Abstract
N-terminal pyroglutamate (pGlu) formation from its glutaminyl (or glutamyl) precursor is required in the maturation of numerous bioactive peptides. The aberrant formation of pGlu may be related to several pathological processes, such as osteoporosis and amyloidotic diseases. This N-terminal cyclization reaction, once thought to proceed spontaneously, is greatly facilitated by the enzyme glutaminyl cyclase (QC). To probe this important but poorly understood modification, we present here the structure of human QC in free form and bound to a substrate and three imidazole-derived inhibitors. The structure reveals an α/β scaffold akin to that of two-zinc exopeptidases but with several insertions and deletions, particularly in the active-site region. The relatively closed active site displays alternate conformations due to the different indole orientations of Trp-207, resulting in two substrate (glutamine t-butyl ester)-binding modes. The single zinc ion in the active site is coordinated to three conserved residues and one water molecule, which is replaced by an imidazole nitrogen upon binding of the inhibitors. Together with structural and kinetic analyses of several active-site-mutant enzymes, a catalysis mechanism of the formation of protein N-terminal pGlu is proposed. Our results provide a structural basis for the rational design of inhibitors against QC-associated disorders.
Keywords: crystallography, intramolecular cyclization, posttranslational modification, Alzheimer's disease, aminopeptidase
N-terminal pyroglutamate (pGlu) formation from its glutaminyl precursor is an important posttranslational or cotranslational event in the processing of numerous bioactive neuropeptides, hormones, and cytokines during their maturation in the secretory pathway. These regulatory peptides require the N-terminal pGlu to develop the proper conformation for binding to their receptors and/or to protect the N termini of the peptides from exopeptidase degradation (1, 2). Previously, this N-terminal cyclization reaction was thought to proceed spontaneously, until the glutaminyl cyclases (QCs) were identified as catalysts that are responsible for this posttranslational modification (3, 4).
QCs (EC 2.3.2.5) are acyltransferases that have been identified in both animal and plant sources (3–5). They are abundant in mammalian neuroendocrine tissues, such as hypothalamus and pituitary (4, 6), and are highly conserved from yeast to human. Animal QCs were shown to have distinct structure and protein stability from plant QCs despite their similar molecular masses [i.e., 33–40 kDa (5, 7)]. No bacterial QCs have been reported thus far; however, the mammalian QCs were predicted to exhibit remarkable homology to the bacterial double-zinc aminopeptidases (8, 9).
In humans, several genetic diseases, such as osteoporosis, a multifactorial hormonal disease that is characterized by reduced bone mass and microarchitectural deterioration of bone tissue (10), appear to result from mutations of the QC gene. The gene encoding QC (QPCT) lies on chromosome 2p22.3. Within the region, 13 SNPs were analyzed and showed a striking correlation with osteoporosis susceptibility in adult women (11). Of these SNPs, R54W presents the most prominent association with osteoporosis statistically, which was proposed to affect the pathogenesis through the hypothalamus–pituitary–gonadal axis (11).
Interestingly, QC also catalyzes the N-terminal glutamate cyclization into pGlu (12). This reaction is probably related to the formation of several plaque-forming peptides, such as amyloid-β (Aβ) peptides and collagen-like Alzheimer amyloid plaque component, which play a pivotal role in Alzheimer's disease (13, 14). Peptides containing N-terminal pGlu [e.g., pGlu3-Aβ(3-40) and pGlu3-Aβ(3-42/43)], are major fractions of the Aβ peptides within the core of neuritic plaques (15–17). The N-terminal pGlu could enhance the hydrophobicity, proteolytic stability, and neurotoxicity of these peptides (17, 18), probably causing a profused accumulation of pGlu-Aβ peptides in several senile plaques and thus accelerating the progression of neurodegenerative disorders.
To date, there remain several arguments concerning the properties and structure of human and animal QCs. In this study, we report the high-resolution crystal structure of QC from human bone marrow. We also show the structures of human QC in complex with a substrate and three imidazole-derived inhibitors. Based on the refined structures, a systematic site-directed mutagenesis and further structural studies were carried out, providing insights into how protein N-terminal pGlu is formed and offering a structural basis for the rational design of inhibitors against QC-associated disorders.
Materials and Methods
Additional experimental details can be found in Supporting Text, which is published as supporting information on the PNAS web site.
Protein Expression and Purification. The cDNA encoding human QC was amplified by PCR from a human bone marrow cDNA library (Clontech); the mature enzyme (residues 33–361) was expressed in Escherichia coli cells by using a pET 32a expression vector (Novagen) with several modifications as described in ref. 19. Selenomethionine-labeled protein was produced in E. coli by using a nonauxotrophic protocol and purified in a manner similar to the native protein. In addition, the mutants of human QC were constructed by using the QuikChange site-directed mutagenesis kit (Stratagene) and were expressed and purified in the same manner as the WT human QC.
Enzyme Kinetic Assay. The enzyme kinetic assay was based on spectrophotometric measurement as described in ref. 19. The assay solutions (500 μl) contained 15 units of glutamate dehydrogenase, 12 mM α-ketoglutarate, 0.6 mM NADH, and varying concentrations of the synthetic substrate (H-Gln-Gln-OH) in 50 mM Tris.HCl (pH 8.0, 7.0, 7.5, 8.5, or 8.8). Reactions were started by adding QC (0.04–2.5 μg), and activity was monitored by recording the decrease of NADH absorbance at 340 nm.
