Abstract
CNS neurons are endowed with the ability to recover from cytotoxic insults associated with the accumulation of proteinaceous aggregates in mouse models of polyglutamine disease, but the cellular mechanism underlying this phenomenon is unknown. Here, we show that autophagy is essential for the elimination of aggregated forms of mutant huntingtin and ataxin-1 from the cytoplasmic but not nuclear compartments. Human orthologs of yeast autophagy genes, molecular determinants of autophagic vacuole formation, are recruited to cytoplasmic but not nuclear inclusion bodies in vitro and in vivo. These data indicate that autophagy is a critical component of the cellular clearance of toxic protein aggregates and may help to explain why protein aggregates are more toxic when directed to the nucleus.
Keywords: autophagy, huntingtin, Huntington's disease
Polyglutamine (polyQ) expansion disorders, such as Huntington's disease (HD) and spinocerebellar ataxia type 1, are caused by the production of mutant proteins, huntingtin (Htt) and ataxin-1 (Atx1), respectively, that adopt cytotoxic, nonnative, oligomeric, or aggregation-prone conformations because of the presence of long homopolymeric tracts of glutamine (1). The length of polyQ tracts in normal humans is polymorphic but always below a threshold of 35–40. PolyQ repeats above this threshold are invariably associated with disease, with a strong inverse correlation between repeat length and age-of-onset of neurological disease (2). These findings, together with studies on Htt fragments expressed in cell culture (3) or polyQ peptides in vitro (4), support a model in which glutamine repeats above the threshold adopt a nonnative conformation that is highly prone to self-associate into high-molecular-weight, stable aggregates. These aggregates accumulate in nuclear or cytoplasmic inclusion bodies (IBs) that are invariably associated with end-stage neurodegenerative disease in patients and animal models (5, 6). Although IBs are probably not directly responsible for cellular toxicity (5) and may even contribute to cytoprotection (7, 8), the toxicity of mutant polyQ-expanded proteins appears to depend strongly on the cellular compartment in which they accumulate (9, 10). Experimental redirection of Atx1, normally a nuclear protein, to the cytoplasm, drastically reduces toxicity and IB formation in a mouse model of spinocerebellar ataxia type 1 (11). Conversely, the addition of a nuclear localization sequence to the N-terminal polyQ-containing fragment of Htt, which normally partitions between the nucleus and cytoplasm, directs it to the nucleus and increases its toxicity (12, 13). These experiments have been interpreted to indicate that the primary target of polyQ toxicity is in the nucleus. However, it also is possible that both the nucleus and cytoplasm contain targets that are equally vulnerable to these toxic proteins, but that the cytoplasm may be better endowed to neutralize or destroy the proteotoxic agent.
Although protein aggregates are highly insoluble (14), animal models using conditional expression of Htt (15) or Atx1 (16) reveal that disease progression and IB formation can be halted or even reversed upon cessation of polyQ protein expression. Thus, neurons are endowed with the ability to eliminate protein aggregates. Lysosomes and autophagic bodies proliferate in HD (17) and Alzheimer's disease brains (18) and in cell-culture models of disease (7, 19–22), suggesting a linkage between degeneration and autophagy (23). It is impossible to discriminate from those observations whether autophagy, a feature of both apoptotic and nonapoptotic cell death (24), is induced in these diseases as a cytoprotective response or a pathogenic manifestation. However, 3-methyladenine, an inhibitor of class III phosphatidylinositol 3-kinases (25), reduces the capacity of cells to eliminate polyQ-expanded Htt, suggesting a role for vesicular processes such as autophagy in cleavage or degradation of aggregation-prone or aggregated Htt (26, 27). Until recently, studies of mammalian autophagy have been hampered by a lack of specific inhibitors and markers for autophagic organelles. The identification of Saccharomyces cerevisiae genes (Atgs) required for starvation-induced autophagy (28) has helped define the molecular mechanisms of autophagocytosis and provides powerful, specific tools to study this process. We have developed antibodies to human orthologs of yeast Atgs and short interfering RNAs (siRNAs), which we exploit to investigate the role of autophagy in the clearance of aggregated pathogenic proteins from the cytoplasmic and nuclear compartments in cellular models of HD and spinocerebellar ataxia type 1. Our data establish that autophagy efficiently degrades polyQ aggregates from the cytoplasm but not the nucleus.
