, Genoa, Italy (221/Cw3 and 221/Cw4), and F. McCann and D. Davis, Imperial College, London (221/Cw6)]. MRC-5 fibroblast cells (HLA typing HLA-Cw5 and HLA-Cw7) were provided by X. de Lamballerie (Hôpital de la Timone, Marseille, France). BL30-B95 is an Epstein–Barr virus (EBV)-converted form of the Burkitt's lymphoma BL30 line (27). The TND-3 [transporter associated with antigen processing (TAP)-1-deficient] and TND-4 (TAP-1-sufficient) B lymphoblastoid cell lines (B-LCLs) type HLA-Cw*150201-positive (28). C1R is a HLA-A-negative, HLA-B-low, but HLA-Cw4-positive B cell line (29). RMA-S/Cw4 are HLA-Cw*0401 transfectants of a TAP-deficient RMA-S line that carries human β2 microglobulin (β2m) (30). Peripheral blood mononuclear cells were obtained from whole blood provided by the Etablissement Français du Sang (Marseille, France). HLA types for donors LB and PG are HLA-Cw*07, HLA-Cw*1202/3, and HLA-Cw*04, HLA-Cw*0602/0607, respectively. Infection of cells was performed as described in Supporting Methods, which is published as supporting information on the PNAS web site.
Antibodies. HLA-A, B, C-FITC (clone G46-2.6), CD19-allophycocyanin (APC; clone HIB19), and isotype control mAbs were from BD Biosciences. Rabbit anti-HSV-1 (catalog no. F0318) was from DAKO Cytomation (Trappes, France). Purified murine IgG1 CD158a/h (clone EB6) and CD56 (clone C218) were from Immunotech (Marseille, France). See Fig. 2 and Supporting Methods for details on anti-HLA class I antibodies.
Fig. 2.
Antibodies recognizing HLA-Cw4 and HLA-Cw6 epitopes block KIR2DS1 and KIR2DL1 tetramer binding. Shown are fluorescence profiles of 221/Cw4 and 221/Cw6 (HLA-C group 2) cells after preincubation with the indicated anti-HLA class I mAb and subsequent incubation with the same antibodies and PE-coupled KIR tetramers. Numbers within histograms indicate MFI. W6/32 preferentially recognizes a monomorphic β2m-dependent epitope on HLA class I heavy chains (51). 6A4 and A6.136 mAbs block interaction of inhibitory KIR with HLA class I molecules in functional assays and do not react with acid-treated misfolded forms of HLA class I (52), B1.23.2 recognizes HLA-B and HLA-C heavy chains with or without β2m. B9.12.1 reacts with the α1 domain of class I heavy chains in complex with β2m (53). HC-10 preferentially recognizes free/misfolded HLA heavy chains (54).
Generation of NK Cell Clones and Cytotoxicity Assays. NK clones were generated and assessed as described in Supporting Methods. Standard 4-h Cr51 assays were performed to measure NK cytotoxicity. Redirected lysis assays using the FcγR-positive target P815 were performed at an effector:target ratio of 3:1. Other assays used a 5:1 effector:target ratio.
Generation of KIR Tetramers and Binding Assays. PLM-1 plasmids containing the entire extracellular domains of KIR2DL1*00301, KIR2DS1*001, KIR2DL3*001, or KIR2DS2*002 (31) were used to produce fluorescent KIR tetramers through biotinylation and streptavidin conjugation as detailed in Supporting Methods. The tetramerized forms of each KIR are hereafter referred to as 2DL1t, 2DS1t, 2DL3t, and 2DS2t, respectively. Unless indicated, binding assays on cell populations were performed by using saturating concentrations of KIR tetramers (1 μg·ml-1 for 2DL1t or 25 μg·ml-1 for all other tetramers) for 1 h at 4°C as previously described (31). For blocking analysis using KIR tetramers, a primary stain with phycoerythrin (PE)-conjugated 2DL1t or 2DS1t tetramers for 1 h at 4°C was followed by addition of APC-conjugated tetramers at the same concentration and additional incubation for 8 min.
Peptide Binding Assay for Stabilization of HLA-C Surface Expression. All of the peptides were purchased from Eurogentec (Liège, Belgium). Peptide Cw4#1 (QYDDAVYKL) is a consensus peptide that binds Cw*0401 (30). Peptide loading of cells was performed by incubating RMA-S/Cw4 (106 ml-1) at 27°C overnight in medium containing 100 μM peptide unless stated otherwise.
Surface Plasmon Resonance (SPR) Analysis. SPR studies were performed at 25°C with a BIAcore 3000 (BIAcore, Paris) in HBS buffer (10 mM Hepes, pH 7.4/150 mM NaCl/3.4 mM EDTA/0.0005% surfactant P20). KIR2DL1, KIR2DS1, or irrelevant GST proteins were covalently coupled via primary amines (AmineCoupling kit, BIAcore) to research grade CM5 sensor chips. Positive and negative controls were CD158a/h antibody and HLA-A2/β2m/peptide (a gift from D. Bossy, Immunotech, Marseille, France), respectively. HLA-Cw4/β2m/peptide samples, produced as described in Supporting Methods, were injected at a flow rate of 20 μl·min-1.