Crystallization and Data Collection. Purified human QC was concentrated to 8–10 mg/ml and crystallized at 25°C by the hanging drop vapor-diffusion method. Rhombohedral crystals for WT, selenomethionine-labeled, and mutant human QC were grown by using equal volumes of the protein solution and the reservoir that contained 1.6–1.8 M (NH4)2SO4, 4% dioxane, and 100 mM Mes (pH 6.5). In the condition of pH 8.0, the Mes buffer in the reservoir was replaced by Tris.HCl. For the inhibitor-bound crystal forms, a 2-μl reservoir was mixed with 1.5 μl of protein solution and 0.5 μl of inhibitor solution (100 mM). In the case of the substrate-bound form, the crystals of the mutant E201Q (grown at pH 7.0) were soaked for 1.5 h in a solution consisting of 75% mother liquor/25% glycerol/1.1 M glutamine t-butyl ester. X-ray diffraction experiments were performed at various synchrotron beamlines (Tables 2 and 3, which are published as supporting information on the PNAS web site). Before mounting on the x-ray machine, crystals were briefly soaked in mother liquor containing 20–25% glycerol (vol/vol) as a cryoprotectant. All diffraction data were processed and scaled by using the hkl package (20) (listed in Tables 2 and 3). The space group of these crystals is R32, with typical unit cells of a = b = 119 Å and c = 333 Å, in which an asymmetric unit comprises two human QC molecules.
Structure Determination and Refinement. The human QC structure at pH 6.5 was solved with the multiwavelength anomalous diffraction (MAD) phasing method with the program solve (21). Using the MAD data at 20- to 2.0-Å resolution range collected at the wavelengths of 0.9792 Å (peak), 0.9794 Å (edge), and 0.9750 Å (high-energy remote) (see Table 2), we successfully located all 14 Se atom sites in the asymmetric unit. Subsequently, performed with resolve (21), the initial electron density was modified by solvent flattening, and up to 83% of the protein model was automatically built by using all MAD data of 50- to 1.8-Å resolution. Manual building of the remaining model and further refinement were carried out with the program o (22) against a 1.66-Å-resolution data set of the WT crystal. The isomorphous structures of R54W, the pH-8.0 crystal form, and the substrate- and inhibitor-bound forms were phased by using the refined model. For each structure, iterative cycles of model building with o and computational refinement with cns (23) were performed. Rfree values were calculated by using 5% reflections. The stereochemical quality of the refined structures was checked with the program procheck (24). The final refined structures each include 323 of the 329 total residues in a human QC molecule, with a small disordered region of residues 183–188. Well ordered water molecules were located and included in the models. The molecular figures were produced by using molscript (25), raster3d (26), and grasp (27).
Results and Discussion
For additional results and discussion, see Supporting Text.
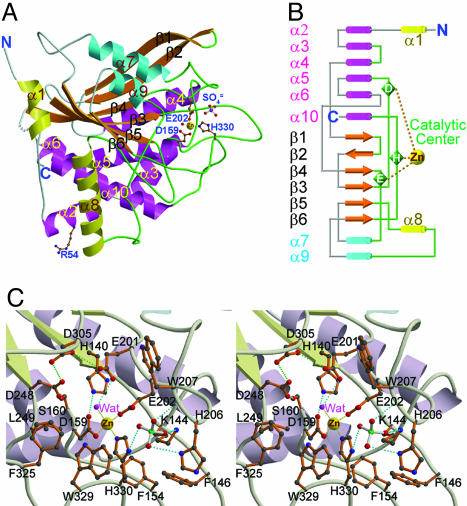
Overall Structure. The mature domain (residues 33–361) of human QC was shown to possess glutaminyl and glutamyl cyclase activities on the putative physiological substrate of human QC (Fig. 6, which is published as supporting information on the PNAS web site). The asymmetric unit of the crystals, grown at pH 6.5, contains two human QC molecules with a rms deviation of 0.386 Å (for all Cα atoms) between them. The globular structure reveals a mixed α/β fold with a size of 63 × 58 × 41 Å3. Up to 36% and 16% of the amino acid residues are involved in the α-helix and β-sheet, respectively, with 6% in the 310-helix regions (Fig. 7, which is published as supporting information on the PNAS web site). The structure has an open-sandwich topology comprising a central six-stranded β-sheet surrounded by two (α7 and α9) and six (α2, α3, α4, α5, α6, and α10) α-helices on opposite sides and flanked by two α-helices (α1 and α8) at one edge of the β-sheet (Fig. 1A). This twisted β-sheet is formed by two antiparallel (β1 and β2) and four parallel (β3, β4, β5, and β6) strands, constituting the hydrophobic core of the molecule. The coil and loop regions of the structure represent 42% of the total residues; about half of them are major components of the active site (Fig. 1B).
Fig. 1.