Experimental Procedures
Cell Lines, Cell Culture, and Transfection. Human embryonic kidney (HEK)-293 cells, HeLa cells, SH-SY5Y, and neuro2a cells were cultured in DMEM supplemented with 10% FBS and antibiotics at 37°C in 95% air/5% CO2. Plasmids were transfected by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Most of the analyses were performed 48 h after transfection unless otherwise noted. Cells were lysed in 1% Nonidet P-40/0.5% sodium deoxycholate/150 mM NaCl/10 mM Tris·HCl (pH 7.5) with protease and phosphatase inhibitors. Clonal cell lines were selected in and maintained at 800 μg/ml G418 (Invitrogen).
Plasmid Constructs. The details of GFP-Q25 and GFP-Q103 Htt and CFTRΔF508 constructs have been published elsewhere (29). The T cell receptor α (TCRα) construct was generated by fusing GFP to mouse TCRα (2B4 clone) (30). The myc-tagged Atx3 construct was a gift from Henry Paulson (University of Iowa, Iowa City). FLAG-tagged Atx1 and the K772T mutant thereof were gifts from Harry Orr (University of Minnesota, Minneapolis). Human Atg5, Atg8-LC3, and Atg12 cDNAs were cloned from a human cDNA library by PCR. Mouse Atg5, Atg8-LC3A, and LC3B cDNA were cloned by RT-PCR from mouse brain RNA. All clones were verified by sequence analysis.
Antibodies and Reagents. Anti-Atg8-LC3 antibody was generated against full-length recombinant human LC3-I produced in Escherichia coli. Immune serum was affinity-purified with recombinant LC3 coupled with Affi-Gel (Bio-Rad). Anti-Atg12 antibody was generated against the N terminus of human Atg12 fused to GST, and the serum was affinity-purified by using this fusion protein. Anti-Atg8-LC3 antibody used for Western blots was a generous gift from Noboru Mizushima (31), and anti-Atg16L antibody was from Tamotsu Yoshimori (32). Anti-GFP antibody was purchased from Roche (Mannheim, Germany). Anti-actin and anti-ubiquitin monoclonal antibodies were from Chemicon International (Temecula, CA). Anti-FLAG M2 antibody was purchased from Sigma. Anti-α-synuclein antibody was purchased from Zymed.
Microscopy. Cells were grown on glass coverslips coated with poly(l-lysine) and collagen, fixed in 4% paraformaldehyde, permeabilized by 0.25% Triton X-100, quenched with 50 mM ammonium chloride, and blocked with 5% BSA in PBS. Primary antibody incubation was done at 4°C overnight, followed by incubation for 1 h at room temperature with Alexa Fluor 488- and Alexa Fluor-546-labeled secondary antibodies (Molecular Probes) and 50 μg/ml bisbenzimide (Sigma). Mouse brains were fixed by perfusion with 4% paraformaldehyde in PBS (pH 7.4) and were paraffin-embedded. The embedded sections were autoclaved for 20 min in 10 mM glycine·HCl (pH 2.5) after deparrafinization. Conventional epifluorescence micrographs were obtained by using an Axiovert 200M microscope with a ×100/numerical aperture 1.4 oil-immersion lens (Zeiss). Digital (12-bit) images were acquired with a cooled charge-coupled device (Roper Scientific, Trenton, NJ) and were processed by using metamorph software (Universal Imaging, Downingtown, PA). The excitation filters used for conventional microscopy were 365WB50 (bisbenzimide), 500AF25 (GFP, Alexa Fluor 488), and 560AF55 (Alexa Fluor 546). The emission filters used were 450 DF65 (bisbenzimide), 480 AF30 (CFP), 545AF35 (GFP, Alexa Fluor 488), 645DF55 (Alexa Fluor 546). The dichroics were: 400 DCLP (bisbenzimide), 455DRLP (CFP), 525 DRLP (GFP, Alexa Fluor 488), and 595 DRLP (Alexa Fluor 546).
Deconvolution Microscopy. For deconvolution microscopy, images were acquired by using an IX-70 inverted microscope (Olympus, Melville, NY) with 490 DF20 (GFP) and 555DF28 (Alexa Fluor 546) filters for excitation and 528 DF38 (GFP) and 617 DF73 (Alexa Fluor 546) filters for emission. Digital images (12-bit) were acquired with a cooled charge-coupled device camera and digitally deconvolved by using deltavision hardware and software (Applied Precision, Issaquah, WA).