Results
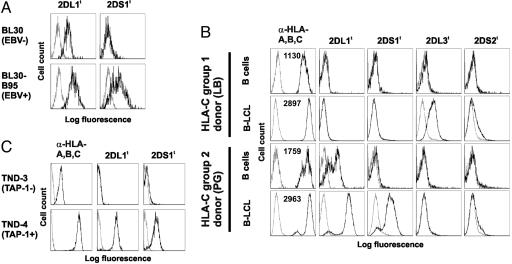
Activating KIR Tetramers Bind Ligands Induced by EBV Infection. The importance of Ly49H in resistance to murine CMV pathology and the increased sensitivity to herpesvirus infection in cases of NK cell deficiency (32) prompted the question of whether human herpesviruses induce ligands for KIR-S. We therefore examined the binding of KIR-S and KIR-L tetramers to cells infected with human herpesviruses from each of the α-, β-, and γ-herpesvirinae subfamilies. Infection of the MRC-5 human fibroblast cell line with HSV-1, HSV-2, or human CMV induced no detectable KIR2DS1 or KIR2DS2 ligand at the cell surface (see Fig. 5, which is published as supporting information on the PNAS web site). In contrast, staining with KIR2DS1 tetramers was brighter on the EBV-positive Burkitt's lymphoma line BL30-B95 than on its EBV-negative parent, BL30 (Fig. 1A), indicating that EBV infection induces the cell surface expression of a ligand for the activating KIR, KIR2DS1. Brighter staining on the EBV-positive BL30-B95 line was also seen using the KIR2DL1 tetramer (Fig. 1A), suggesting that the increased binding of both KIR2DS1 and KIR2DL1 might be due to higher levels of HLA class I expression, which occur because of the EBV latency protein LMP1 (33).
Fig. 1.
KIR-S binding to EBV-positive cells is HLA-C allotype-specific and TAP-dependent. Flow cytometry using PE-coupled KIR tetramers on EBV-positive and parental cells is shown. Unless stated otherwise, fluorescence profiles (black) are overlayed on streptavidin-PE only or isotype-matched antibody control profiles (gray) as appropriate. (A) EBV-negative Burkitt's lymphoma line BL30 and its EBV-positive BL30-B95 derivative. Mean fluorescence intensities (MFI) of W6/32 HLA-class I staining were 992 (BL30) and 3,849 (BL30-B95). (B) Peripheral blood CD19+ B cells and EBV-transformed B-LCL from HLA-C1 and HLA-C2 homozygous donors. Numbers within anti-HLA-A,B,C histograms give the MFIs of G46-2.6 staining. (C) TND-4 TAP-sufficient and TND-3 TAP-deficient B-LCLs from HLA-C2-positive sibling donors. KIR tetramer fluorescence profiles are overlaid on unstained cells.
Classical MHC Class I Molecules Are Activating KIR Ligands. To confirm that recognition of EBV-infected cells by KIR-S was due to the binding of HLA-C molecules as occurs for inhibitory KIR, experiments were performed with KIR tetramers on cells from HLA-C-genotyped individuals, TAP-deficient individuals, and HLA-C-transfected cell lines. KIR tetramer staining was performed on pairs of peripheral blood CD19+ B cells and EBV-transformed B-LCL from various donors (Fig. 1B). Peripheral blood B cells were stained with KIR2DL1 tetramers only when the donor's HLA typed positive for HLA-C2 alleles. No staining was observed using other KIR tetramers on peripheral blood B cells (Fig. 1B and data not shown). As expected, HLA class I expression, as assessed by G46-2.6 staining, was higher on the B-LCLs than peripheral blood mononuclear cell B cells from the same donor. KIR2DL3 and, to some extent, KIR2DS2 tetramers stained the HLA-C1-positive B-LCL, and KIR2DL1 and KIR2DS1 tetramers stained the HLA-C2 B-LCL. In all cases, the intensity of fluorescence obtained by using activating KIR tetramers was lower than that of the corresponding inhibitory KIR. These data suggest that the pattern of HLA recognition by KIR2DS1 and KIR2DS2 follows that of their highly homologous inhibitory counterparts (KIR2DL1 and KIR2DL2/3, respectively).
B-LCL from TAP-sufficient and TAP-deficient donors were stained with KIR tetramers to test the requirement for the HLA class I processing and presentation pathway. Whereas the TND-4 TAP-sufficient line stained brightly with tetramers of KIR2DL1 and KIR2DS1, the TND-3 TAP-1-deficient line bound neither (Fig. 1C), indicating that the presence of KIR ligand on these cells depends on TAP.
To confirm that the binding of KIR-S tetramers depended on HLA-C expression, cell lines expressing restricted sets of HLA class I alleles were stained with KIR tetramers. HLA-Cw4 and HLA-Cw6 transfectants of the classical class I-negative L721.221 cell line stained positive with both KIR2DL1 and KIR2DS1 probes, but no shift was observed on HLA-Cw3 transfectants (see Fig. 6A, which is published as supporting information on the PNAS web site). In addition, HLA-Cw4-positive line C1R was stained brightly with 2DL1t, but only weakly, with the 2DS1t (Fig. 6A). No convincing staining was observed using the KIR2DS2 tetramer on any of the cell lines (data not shown). These data indicate that KIR2DS1 specificity for HLA-C alleles follows the group 2 HLA class I specificity of KIR2DL1.