Structure of human QC. (A) A ribbon diagram of the overall structure of human QC. The central six β-strands are colored orange. The α-helices located on the top, bottom, and edge are colored cyan, magenta, and yellow, respectively. The zinc ion is shown as a yellow sphere. The zinc-coordinated residues, Arg-54 (genetic mutation to Trp residue occurred frequently in adult women with osteoporosis), and a sulfate ion are depicted with a ball-and-stick model. The coils and loops adjacent to the catalytic center are painted green, whereas those distant from the active site are colored gray. Gray dots represent the disordered region of residues 183–188. (B) A topology diagram of the human QC structure. The color codes for secondary structural elements are identical to those in A.(C) A stereoview of the human QC catalytic region. The active-site residues in conf-A are shown and labeled. Possible hydrogen and coordination bonds are represented with dotted lines colored cyan and yellow, respectively. The green dotted lines depict the possibly unusual hydrogen bonds between D305 and E201 (3.06 Å) and between D305 and D248 (2.53 Å).
The structures at pH 6.5 and 8.0 are essentially similar, with a rms deviation (rmsd) of 0.155 Å (for all Cα atoms) between them. The catalytic activity of the enzyme under pH 8.0 was better than that analyzed at lower pH values (Table 1). This observation agrees with the proposal that human QC activity requires an unprotonated α-amino group of the substrate (28). Interestingly, the structure of the genetic mutant R54W showed only a slight movement of the residues adjacent to W54 (rmsd = 0.119 Å for all Cα atoms compared with the pH-6.5 structure), which is ≈34 Å away from the active site (see Fig. 8, which is published as supporting information on the PNAS web site). This mutant retains ≈70% of the catalytic activity of the WT enzyme (Table 1), and its association with osteoporosis may be attributed to some unknown interactions with other molecules rather than its activity.
Table 1. Kinetic parameters of WT and mutant human QC.
| QC | Km, mM | kcat, S-1 | kcat/Km, mM-1·S-1 |
|---|---|---|---|
| Wild | |||
| pH 7.0 | 0.79 ± 0.13* | 7.30 ± 0.01 | 9.459 ± 1.544 |
| pH 7.5 | 0.90 ± 0.09 | 9.76 ± 1.47 | 11.104 ± 2.716 |
| pH 8.0 | 0.63 ± 0.01 | 8.63 ± 0.48 | 13.663 ± 0.497 |
| pH 8.5 | 0.90 ± 0.05 | 9.93 ± 0.30 | 11.044 ± 0.331 |
| pH 8.8 | 2.06 ± 0.62 | 8.56 ± 1.55 | 4.319 ± 0.544 |
| Mutant† | |||
| R54W | 0.76 ± 0.04 | 7.35 ± 0.26 | 9.704 ± 0.824 |
| K144A | 1.47 ± 0.02 | 11.67 ± 0.34 | 7.944 ± 0.368 |
| F146A | 0.82 ± 0.16 | 7.91 ± 2.14 | 9.536 ± 0.769 |
| E201D | 12.62 ± 2.98 | 0.87 ± 0.28 | 0.068 ± 0.007 |
| E201Q‡ | ND | ||
| W207L | 1.77 ± 0.07 | 0.43 ± 0.01 | 0.243 ± 0.002 |
| W207F | 0.59 ± 0.05 | 2.32 ± 0.07 | 3.943 ± 0.189 |
| D248A‡ | ND | ||
| Q304L | 1.16 ± 0.09 | 9.39 ± 1.18 | 8.028 ± 0.386 |
| D305L‡ | ND | ||
| F325A | 4.67 ± 0.24 | 12.91 ± 0.06 | 2.772 ± 0.132 |
| W329A | 29.53 ± 2.29 | 1.35 ± 0.07 | 0.046 ± 0.001 |
ND, not detectable.
Values are represented as mean ± SD (n = 2 or 3).
The assays for mutants were carried out under pH 8.0.
E201Q, D248A, and D305L were shown to possess ≈0.001%, ≈0.1%, and ≈0.03% of the activity of the WT enzyme, respectively.
A structural homology search using the CE/CL server (http://cl.sdsc.edu) revealed that human QC bears some degrees of similarity to the exopeptidase family (Table 4, which is published as supporting information on the PNAS web site). Among these, ApAP (Aeromonas proteolytica aminopeptidase) (29), SgAP (Streptomyces griseus aminopeptidase) (30), and CPG2 (carboxypeptidase G2) (31) share the most conserved scaffold with human QC (see Fig. 9, which is published as supporting information on the PNAS web site), consistent with the previous predictions (8, 9). The key structural difference of human QC with respect to these exopeptidases is in the coil and loop regions attributed to several insertions and deletions (see Fig. 10, which is published as supporting information on the PNAS web site), especially the loops surrounding the active-site pocket. Consequently, human QC structure shows a relatively less compact fold with a larger surface area, in contrast to these exopeptidases.
Structure of the Active Site. The active site is mainly created by six loops between α3 and α4, β3 and α5, β4 and α7, β5 and α8, α8 and α9, and β6 and α10 (Fig. 1B). The catalytic pocket is near the C-terminal edge of the central parallel strands β3, β4, and β5 (Fig. 1 A). It is relatively narrow but accessible to the bulk solvent by means of a solvent channel. The single zinc ion of human QC (19) lies at the bottom of the active-site pocket and is tetrahedrally coordinated to D159 Oδ2, E202 Oε1, H330 Nε2, and a water molecule. In addition, several other completely conserved residues, including E201, W207, D248, D305, F325, and W329 (see Fig. 7), abut the zinc environment (see Fig. 1C), suggesting some roles in catalysis. Mutations of those amino acids decreased enzyme activity significantly (Table 1), in agreement with some results by Bateman et al. (32). Interestingly, the acidic E201, D248, and D305 are pointing to each other at pH 6.5 and 8.0, likely forming hydrogen bonds between them. The peptide bond between the zinc-coordinated D159 and the following S160 adopts a cis-configuration stabilized by a network of hydrogen bonds, including D159 Oδ1-H140 Nε2 (2.70 Å), S160 Oγ-D248 Oδ1 (2.66 Å), D159 O-water (2.65 Å), and S160 N-water (2.80 Å).