RNA Interference. Autophagin short interfering RNAs (siRNAs) were generated in vitro by using recombinant DICER (33). siRNA was generated against full-length human LC3 (GenBank accession no. NM_022818), mouse LC3 (GenBank accession nos. AK003122 and NM_026160), a fragment of human Atg5 (base pairs 215–780; GenBank accession no. NM_004849), and mouse Atg5 (base pairs 215–780; GenBank accession no. NM_053069). For control RNA, sequence from Pseudomonas aeruginosa β-galactosidase (base pairs 293–890; GenBank accession no. AF140579) was used. Purified siRNAs were introduced by using Gene Silencer (Gene Therapy Systems, San Diego) according to the manufacturer's instructions.
Filter-Trap Assay. PolyQ filter-trap assay was done according to the published protocol (3, 34) by using anti-GFP or anti-FLAG antibody. The total protein load was normalized to the volume of the soluble fraction. For the Atx1 filter-trap assay, DNase I was included from the beginning of the solubilization procedure.
Promoter Shutdown Experiment. Neuro2a cells, which express GFP-Htt-Q150 under the control of an ecdysone promoter (35), were induced with 1 μM ponasterone A for 72 h, after which time the medium was replaced with DMEM lacking ponasterone A. Five millimolar dibutyryl-cAMP (N6,2′-O-dibutyryladenosine-3′,5′-cyclic monophosphate sodium salt, Sigma) was present throughout the experiment.
Data Analysis. Image analysis was performed with image j software (version 1.30, http://rsb.info.nih.gov/ij). Statistical analyses were performed with statview (version 5.0, SAS Institute, Cary, NC) by using Student's t test.
Results and Discussion
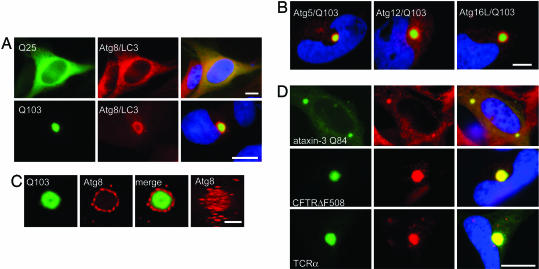
To investigate the role of autophagy in the cellular response to protein aggregation, the distribution of endogenous Atgs was assessed by immunofluorescence microscopy in HeLa cells transiently overexpressing a GFP–Htt exon 1 fusion (Fig. 1). Cytoplasmic IBs of GFP–Htt(Q103) were strongly labeled with antibody to Atg8/LC3, the human ortholog of Atg8, a lipid-linked intrinsic marker of autophagic membranes (36) (Fig. 1 A). An indistinguishable pattern of labeling was observed by using antibodies to Atg12, Atg5, and Atg16L (Fig. 1B). Htt IBs also colocalized with yellow fluorescent protein fluorescence in cells cotransfected with myc-Htt(Q103) and yellow fluorescent protein–Atg8/LC3 (data not shown). By contrast, the distribution of endogenous Atgs remained diffusely cytoplasmic in cells overexpressing GFP-Htt(Q25), which is unable to aggregate even when massively overexpressed. Digital deconvolution microscopy (37) of Atg8/LC3 immunofluorescence revealed that GFP-Htt(Q103) IBs were surrounded by many punctate structures with dimensions consistent with conventional mammalian autophagosomes (Fig. 1C). Similar colocalization between endogenous Atgs and IBs was observed in SH-SY5Y, HEK-293, and neuro2a cells lines expressing expanded polyQ-bearing Htt (data not shown), indicating that Atg recruitment to IBs is not unique to HeLa cells. Robust colocalization also was observed between Atg8/LC3 and IBs composed of other aggregation-prone proteins, including Atx3, a cytoplasmic protein containing an expanded polyQ repeat (Q84) (38), CFTRΔF508, a folding-defective mutant of the cystic fibrosis transmembrane conductance regulator (39), TCRα, which also is prone to aggregate when expressed in the absence of its oligomeric partners (40) (Fig. 1D), and P23H rhodopsin, a folding-defective G protein-coupled receptor (40) (data not shown). Thus, recruitment of autophagins to IBs is a general response to the production of cytoplasmic protein aggregates.
Fig. 1.