Blocking experiments using a panel of anti-HLA class I mAb were performed to confirm the interaction of KIR2DL1 and KIR2DS1 with HLA class I and to investigate the requirements of this interaction (Fig. 2). The staining of 221 transfectants expressing two distinct HLA-C2 alleles, 221/Cw4 and 221/Cw6, with 2DL1t and 2DS1t was increased when the cells had been preincubated with W6/32 or G46-2.6 mAbs, indicating that these antibodies stabilize or otherwise aid the binding of KIR2DS1 and KIR2DL1 to the cells. Increased KIR tetramer binding was possibly due to aggregation and thus higher effective avidity of cell surface HLA class I molecules. Blocking of KIR tetramer staining was observed when the mAbs 6A4, A6.136, and B1.23.2 were preincubated with the HLA-C transfectants, reducing the fluorescence to levels observed using streptavidin-PE alone. Interestingly, the mAb B9.12.1 produced disparate effects between 221/Cw4 and 221/Cw6. On 221/Cw4 cells, preincubation with B9.12.1 did not change the intensity of KIR tetramer staining. However, on 221/Cw6 cells, the same preincubation decreased the levels of KIR tetramer staining (Fig. 2). This specific blocking effect was titrated by using concentrations of the B9.12.1 antibody from 1 to 100 μg·ml-1 showing progressive loss of staining of both KIR2DS1 and KIR2DL1 tetramers on 221/Cw6 cells, but no effect on 221/Cw4, despite both lines staining brightly with the antibody (data not shown). Of those mAbs affecting the binding of 2DS1t, W6/32, 6A4, A6.136, and B9.12.1 have varying degrees of preference for β2m-associated HLA heavy chains (see Fig. 2 legend). In contrast, HC-10, which preferentially reacts with free/misfolded HLA heavy chains, did not affect 2DS1t binding. Furthermore, 2DL1t and 2DS1t staining of HLA-C2-positive PHA blasts, which express high levels of free/misfolded heavy chains (34), correlated more closely with W6/32 mAb β2m-dependent staining than that of HC-10 (data not shown). Together these data show that binding of KIR2DS1 and KIR2DL1 depends on the availability of HLA-C2 epitopes on the cell surface and suggest that these HLA-C2 epitopes are β2m-dependent.
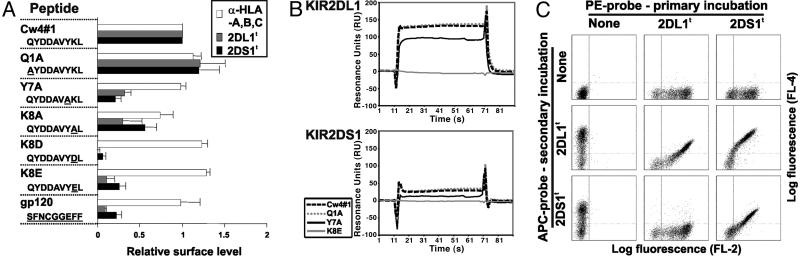
KIR2DS1 Binding to HLA-C Depends on the Presented Peptide. The inhibitory counterpart of KIR2DS1, KIR2DL1, has a level of restriction in its binding to HLA-C dependent on the peptide present in the peptide binding groove (30). To determine whether KIR2DS1 exhibits the same peptide dependency, we used TAP-negative RMA-S transfectants of HLA-Cw4 (RMA-S/Cw4) that can be loaded with peptides to present single-peptide–HLA-Cw4 complexes. Loading of these cells with the canonical QYDDAVYK (Cw4#1) peptide that is permissive to KIR2DL1 binding (30) permitted staining with anti-HLA class I antibody, KIR2DL1, and KIR2DS1 in a peptide concentration-dependent manner (see Fig. 7, which is published as supporting information on the PNAS web site), indicating that the HLA-Cw4–Cw4#1 complex is also a ligand for KIR2DS1. RMA-S/Cw4 cells were then loaded with a panel of peptides containing amino acid substitutions from the Cw4#1 sequence. We restricted our analysis to peptides that reconstituted at least 60% of HLA-Cw4 expression at the cell surface as assessed by the binding of the G46-2.6 anti-HLA class I mAb, thus eliminating peptide substitutions at positions 2–6. As shown in Fig. 3A, the Y7A, K8D, and K8E substitutions abolish most KIR2DS1 binding. In contrast, the substitution K8A led to a modest reduction in binding, and that of Q1A increased the interaction with both the anti-HLA class I mAb and KIR2DS1. In all cases, substitutions that had an effect on the binding of KIR2DS1 had similar effects on KIR2DL1 binding.
Fig. 3.
Influence of peptide on recognition of HLA-Cw4 by KIR2DS1. (A) Relative binding levels of anti-HLA class I antibody (G46-2.6), KIR2DL1 (2DL1t), and KIR2DS1 (2DS1t) tetramers to RMA-S/Cw4 cells loaded with the indicated peptides. Peptide names and amino acid sequences are given with differences from the canonical Cw4#1 peptide underlined. Relative surface level is calculated as (MFI with test peptide - MFI no peptide)/(MFI with Cw4#1 peptide - MFI no peptide). Error bars show standard deviations. (B) SPR profiles of HLA-Cw4–peptide complexes passed over immobilized KIR2DL1 and KIR2DS1. HLA-Cw4 was refolded with the peptides indicated and used at a final concentration of 20 mM. Curves show signals after subtraction of background (obtained using immobilized GST). (C) Effects of masking KIR ligands on KIR tetramer binding. 221/Cw4 cells were incubated for 1 h with the indicated PE-coupled KIR tetramers followed by addition of APC-coupled KIR tetramers and incubation for a further 8 min.