The hydrophobic active-site pocket is lined by residues K144, F146, F154, L249, I303, I321, F325, and W329, having approximate dimensions of 13 × 11 × 7 Å3. There are six water molecules located inside the pocket, including the water coordinated to the zinc ion. In addition, a sulfate ion is located near the opening of the pocket, hydrogen-bonded to K144 Nζ, H206 Nδ1, W207 N, H330 Nδ1, and several water molecules (Fig. 1C). Surprisingly, the two independent QCs in the asymmetric unit have different active-site conformations (denoted as “conf-A” and “conf-B”), particularly at the segment of L205-H206-W207 (see Fig. 11, which is published as supporting information on the PNAS web site). The W207 indole ring in conf-A is directed toward the surface of the molecule, whereas that in conf-B is oriented closer to the zinc ion.
Comparison of the Active-Site Structures of Mononuclear Human QC with the Binuclear Exopeptidases. The five zinc-coordinated residues on the active site of ApAP, SgAP, and CPG2, except for E200 in CPG2, are completely conserved in animal QCs (Figs. 7 and 10A). The orientations of these residues appear to superpose well with those in human QC structure (Fig. 2A). However, human QC has only a single zinc ion, structurally equivalent to the first and higher-affinity zinc (Zn1) in these two-zinc proteases. Presumably, this difference is due to the slight rotation of the H140 and D159 side chains in human QC, which creates a new hydrogen bond between the two amino acids. The new juxtaposition of H140 and D159 does not favor the binding of the second zinc ion (Zn2). Such evolutional conversion of the metal center (and thus the active-site pocket) between the proteases and human QC might play some role in the reorientation of the substrate necessary for these functionally distinct enzymes.
Fig. 2.
Comparison of the active-site structures of human QC and the exopeptidases. (A) Superposition (stereoview) of the conserved active-site residues in human QC (magenta), ApAP (cyan), SgAP (yellow), and CPG2 (green). The orange circles indicate the slight rotation of human QC residues compared with those in the aminopeptidases. The dotted line (distance is in Å) represents a hydrogen bond between His-140 and Asp-159 in human QC. The tightly and weakly bound zinc ions in the active site of ApAP, SgAP, and CPG2 are marked as Zn1 and Zn2, respectively. (B) Molecular surface of the human QC active site with bound 1-benzylimidazole. To enhance the clarity, Trp-329 was sliced out of view. The region structurally corresponding to the S1 pocket of aminopeptidases is colored. The conserved active-site residues are colored green, the inhibitor is pink, and the zinc ion is gray. (C) Molecular surface of the ApAP active site with bound p-iodo-d-Phe hydroxamate (Protein Data Bank ID code 1IGB). The residues Tyr-225 and Phe-248 have been sliced out of view. The region of S1 pocket is colored in a fashion similar to B. The zinc-binding residues are colored green, p-iodo-d-Phe hydroxamate is pink, and zinc ions are gray.
As discussed above, the major structural differences between human QC and the proteases lie in the active-site region created by several flexible loops. Consequently, the specific S1 pocket of the aminopeptidases was not found in the active site of human QC. As shown in Fig. 2 B and C, Y251 of ApAP, lying at the opening of the S1 site, is structurally replaced by F325 in human QC in a drastically different orientation. The resulting more closed conformation of the human QC active site probably induces the γ-amide group of the substrate N-terminal Gln residue to direct toward the zinc-catalytic center in favor of an intramolecular cyclization.
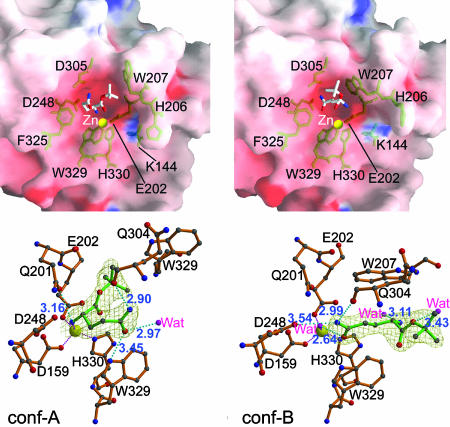
Structure of the Enzyme–Substrate Complex. The mutant E201Q, which is almost totally inactive (Table 1), was used to produce the substrate–enzyme complex. Compared with the WT structure, E201Q showed little conformational change, except for an additional hydrogen bond between Q201 Nε2 and D305 Oδ1 (3.11 Å). The bound substrate, glutamine t-butyl ester, in conf-A and conf-B of E201Q adopts two binding modes (Fig. 3), likely due to the different indole orientations of W207. In conf-A, the bound substrate uses its α-amino nitrogen of the Gln residue to coordinate to the zinc ion by displacing the zinc-ligated water molecule. This α-amino nitrogen is also hydrogen-bonded to Q201 Oε1. The γ-amide of the substrate Gln is stabilized in conf-A by hydrogen-bonding to Q304 O and W329 Nε1; the t-butyl group is embedded between E202 and W207 of the enzyme with few specific interactions. In conf-B, the substrate t-butyl group is oriented toward the surface of the enzyme, likely due to the crowding of the bulky indole ring of W207. Surprisingly, the γ-amide carbonyl group of the substrate did not displace the zinc-coordinated water molecule; instead, it hydrogen-bonded to it. The γ-amide amino group is stabilized by two hydrogen bonds to Q304 O and D248 Oδ2 of the enzyme. In contrast to that in conf-A, the α-amino group of the substrate shows no direct contact with human QC.