Atgs are recruited to Htt IBs. (A) IBs in Q103-expressing cells are surrounded by Atg8/LC3 staining material. Shown are micrographs of HeLa cells transiently transfected with GFP-Htt(Q25) (Upper) or GFP-Htt(Q103) (Lower, green) and stained with antibodies to Atg8/LC3 (red) or bisbenzimide to label nuclei (blue). (Scale bar, 10 μm.) (B) Htt IBs are immunopositive for other Atg proteins. Shown are micrographs of HeLa cells transiently transfected with GFP-Htt-Q103 (green) and stained with antibodies to the indicated Atgs (red) or bisbenzimide to label nuclei (blue). (Scale bar, 10 μm.) (C) Analysis of Atg8/LC3 localization to a GFP-Htt(Q103) IB by digital deconvolution microscopy. (Right) A projection from a reconstruction of optical sections. (Scale bar, 1 μm.) (D) Colocalization of Atg8/LC3 (Center, red) in HeLa cells transiently transfected with myc-tagged Atx3(Q84) (Top) or HEK-293 cells stably expressing GFP-CFTRΔF508 (Middle) or TCRα-GFP (Bottom). CFTR and TCRα cells were treated overnight with the proteasome inhibitor N-acetyl-Leu-Leu-norleucinal (ALLN; 10 μg/ml) to enhance aggregation. (Scale bar, 2 μm.)
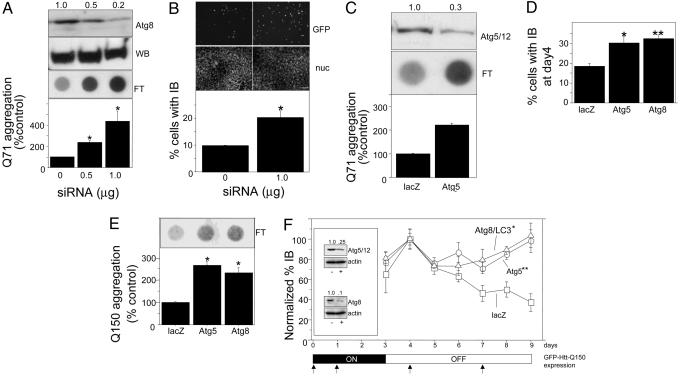
To test whether autophagy suppresses protein aggregate accumulation, HEK-293 cells stably expressing GFP-Htt(Q71) were exposed to siRNA to Atg8/LC3 (Fig. 2 A and B) or Atg5 (Fig. 2C). Steady-state levels of aggregated cellular Htt, assessed by a filter-trap assay (Fig. 2 A) or by the frequency of IBs (Fig. 2B), increased in inverse proportion to Atg8/LC3 levels after siRNA knockdown. Similarly, Atg5 knockdown resulted in a significant increase in filter-retained GFP-Htt(Q71) aggregates (Fig. 2C). Thus, autophagin knockdown increases aggregate burden either by increasing the aggregation of soluble Htt or by suppressing the removal of aggregated Htt. To discriminate between these two possibilities, a promoter shutoff experiment was used to assess the requirement for autophagins in clearing preexisting Htt aggregates (Fig. 2 D–F). GFP-Htt(Q150) expression in differentiated, postmitotic mouse neuro2a cells (35) was induced for 3 days in the presence of siRNA, followed by withdrawal of the inducer in the continued presence of siRNA. By day 4, the vast majority of cellular Htt was in IBs, which were twice as abundant in cells exposed to autophagin siRNA, compared with cells treated with control (lacZ) siRNA, suggesting that the autophagy pathway was actively eliminating GFP-Htt(Q150) aggregates during the induction period (Fig. 2D). The frequency of IBs in cells exposed to control siRNA increased slightly on the first day after gene shutoff and declined steadily over the subsequent 5 days, demonstrating that neuro2a cells are endowed with the capacity to eliminate GFP-Htt(Q150) aggregates (Fig. 2E). By contrast, reducing hAtg5/12 or Atg8/LC3 levels by 75% and 90%, respectively (Fig. 2F Inset), completely abrogated the observed decrease in IBs (Fig. 2F) and in filter-trapped GFP-Htt (Fig. 2E). These data unambiguously establish that autophagy plays an essential role in the removal of preexisting aggregates from cells.
Fig. 2.