Affinities of KIR2DS1 and KIR2DL1 for Various Peptide–HLA-Cw4 Complexes. To investigate whether the peptide “selectivity” of KIR2DL1 and KIR2DS1 is completely analogous, we used SPR of selected HLA-Cw4–peptide complexes on immobilized KIR molecules. Because interactions between inhibitory KIR and HLA are known to have fast association and dissociation kinetics (35, 36), response at equilibrium (REq) analysis was used for KD determination. A linear relationship was found between REq and REq/concentration for KIR2DL1 and HLA-Cw4–Cw4#1, testifying for the validity of this analysis (see Fig. 8, which is published as supporting information on the PNAS web site) (37). Furthermore, the affinity measurement for the interaction between KIR2DL1 and HLA-Cw4–Cw4#1 complexes with a dissociation constant KD of 7.2 μM was consistent with the value of ≈10 μM obtained previously using KIR2DL1 and HLA-Cw6 as a binding partner (38). HLA-Cw4 complexes refolded with peptides Cw4#1 and Q1A showed highest affinity binding of both KIR2DL1 and KIR2DS1, those with K8E showed no detectable binding, and those with Y7A had an intermediate phenotype (Fig. 3B and Table 1). Where measurable, the dissociation constant of KIR2DS1 was ≈3.5 times that of KIR2DL1 for the same ligand (the binding of KIR2DS1 to Y7A-containing complexes was of too low affinity to obtain a value for the KD).
Table 1. Binding affinities of KIR2DS1 and KIR2DL1 for HLA-Cw4/peptide complexes.
| HLA-Cw4 bound peptide | KIR2DS1 KD, μM | KIR2DL1 KD, μM |
|---|---|---|
| QYDDAVYKL (Cw4#1) | 30 | 7.2 |
| AYDDAVYKL (Q1A) | 22.8 | 7.6 |
| QYDDAVAKL (Y7A) | >50 | 14.7 |
| QYDDAVYEL (K8E) | No measurable binding | |
Dissociation constants (KD) for KIR2DS1 and KIR2DL1 to the indicated HLA-Cw4/peptide complexes were calculated from response at equilibrium analysis of SPR readings as described in Fig. 3B and Methods.
KIR2DS1 Shares Most HLA-Cw4–Peptide Binding Partners with KIR2DL1. The above data indicate that KIR2DS1 is capable of binding peptide–HLA complexes that also provide a ligand for KIR2DL1. We wished to test this for a broader set of peptides. 221/Cw4 cells presenting a repertoire of peptides were stained with KIR tetramers with or without prior incubation with KIR tetramers coupled to a different fluorochrome (Fig. 3C). Staining using KIR2DL1-APC or KIR2DS1-APC was reduced, but not eliminated, after preincubation of cells with the PE-conjugated tetramers of the same KIR. This finding suggests that enough free KIR ligand was available during the secondary incubation to permit binding of the second conjugate. Despite this free KIR ligand, the primary incubation of 221/Cw4 cells with KIR2DL1 almost completely abolished secondary staining with KIR2DS1. These data suggest that all of the detectable repertoire of HLA-Cw4–peptide complexes that binds KIR2DS1 also binds KIR2DL1.
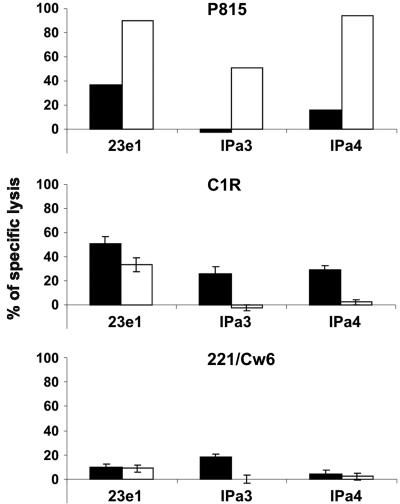
KIR2DS1–HLA-C Interactions Activate NK Cell Cytotoxicity Under Restricted Conditions. To assess whether activation of NK cells could be conferred through the KIR2DS1–HLA-C–peptide ligation, we examined KIR2DS1-dependent NK cell cytotoxicity against targets expressing HLA-C2 molecules (Fig. 4). We selected NK clones that killed P815 cells in a redirected cytotoxicity assay in the presence of CD158a/h mAb (recognizing both KIR2DL1 and KIR2DS1). Blocking concentrations of CD158a/h mAb reduced or abolished NK cytolysis of C1R targets that express HLA-Cw4, indicating that NK cytolytic activity can be KIR2DS1-dependent. This finding is in line with a previous report showing reduction in killing of C1R by KIR2DS1+ NK clones after the addition of blocking antibodies against KIR2DS1 or MHC class I (16). However, only one clone was capable of low-level KIR2DS1-dependent cytotoxicity against 221/Cw6 targets (Fig. 4), despite the higher level of KIR2DS1 ligand on 221/Cw6 than C1R (Fig. 6A). In addition, the expression of HLA-C2 on peptide-loaded RMA-S/Cw4 cells was not sufficient to induce cytotoxicity or IFN-γ production by NK clones expressing KIR2DS1 (data not shown). These data suggest that the engagement of KIR2DS1 with HLA-C2 is not sufficient to drive NK cell cytotoxicity or IFN-γ production. In particular, it is possible that other NK cell-activating receptors engage ligands that are absent from 221/Cw6 but present on C1R and depend on the simultaneous engagement of KIR2DS1. It is equally possible that KIR2DS1-dependent signals are highly sensitive to inhibition through receptors such as CD94/NKG2A, the ligands of which (HLA-E) are likely to be more brightly expressed on 221/Cw6 than on C1R.