Fig. 3.
Comparison of conf-A (Left) and conf-B (Right) of the mutant E201Q bound to the substrate glutamine t-butyl ester. (Upper) Properties of the molecular surface. The charge potentials are calculated by using grasp (27) with a range of –20 to +20 kBT (where kB is the Boltzmann constant and T is temperature in kelvin), colored from red to blue. The stick models for substrate and some active-site residues are colored white and green, respectively. The zinc ions are shown as yellow balls. Note that the different orientations of His-206 and Trp-207 in conf-A and conf-B result in the different active-site conformations and different substrate-binding modes. (Lower) A close-up view of the binding of glutamine t-butyl ester to the active site. The substrate and human QC residues are represented with a ball-and-stick model colored green and orange, respectively. The 2Fo – Fc electron density maps (contoured at 1.0σ) for substrate are shown. Dotted lines in cyan and magenta depict the hydrogen bonds (distance is in Å) and coordination bonds, respectively.
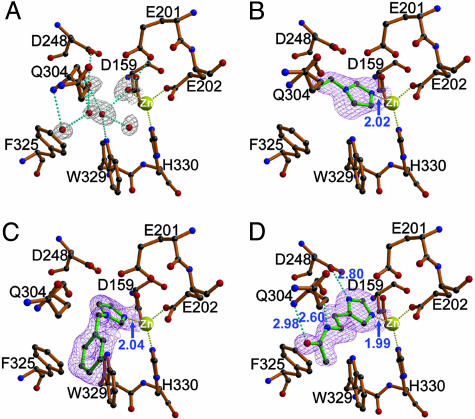
Structure of Enzyme–Inhibitor Complexes. The binding mode of the imidazole-derived inhibitors (for chemical structures, see Fig. 12 A, which is published as supporting information on the PNAS web site) has no obvious difference between conf-A and conf-B. As shown in Fig. 4, binding of the inhibitors results in the removal of six water molecules within the active-site pocket, including the zinc-coordinated one, which is replaced by an imidazole nitrogen of the inhibitors. The inhibitors adopt different orientations because of their different modifications on the imidazole ring. The small vinyl moiety of 1-vinylimidazole shows no interaction with the active site of human QC, leaving a large space in the catalytic pocket after its binding (Fig. 4B). However, the bulky hydrophobic phenyl ring on 1-benzylimidazole is closely surrounded and stabilized by the phenyl and indole groups of F325 and W329, respectively (Fig. 4C). In contrast, the substituent of N-ω-acetylhistamine is oriented almost parallel to the backbone of segment G301-Q304, stabilized mainly by three additional hydrogen bonds to D248 Oδ2, Q304 N, and Q304 O of the enzyme (Fig. 4D). In general, the binding of the inhibitors does not induce significant conformational changes in the enzyme, likely because of their smaller sizes compared with those of the active-site pocket (Fig. 12 B and C).
Fig. 4.
Structures of human QC bound to imidazole-derived inhibitors. (A) The zinc-binding environment of the free-form human QC. The 2Fo – Fc electron density maps (contoured at 1.0σ) (gray) corresponding to the water molecules inside the active-site pocket are shown. Representations of the models, hydrogen bonds, and coordination bonds are identical to those in Fig. 1C. (B–D) Structures of human QC bound to 1-vinylimidazole (1.68-Å resolution), 1-benzylimidazole (1.64-Å resolution), and N-ω-acetylhistamine (1.56-Å resolution), respectively. The 2Fo – Fc maps (contoured at 1.0σ) (magenta) for the inhibitors are overlaid with the final refined models. Distances for enzyme–inhibitor interaction are indicated in Å.
Recently, several imidazole N1 derivatives were reported to be potent inhibitors of human QC, whereas substitution on the carbon atoms of imidazole resulted in a lower inhibitory effect (8). The only exception was N-ω-acetylhistamine, whose Ki (17 μM) is 50-fold lower than that of l-histamine (Ki = 850 μM) (8). From our structures, this difference is likely due to the additional hydrogen bonds. Interestingly, despite its hydrogen bondings, the inhibitory activity of N-ω-acetylhistamine is still weaker than that of 1-benzylimidazole (Ki = 7.1 μM), which binds to the enzyme mostly by hydrophobic interaction. The substituent on the C2 atom of the imidazole ring might have steric hindrance by means of this carbon atom adjacent to the zinc-coordinated N1 or N3 atoms. In addition, the N1 or C4 substituents of imidazole are primarily surrounded by hydrophobic L249, F325, and W329. Thus, the charged or hydrophilic groups on these positions of imidazole are likely unfavorable, unless they adopt an orientation that is preferable for the formation of salt bridges or hydrogen bonds to the enzyme, like the binding mode of N-ω-acetylhistamine. These observations are consistent with several results from Schilling's (8) studies [e.g., the significant decrease of the inhibition activities of N-benzoylimidazole (Ki = 174 μM) and l-histidine (Ki = 4,400 μM) compared with 1-benzylimidazole and histidinamide (Ki = 560 μM), respectively]. We conclude that an electron-rich nucleophile with a good ability to ligate the zinc ion of human QC, combined with bulky hydrophobic substituents, is likely the structural basis of a potent QC inhibitor.
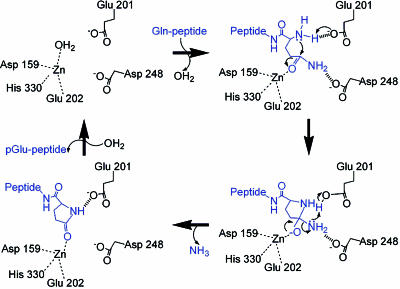
Proposed Substrate-Binding and Catalysis Mechanism. It has been established that zinc-dependent exopeptidases employ a catalytic glutamyl residue, such as E270, E151, and E131 in carboxypeptidase A, ApAP, and SgAP, respectively (33–36). The glutamyl residue accepts a proton from a zinc-bound water molecule and transfers the proton to the leaving NH group of the scissile peptide bond during the catalysis process. Surprisingly, such a glutamyl residue is also conserved in the structures of human QC and other animal QCs. As shown in Fig. 2 A, E201 of human QC adopts an almost identical orientation to E151 and E131 of ApAP and SgAP, respectively, and is absolutely conserved from yeast to human QCs (Fig. 7), suggesting an analogous role in catalysis. This proposal is strengthened by the mutants E201D and E201Q, which showed a drastic decrease of QC activity compared with that of the WT enzyme (Table 1).
Moreover, because QC catalyzes an intramolecular cyclization, the proper positioning of the N-terminal α-amino group of the substrate needs to be in close proximity to the γ-carbonyl carbon. Based on the proposed catalysis mechanism of ApAP and SgAP (35, 36), it is reasonable to assume that the zinc ion of QC acts by polarizing the γ-amide group of the substrate Gln residue and simultaneously stabilizing the oxyanion formed by the nucleophilic attack of the α-nitrogen on the scissile γ-carbonyl carbon. E201 of human QC may transfer a proton from the α-amine of the substrate to the leaving amino group on the scissile γ-amide (Fig. 5). Such a catalysis mechanism is supported by several lines of evidence: (i) It does not require the presence of the second zinc ion, because Zn2 in aminopeptidases was thought to play a role in stabilizing the terminal α-amino group of the substrate (34, 35). (ii) The mutant E201D showed an ≈20-fold higher Km value compared with that of the WT enzyme. (iii) The mutant E201Q showed almost no catalysis activity. In addition, the leaving amino group on the scissile γ-amide of the substrate is probably stabilized by D248 during the catalysis process, which is suggested by the plausible orientation of D248 (see Figs. 1C and 3) and the drop of D248A activity. The proposed mechanism is very similar to the one proposed for ApAP where only Zn1 is present in the active site of the protease (37).
Fig. 5.
Proposed catalysis mechanism of human QC. The conserved Glu-201 acts as the general base and acid to transfer a proton from the α-amino group of the substrate (blue) to the leaving amino group on the scissile γ-amide. The zinc ion polarizes the γ-amide carbonyl group of the substrate and simultaneously stabilizes the oxyanion formed by the nucleophilic attack of the α-nitrogen. Asp-248 probably stabilizes the leaving γ-amide amino group during the catalysis process. In respect of the mechanism of glutamyl cyclase activity, this leaving amino group is replaced by a hydroxyl group, and the reaction is favored at pH 6.0 (12).
Human QC substrates were shown to prefer a large hydrophobic group on their penultimate position, such as Trp, Phe, or Tyr residues (28). From our structures, this preference is likely due to the stacking effect of W207. Mutations on this residue lowered the kcat value of the enzyme remarkably (Table 1), suggesting that W207 also participates in the catalysis process. By superposition, W329 of human QC corresponds well to Y246 in SgAP, which was proposed to involve in the catalysis of the protease (36). Similarly, our structure of enzyme–substrate complex as well as the significantly increased Km value of W329A indicate the importance of W329 in the binding of the substrate. Like W329, F325 also plays some role in the substrate binding, as supported by an ≈7.5-fold increase of the Km value of the mutant F325A. Moreover, as shown in Fig. 1C, D305 is closely contiguous to the catalysis-essential E201 and D248 and contributes to the acidity of the catalytic environment. Drop of the D305L activity suggests a role of D305 in stabilizing the unusual hydrogen bondings between D305 and E201 and between D305 and D248. Conceivably, the decrease in QC activity above pH 8.0 (see Table 1) may be linked to a breakdown of this hydrogen-bond network, and the drop in D248A activity by three orders of magnitude may be a consequence of partial disruption of this network. In contrast, K144, F146, and the side chain of Q304 in the active site appear to be away from the bound substrate, consistent with the results from kinetic assays of the mutants K144A, F146A, and Q304L (Table 1).