Fig. 4.
KIR2DS1 triggers NK cytolytic activity. Cr51 release assays of CD158a/h-expressing NK clones against P815, C1R, and 221/Cw6 targets. For each clone, percentage specific lysis is shown for isotype-matched control CD56 (black bars) and test CD158a/h (white bars) antibodies. Antibody was added at 0.5 μg·ml-1 for redirected killing assays against P815 or 10 μg·ml-1 for blocking assays against C1R and 221/Cw6. P815 assays were performed in duplicate with <5% deviation in values. Error bars show standard deviations of triplicates.
Discussion
Here we describe the use of KIR tetramers to detect the presence of ligands for activating KIR. From a range of human herpesviruses tested, EBV infection was found to induce up-regulation of activating and inhibitory KIR ligands. This up-regulation correlated with that of MHC class I expression by these cells and was TAP-dependent. Transfectant and HLA-genotyped donor-based assays showed that the binding of KIR2DS1 tetramers follows the rules of HLA-C allelic specificity of its “corresponding” inhibitory KIR, KIR2DL1. Analysis of peptide specificity of this interaction indicated that the peptide requirement for KIR2DS1 binding to HLA-Cw4 closely resembles that for KIR2DL1. We finally showed that KIR2DS1 induces functional activity in response to HLA-C under some, but not all, circumstances.
The roles and functions of activating KIR receptors pose a perplexing question in our understanding of NK cell biology. Variation in KIR locus gene content is foremost a result of the presence or absence of KIR-S genes rather than that of KIR-L genes (39). Disease-association studies have implicated KIR-S in control of viral infections and malignancy, susceptibility to autoimmunity, and reproductive success (for an overview, see ref. 40). In a number of these studies, the association is found with both KIR-S genes and HLA alleles encoding ligands for “corresponding” inhibitory KIR molecules (see below). However, previous studies directly addressing whether activating KIR have the same ligands as their inhibitory homologues have identified only low-level binding of KIR2DS1 and KIR2DS4 to HLA-C2 (17, 41).
We confirm previous data on binding of KIR2DS1 to HLA-C group 2 molecules (17) and characterize this interaction in detail. We find no evidence to support the hypothesis that free/misfolded HLA-C heavy chains are involved in KIR2DS1 recognition by NK cells (18). Another hypothesis for KIR-S function states that increased levels of HLA class I, induced, for example, by IFN-γ during infections, may be required for KIR-S function. Contrary to this hypothesis, we find that the lower levels of HLA-C2 expression observed on C1R compared with 221/Cw6 are effective inducers of KIR2DS1 function. On the role of HLA-C-bound peptide in KIR binding, our data confirm those previously reported for KIR2DL1–peptide selectivity at positions 7 and 8 (30) and extend these to the activating receptor KIR2DS1. In addition, blocking of 2DS1t binding to 221/Cw4 by preincubation with 2DL1t (Fig. 3C) further argues against the hypothesis that certain HLA–peptide complexes are specific for KIR2DS1 receptors over KIR2DL1.
To date, it has proven difficult to demonstrate the binding of KIR2DS2 to HLA-C group 1 molecules (31, 35, 36, 42). Low levels of specific binding of 2DS2t to HLA-C1-expressing cells occurs under certain circumstances (Fig. 1B), but we were unable to consistently observe binding to 221/Cw3 cells (data not shown), possibly because of an extremely low affinity of this interaction. This finding fits with the suggested trend for lower-affinity interactions of HLA-C1 receptors (KIR2DL2 and KIR2DL3) than the HLA-C2 receptor KIR2DL1 (40). SPR analysis was also performed to examine KIR2DS2, KIR2DL2, and KIR2DL3 binding to various HLA-Cw3–peptide complexes (see Fig. 9, which is published as supporting information on the PNAS web site) as described previously for KIR2DL2 (43). The strength of binding followed the pattern KIR2DL2→KIR2DL3→KIR2DS2 when permissive peptides were bound to HLA-Cw3. Whereas KIR2DL2 and KIR2DL3 binding was highly sensitive to peptide mutations, the low KIR2DS2 binding appeared relatively peptide-indifferent. Previous crystallographic data suggested that Gly71, which forms a hydrogen bond with Ala8 of the GAV peptide in the KIR2DL2–HLA-Cw3 interaction, would be unable to form this hydrogen bond if provided by KIR2DS2 (31). These data are therefore consistent with a very-low-affinity KIR2DS2–HLA-C interaction that does not have the same degree of peptide selectivity as other KIR–HLA interactions.