Because the lone-pair electrons on the α-amino nitrogen of the substrate are necessary for the cyclization reaction, the bound glutamine t-butyl ester in conf-A seems to be in a substrate-inhibition state, and the enzyme reaction cannot proceed further. However, in conf-B, the substrate α-amino group is ≈5.05 Å from the γ-amide, perhaps because of the Glu-to-Gln mutation. In contrast, the α-amino nitrogen of the substrate in conf-A is hydrogen-bonded to Q201, and the γ-amide in conf-B is pointing to the active-site zinc ion. These findings support our proposed catalysis mechanism. It is notable that conf-A has a larger active-site pocket than conf-B (see Fig. 3); thus, conf-A seems preferable for accommodating the cyclized intermediate and product during the catalysis process. Interestingly, in conf-B, the orientation of the substrate t-butyl group is in agreement with the fact that the specificity of a human QC substrate depends on its first three residues (28). These observations raise the question of whether interconversion between conf-A and conf-B occurs during catalysis.
In conclusion, many peptides that are involved in the progression of amyloid plaque, such as collagen-like Alzheimer amyloid plaque component (14) and ABri peptide (38), in addition to the Aβ peptides, have been reported to contain the N-terminal pGlu residue that is derived from its Glu precursor. Here we show that human QC can time-dependently convert the Glu-Aβ peptide into pGlu-Aβ, despite a significantly lower rate compared with the QC activity. Because QCs are abundant in mammalian brain tissues (4, 6), QC may be responsible for the formation of N-terminal pGlu on several amyloid-related peptides. Here, we present the atomic-resolution crystal structures of human QC and its complexes with substrate and inhibitors. The data of our systematic site-directed mutagenesis corroborate the results from the structural analysis. Our study provides insights into the mechanism of protein N-terminal pGlu formation and forms a firm basis for the rational design of inhibitors against QC-associated disorders.
Supplementary Material
Acknowledgments
We thank Dr. Noriyuki Igarashi (Photon Factory, Tsukuba, Japan) and Dr. Yu-San Huang (National Synchrotron Radiation Research Center, Hsinchu, Taiwan) for x-ray data collections and Dr. Hui-Ming Yu (Academia Sinica) for the chemical synthesis of QC substrates. We are grateful to the National Core Facility of Proteomics (Taipei, Taiwan) (Grant NSC 93-3112-B-001-010-Y) for mass spectrometry analysis. This work was supported by grants from Academia Sinica and National Core Facility of High-Throughput Protein Crystallography Grant NSC 93-3112-B-001-011-Y (to A.H.-J.W.).
Author contributions: K.-F.H. and A.H.-J.W. designed research; K.-F.H., Y.-L.L., and W.-J.C. performed research; K.-F.H., Y.-L.L., W.-J.C., and T.-P.K. analyzed data; and K.-F.H. and A.H.-J.W. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: QC, glutaminyl cyclase; pGlu, pyroglutamate; ApAP, Aeromonas proteolytica aminopeptidase; SgAP, Streptomyces griseus aminopeptidase; CPG2, carboxypeptidase G2;Aβ, amyloid-β.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2AFM, 2AFO, 2AFU, 2AFW, 2AFX, 2AFZ, and 2AFS).
References
- 1.Van Coillie, E., Proost, P., Van Aelst, I., Struyf, S., Polfliet, M., De Meester, I., Harvey, D. J., Van Damme, J. & Opdenakker, G. (1998) Biochemistry 37, 12672–12680. [DOI] [PubMed] [Google Scholar]
- 2.Hinke, S. A., Pospisilik, J. A., Demuth, H. U., Mannhart, S., Kühn-Wache, K., Hoffmann, T., Nishimura, E., Pederson, R. A. & McIntosh, C. H. S. (2000) J. Biol. Chem. 275, 3827–3834. [DOI] [PubMed] [Google Scholar]
- 3.Fischer, W. H. & Spiess, J. (1987) Proc. Natl. Acad. Sci. USA 84, 3628–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Busby, W. H. J., Quackenbush, G. E., Humm, J., Youngblood, W. W. & Kizer, J. S. (1987) J. Biol. Chem. 262, 8532–8536. [PubMed] [Google Scholar]
- 5.Oberg, K. A., Ruysschaert, J. M., Azarkan, M., Smolders, N., Zerhouni, S., Wintjens, R., Amrani, A. & Looze, Y. (1998) Eur. J. Biochem. 258, 214–222. [DOI] [PubMed] [Google Scholar]
- 6.Sykes, P. A., Watson, S. J., Temple, J. S. & Bateman, R. C., Jr. (1999) FEBS Lett. 455, 159–161. [DOI] [PubMed] [Google Scholar]
- 7.Schilling, S., Hoffmann, T., Rosche, F., Manhart, S., Wasternack, C. & Demuth, H. U. (2002) Biochemistry 41, 10849–10857. [DOI] [PubMed] [Google Scholar]
- 8.Schilling, S., Niestroj, A. J., Rahfeld, J. U., Hoffmann, T., Wermann, M., Zunkel, K., Wasternack, C. & Demuth, H. U. (2003) J. Biol. Chem. 278, 49773–49779. [DOI] [PubMed] [Google Scholar]
- 9.Booth, R. E., Lovell, S. C., Misquitta, S. A. & Bateman, R. C., Jr. (2004) BMC Biol. 2, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart, T. L. & Ralston, S. H. (2000) J. Endocrinol. 166, 235–245. [DOI] [PubMed] [Google Scholar]
- 11.Ezura, Y., Kajita, M., Ishida, R., Yoshida, S., Yoshida, H., Suzuki, T., Hosoi, T., Inoue, S., Shiraki, M., Orimo, H., et al. (2004) J. Bone Miner. Res. 19, 1296–1301. [DOI] [PubMed] [Google Scholar]
- 12.Schilling, S., Hoffmann, T., Manhart, S., Hoffmann, M. & Demuth, H. U. (2004) FEBS Lett. 563, 191–196. [DOI] [PubMed] [Google Scholar]
- 13.Morgan, C., Colombres, M., Nuñez, M. T. & Inestrosa, N. C. (2004) Prog. Neurobiol. 74, 323–349. [DOI] [PubMed] [Google Scholar]
- 14.Hashimoto, T., Wakabayashi, T., Watanabe, A., Kowa, H., Hosoda, R., Nakamura, A., Kanazawa, I., Arai, T., Takio, K., Mann, D. M. A., et al. (2002) EMBO J. 21, 1524–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saido, T. C., Iwatsubo, T., Mann, D. M., Shimada, H., Ihara, Y. & Kawashima, S. (1995) Neuron 14, 457–466. [DOI] [PubMed] [Google Scholar]
- 16.Kuo, Y. M., Emmerling, M. R., Woods, A. S., Cotter, R. J. & Roher, A. E. (1997) Biochem. Biophys. Res. Commun. 237, 188–191. [DOI] [PubMed] [Google Scholar]
- 17.Russo, C., Violani, E., Salis, S., Venezia, V., Dolcini, V., Damonte, G., Benatti, U., D'Arrigo, C., Patrone, E., Carlo, P., et al. (2002) J. Neurochem. 82, 1480–1489. [DOI] [PubMed] [Google Scholar]
- 18.Harigaya, Y., Saido, T. C., Eckman, C. B., Prada, C. M., Shoji, M. & Younkin, S. G. (2000) Biochem. Biophys. Res. Commun. 276, 422–427. [DOI] [PubMed] [Google Scholar]
- 19.Huang, K. F., Liu, Y. L. & Wang, A. H. J. (2005) Protein Expression Purif., in press.
- 20.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 21.Terwilliger, T. C. (2003) Methods Enzymol. 374, 22–37. [DOI] [PubMed] [Google Scholar]
- 22.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 23.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 24.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 25.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24, 946–950. [Google Scholar]
- 26.Merrit, E. A. & Bacon, D. J. (1997) Methods Enzymol. 277, 505–524. [DOI] [PubMed] [Google Scholar]
- 27.Nicholls, A., Sharp, K. A. & Honig, B. (1991) Proteins 11, 281–296. [DOI] [PubMed] [Google Scholar]
- 28.Schilling, S., Manhart, S., Hoffmann, T., Ludwig, H. H., Wasternack, C. & Demuth, H. U. (2003) Biol. Chem. 384, 1583–1592. [DOI] [PubMed] [Google Scholar]
- 29.Chevrier, B., Schalk, C., D'Orchymont, H., Rondeau, J. M., Moras, D. & Tarnus, C. (1994) Structure 2, 283–291. [DOI] [PubMed] [Google Scholar]
- 30.Greenblatt, H. M., Almog, O., Maras, B., Spungin-Bialik, A., Barra, D., Blumberg, S. & Shoham, G. (1997) J. Mol. Biol. 265, 620–636. [DOI] [PubMed] [Google Scholar]
- 31.Rowsell, S., Pauptit, R. A., Tucker, A. D., Melton, R. G., Blow, D. M. & Brick, P. (1997) Structure 5, 337–347. [DOI] [PubMed] [Google Scholar]
- 32.Bateman, R. C., Jr., Temple, J. S., Misquitta, S. A. & Booth, R. E. (2001) Biochemistry 40, 11246–11250. [DOI] [PubMed] [Google Scholar]
- 33.Lipscomb, W. N. & Sträter, N. (1996) Chem. Rev. 96, 2375–2434. [DOI] [PubMed] [Google Scholar]
- 34.Lowther, W. T. & Matthews, B. W. (2002) Chem. Rev. 102, 4581–4607. [DOI] [PubMed] [Google Scholar]
- 35.Bzymek, K. P. & Holz, R. C. (2004) J. Biol. Chem. 279, 31018–31025. [DOI] [PubMed] [Google Scholar]
- 36.Fundoiano-Hershcovitz, Y., Rabinovitch, L., Langut, Y., Reiland, V., Shoham, G. & Shoham, Y. (2004) FEBS Lett. 571, 192–196. [DOI] [PubMed] [Google Scholar]
- 37.Holz, R. C. (2002) Coord. Chem. Rev. 232, 5–26. [Google Scholar]
- 38.Vidal, R., Frangione, B., Rostagno, A., Mead, S., Révész, T., Plant, G. & Ghiso, J. (1999) Nature 399, 776–781. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.