A recent report indicating that activating members of the KIR and Ly49 families evolved from their inhibitory homologues and that their genes are comparatively young suggests that both positive and negative evolutionary pressures have acted on them (44). One possibility is that HLA-C2 molecules provided the only ligand on which KIR2DS1 has been selected. A potential mechanism for the positive selection of interactions involving KIR-S and normal HLA class I molecules is due to an enhanced activation state of the NK cell population. This enhanced activation state of NK cells has been suggested as the reason for genetic association of the “weak” inhibitory KIR, KIR2DL3, and HLA-C1 with better resolution of hepatitis C virus infection, along with the association of certain maternal KIR and fetal HLA-C combinations with reduced risk of preeclampsia during pregnancy (11, 45, 46). A higher state of NK cell activation may not always lead to overt NK cell responses, but rather increase the propensity of the NK cell population to perform cytolytic functions and produce proinflammatory cytokines. Our data on the insufficiency of KIR2DS1 ligation to induce NK cell responses support this modulating role, because they suggest a requirement of additional activating or reduced inhibitory signals for the comparatively strong KIR2DS1-mediated activation seen against C1R targets (Fig. 4).
Higher basal NK cell activation states due to ligation of KIR-S molecules may also explain other genetic disease associations involving both KIR-S and HLA ligands. These include the slower progression to AIDS associated with the combination of KIR3DS1 and HLA-Bw4 alleles containing Ile-80 (47), type I diabetes risk associated with KIR2DS2 combined with presence of HLA-C1 but lack of HLA-C2 and HLA-Bw4 alleles (48), and psoriasis vulgaris associated with KIR2DS1 and HLA-Cw*06 (49, 50). If so, why should some diseases be associated with only certain combinations of genes? One possibility is differential expression of HLA class I genes in different tissues, for example, the selective expression of HLA-C, HLA-E, and HLA-G but not HLA-A and HLA-B on extravillous trophoblast cells (see 46). Selective interactions of receptor–ligand pairs may also be affected by their different affinities. Another is that the peptide preferences of KIR-S–HLA interactions, such as those described here for KIR2DS1, may define which interactions are important in particular tissues. Alternatively, KIR-S may play additional roles when expressed by subsets of T cells. The large number of KIR-S receptors, their high level of polymorphism, their associations with health and disease, and the subtleties of their activation demonstrate the complexity and plasticity of NK cells as effectors of the innate immune system.
Supplementary Material
Acknowledgments
We thank Hsiang-Ting Hsu, L. Gastinel, and A. Necker for their help. We thank Q. R. Fan (Columbia University, New York), D. Bossy, H. Vié, X. de Lamballerie, V. Pitard, J. Déchanet-Merville, E. Long (National Institute of Allergy and Infectious Diseases, Rockville, MD), H. de la Salle (Establissement Français du Sang, Strasbourg, France), R. Biassoni, G. Lenoir (Hôpital Necker–Enfants Malades, Paris), D. Davis, and F. McCann for providing cells, virus stocks, antibodies, and proteins as detailed or referenced in the text. We thank D. Reviron (Etablissement Français du Sang, Marseille, France) for providing HLA typing data of blood donors. This work was supported in part by institutional grants from Institut National de la Santé et de la Recherche Médicale (INSERM), Centre National de la Recherche Scientifique, and Ministère de l'Enseignement Supérieur et de la Recherche and specific grants from the European Union (AlloStem), Ligue Nationale Contre le Cancer (“Equipe labellisée La Ligue” to E.V.), INSERM Région Provence Alpes-Côte d'Azur (to F.L.-A.), and la Fondation pour la Recherche Médicale (to C.A.S.).
Author contributions: C.A.S., F.L.-A., F.V., P.D.S., S.U., and E.V. designed research; C.A.S., F.L.-A., F.V., X.S., J.R., A.T., L.G., F.R., G.F., and F.A.A., performed research; C.A.S., F.L.-A., F.V., J.R., G.F., A.M., P.D.S., S.U., and E.V. analyzed data; C.A.S. and E.V. wrote the paper; and F.L.-A., X.S., A.T., L.G., F.R., G.F., F.A.A., and A.M. contributed new reagents/analytic tools.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: KIR, killer Ig-like receptor; EBV, Epstein–Barr virus; TAP, transporter associated with antigen processing; B-LCL, B lymphoblastoid cell line; NK, natural killer; PE, phycoerythrin; APC, allophycocyanin; β2m, β2 microglobulin; SPR, surface plasmon resonance; MFI, mean fluorescence intensity.
References
- 1.Raulet, D. H. (2004) Nat. Immunol. 5, 996-1002. [DOI] [PubMed] [Google Scholar]
- 2.Lanier, L. L. (2005) Annu. Rev. Immunol. 23, 225-274. [DOI] [PubMed] [Google Scholar]
- 3.Moretta, A. (2002) Nat. Rev. Immunol. 2, 957-964. [DOI] [PubMed] [Google Scholar]
- 4.Cooper, M. A., Fehniger, T. A., Fuchs, A., Colonna, M. & Caligiuri, M. A. (2004) Trends Immunol. 25, 47-52. [DOI] [PubMed] [Google Scholar]
- 5.Zingoni, A., Sornasse, T., Cocks, B. G., Tanaka, Y., Santoni, A. & Lanier, L. L. (2004) J. Immunol. 173, 3716-3724. [DOI] [PubMed] [Google Scholar]
- 6.Martin-Fontecha, A., Thomsen, L. L., Brett, S., Gerard, C., Lipp, M., Lanzavecchia, A. & Sallusto, F. (2004) Nat. Immunol. 5, 1260-1265. [DOI] [PubMed] [Google Scholar]
- 7.Andrews, D. M., Andoniou, C. E., Scalzo, A. A., van Dommelen, S. L., Wallace, M. E., Smyth, M. J. & Degli-Esposti, M. A. (2005) Mol. Immunol. 42, 547-555. [DOI] [PubMed] [Google Scholar]
- 8.Blanca, I. R., Bere, E. W., Young, H. A. & Ortaldo, J. R. (2001) J. Immunol. 167, 6132-6139. [DOI] [PubMed] [Google Scholar]
- 9.Karre, K., Ljunggren, H. G., Piontek, G. & Kiessling, R. (1986) Nature 319, 675-678. [DOI] [PubMed] [Google Scholar]
- 10.Yokoyama, W. M. & Plougastel, B. F. (2003) Nat. Rev. Immunol. 3, 304-316. [DOI] [PubMed] [Google Scholar]
- 11.Parham, P. (2005) Nat. Rev. Immunol. 5, 201-214. [DOI] [PubMed] [Google Scholar]
- 12.Vivier, E., Nunes, J. A. & Vely, F. (2004) Science 306, 1517-1519. [DOI] [PubMed] [Google Scholar]
- 13.Natarajan, K., Dimasi, N., Wang, J., Mariuzza, R. A. & Margulies, D. H. (2002) Annu. Rev. Immunol. 20, 853-885. [DOI] [PubMed] [Google Scholar]
- 14.Boyington, J. C. & Sun, P. D. (2002) Mol. Immunol. 38, 1007-1021. [DOI] [PubMed] [Google Scholar]
- 15.Raulet, D. H., Vance, R. E. & McMahon, C. W. (2001) Annu. Rev. Immunol. 19, 291-330. [DOI] [PubMed] [Google Scholar]
- 16.Moretta, A., Sivori, S., Vitale, M., Pende, D., Morelli, L., Augugliaro, R., Bottino, C. & Moretta, L. (1995) J. Exp. Med. 182, 875-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biassoni, R., Pessino, A., Malaspina, A., Cantoni, C., Bottino, C., Sivori, S., Moretta, L. & Moretta, A. (1997) Eur. J. Immunol. 27, 3095-3099. [DOI] [PubMed] [Google Scholar]
- 18.Allen, R. L., Raine, T., Haude, A., Trowsdale, J. & Wilson, M. J. (2001) J. Immunol. 167, 5543-5547. [DOI] [PubMed] [Google Scholar]
- 19.Katz, G., Gazit, R., Arnon, T. I., Gonen-Gross, T., Tarcic, G., Markel, G., Gruda, R., Achdout, H., Drize, O., Merims, S., et al. (2004) J. Immunol. 173, 1819-1825. [DOI] [PubMed] [Google Scholar]
- 20.Lee, S. H., Girard, S., Macina, D., Busa, M., Zafer, A., Belouchi, A., Gros, P. & Vidal, S. M. (2001) Nat. Genet. 28, 42-45. [DOI] [PubMed] [Google Scholar]
- 21.Brown, M. G., Dokun, A. O., Heusel, J. W., Smith, H. R., Beckman, D. L., Blattenberger, E. A., Dubbelde, C. E., Stone, L. R., Scalzo, A. A. & Yokoyama, W. M. (2001) Science 292, 934-937. [DOI] [PubMed] [Google Scholar]
- 22.Arase, H., Mocarski, E. S., Campbell, A. E., Hill, A. B. & Lanier, L. L. (2002) Science 296, 1323-1326. [DOI] [PubMed] [Google Scholar]
- 23.Smith, H. R., Heusel, J. W., Mehta, I. K., Kim, S., Dorner, B. G., Naidenko, O. V., Iizuka, K., Furukawa, H., Beckman, D. L., Pingel, J. T., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 8826-8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krug, A., French, A. R., Barchet, W., Fischer, J. A., Dzionek, A., Pingel, J. T., Orihuela, M. M., Akira, S., Yokoyama, W. M. & Colonna, M. (2004) Immunity 21, 107-119. [DOI] [PubMed] [Google Scholar]
- 25.Carayannopoulos, L. N. & Yokoyama, W. M. (2004) Curr. Opin. Immunol. 16, 26-33. [DOI] [PubMed] [Google Scholar]
- 26.Desrosiers, M. P., Kielczewska, A., Loredo-Osti, J. C., Adam, S. G., Makrigiannis, A. P., Lemieux, S., Pham, T., Lodoen, M. B., Morgan, K., Lanier, L. L., et al. (2005) Nat. Genet. 37, 593-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baran-Marszak, F., Fagard, R., Girard, B., Camilleri-Broet, S., Zeng, F., Lenoir, G. M., Raphael, M. & Feuillard, J. (2002) Lab. Invest. 82, 1463-1479. [DOI] [PubMed] [Google Scholar]
- 28.de la Salle, H., Zimmer, J., Fricker, D., Angenieux, C., Cazenave, J. P., Okubo, M., Maeda, H., Plebani, A., Tongio, M. M., Dormoy, A., et al. (1999) J. Clin. Invest. 103, R9-R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zemmour, J., Little, A. M., Schendel, D. J. & Parham, P. (1992) J. Immunol. 148, 1941-1948. [PubMed] [Google Scholar]
- 30.Rajagopalan, S. & Long, E. O. (1997) J. Exp. Med. 185, 1523-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saulquin, X., Gastinel, L. N. & Vivier, E. (2003) J. Exp. Med. 197, 933-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orange, J. S. (2002) Microbes Infect. 4, 1545-1558. [DOI] [PubMed] [Google Scholar]
- 33.Rowe, M., Khanna, R., Jacob, C. A., Argaet, V., Kelly, A., Powis, S., Belich, M., Croom-Carter, D., Lee, S., Burrows, S. R., et al. (1995) Eur. J. Immunol. 25, 1374-1384. [DOI] [PubMed] [Google Scholar]
- 34.Santos, S. G., Powis, S. J. & Arosa, F. A. (2004) J. Biol. Chem. 279, 53062-53070. [DOI] [PubMed] [Google Scholar]
- 35.Vales-Gomez, M., Reyburn, H. T., Erskine, R. A. & Strominger, J. (1998) Proc. Natl. Acad. Sci. USA 95, 14326-14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maenaka, K., Juji, T., Nakayama, T., Wyer, J. R., Gao, G. F., Maenaka, T., Zaccai, N. R., Kikuchi, A., Yabe, T., Tokunaga, K., et al. (1999) J. Biol. Chem. 274, 28329-28334. [DOI] [PubMed] [Google Scholar]
- 37.Vely, F., Trautmann, A. & Vivier, E. (2000) Methods Mol. Biol. 121, 313-321. [DOI] [PubMed] [Google Scholar]
- 38.Vales-Gomez, M., Reyburn, H. T., Mandelboim, M. & Strominger, J. L. (1998) Immunity 9, 337-344. [DOI] [PubMed] [Google Scholar]
- 39.Hsu, K. C., Chida, S., Geraghty, D. E. & Dupont, B. (2002) Immunol. Rev. 190, 40-52. [DOI] [PubMed] [Google Scholar]
- 40.Rajagopalan, S. & Long, E. O. (2005) J. Exp. Med. 201, 1025-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katz, G., Markel, G., Mizrahi, S., Arnon, T. I. & Mandelboim, O. (2001) J. Immunol. 166, 7260-7267. [DOI] [PubMed] [Google Scholar]
- 42.Winter, C. C., Gumperz, J. E., Parham, P., Long, E. O. & Wagtmann, N. (1998) J. Immunol. 161, 571-577. [PubMed] [Google Scholar]
- 43.Boyington, J. C., Motyka, S. A., Schuck, P., Brooks, A. G. & Sun, P. D. (2000) Nature 405, 537-543. [DOI] [PubMed] [Google Scholar]
- 44.Abi-Rached, L. & Parham, P. (2005) J. Exp. Med. 201, 1319-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khakoo, S. I., Thio, C. L., Martin, M. P., Brooks, C. R., Gao, X., Astemborski, J., Cheng, J., Goedert, J. J., Vlahov, D., Hilgartner, M., et al. (2004) Science 305, 872-874. [DOI] [PubMed] [Google Scholar]
- 46.Hiby, S. E., Walker, J. J., O'Shaughnessy K. M., Redman, C. W., Carrington, M., Trowsdale, J. & Moffett, A. (2004) J. Exp. Med. 200, 957-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin, M. P., Gao, X., Lee, J. H., Nelson, G. W., Detels, R., Goedert, J. J., Buchbinder, S., Hoots, K., Vlahov, D., Trowsdale, J., et al. (2002) Nat. Genet. 31, 429-434. [DOI] [PubMed] [Google Scholar]
- 48.van der Slik, A. R., Koeleman, B. P., Verduijn, W., Bruining, G. J., Roep, B. O. & Giphart, M. J. (2003) Diabetes 52, 2639-2642. [DOI] [PubMed] [Google Scholar]
- 49.Luszczek, W., Manczak, M., Cislo, M., Nockowski, P., Wisniewski, A., Jasek, M. & Kusnierczyk, P. (2004) Hum. Immunol. 65, 758-766. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki, Y., Hamamoto, Y., Ogasawara, Y., Ishikawa, K., Yoshikawa, Y., Sasazuki, T. & Muto, M. (2004) J. Invest. Dermatol. 122, 1133-1136. [DOI] [PubMed] [Google Scholar]
- 51.Parham, P., Barnstable, C. J. & Bodmer, W. F. (1979) J. Immunol. 123, 342-349. [PubMed] [Google Scholar]
- 52.Ciccone, E., Pende, D., Nanni, L., Di Donato, C., Viale, O., Beretta, A., Vitale, M., Sivori, S., Moretta, A. & Moretta, L. (1995) Int. Immunol. 7, 393-400. [DOI] [PubMed] [Google Scholar]
- 53.Rebai, N. & Malissen, B. (1983) Tissue Antigens 22, 107-117. [DOI] [PubMed] [Google Scholar]
- 54.Stam, N. J., Spits, H. & Ploegh, H. L. (1986) J. Immunol. 137, 2299-2306. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.