Autophagy is essential for aggregate clearance. (A) Atg8/LC3 knockdown increases aggregate burden. Effect of Atg8/LC3 siRNA on GFP-Htt(Q71) aggregation assessed by filter retardation is shown. HEK-293 cells stably expressing GFP-Htt(Q71) were treated with the indicated amount of siRNA and evaluated for Atg8/LC3 or GFP-Htt(Q71) expression by Western blot (WB) or filter-trapped GFP-Htt(Q71) (FT). Intensities (relative to 0 siRNA) of filter-retained spots were quantified by image analysis (Upper, corresponding spots) and plotted (Lower). *, P < 0.0001 vs. control. n = 3. (B) Effect of Atg8/LC3 siRNA on GFP-Htt(Q71) aggregation assessed by IB frequency. Cells with IBs (representative field, GFP, Top) were counted (n = 300 from three independent trials) and quantified (Bottom). Bisbenzimide staining (nuc, Middle) shows the total number of nuclei in representative microscopic fields. *, P = 0.0049. (Scale bar, 50 μm.) (C) Atg5 knockdown increases aggregated burden. Effect of HAtg5 siRNA on GFP-Htt(Q71) aggregation assessed by filter retardation is shown. HEK-293 cells stably expressing GFP-Htt(Q71) were treated with 0.8 μg of siRNA and evaluated for HAtg5/12 conjugate expression (Top), or filter-trapped GFP-Htt(Q71) (Middle). Intensities (relative to lacZ siRNA) of filter-retained spots were quantified by image analysis (of the corresponding spots) and plotted (Bottom). *, P < 0.0001. n = 3. (D) Effect of Atg knockdown on IB frequency in differentiated neuro2a cells induced for 4 days to express GFP-Htt(Q150). Cells were transfected with the indicated siRNA on 2 consecutive days as indicated by the timeline in F and counted (n = 100 per condition) for the presence of IBs. *, P = 0.0064; **, P = 0.0032 vs. control. (E) Effect of siRNA on GFP-Htt(Q150) aggregation assessed by filter retardation. Filter retardation assay was performed on lysates of neuro2a cells treated with the indicated siRNAs 5 days after removal of inducer. *, P < 0.0001 vs. control. (F) Atg knockdown impairs clearance of preformed aggregates. Differentiated neuro2a cells were induced to express GFP-Htt(Q150) for 3 days (ON, timeline at bottom), followed by 6 days in culture without inducer (OFF). Cells were transfected with indicated siRNA on days 0, 1, 4, and 7 (upward arrows) and were scored for the presence of IBs by microscopy from days 3–9. Data are normalized to 100% on day 4 for each condition to permit comparison of rates. *, P = 0.0006; **, P = 0.0007 vs. control. Absolute levels on day 4 are shown in D.(Inset) Effect of siRNA on steady-state HAtg5/12 (Upper) and Atg8/LC3 (Lower) levels. Cell lysates from cells transfected with Atg (+) or lacZ (–) siRNA on day 5 were analyzed by immunoblotting using antibodies to Atg12 or Atg8/LC3 or actin for loading control.
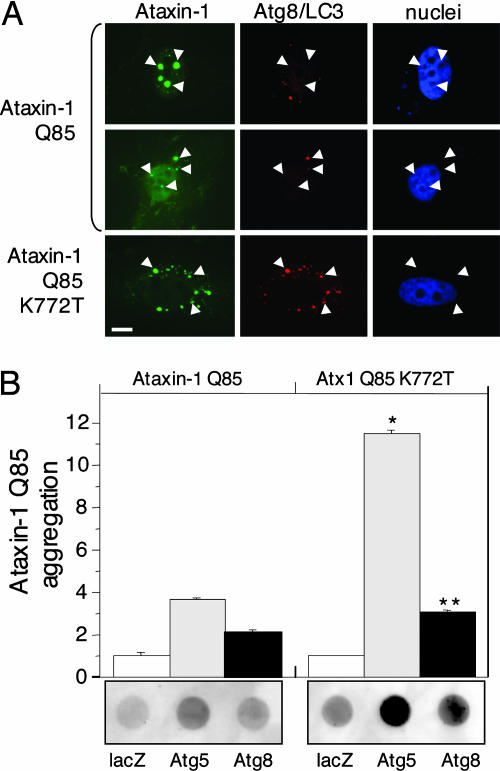
To evaluate the role of autophagy in the clearance of nuclear aggregates, we transfected HeLa cells with Atx1(Q85), the product of the gene responsible for spinocerebellar ataxia type 1 (9). The Atx1 protein contains a robust nuclear localization sequence that contributes to its toxicity (9). The majority of cells expressing Atx1(Q85) displayed prominent multiple nuclear inclusions, although occasional cytoplasmic inclusions were observed (Fig. 3A). Immunolabeling of endogenous Atg8/LC3 revealed only diffuse background staining in cells containing Atx1 nuclear inclusions. Strikingly, the occasional cytoplasmic Atx1(Q85)-positive inclusion was strongly labeled with Atg8/LC3 antibody (Fig. 3A, arrow). The Atx1 mutant Atx1(Q85)-K772T, containing a defective nuclear localization sequence (11), formed exclusively cytoplasmic inclusions that were strongly decorated with Atg8/LC3 antibody. Similarly, although Atg8/LC3 antibodies consistently label cytoplasmic Htt IBs, the occasional nuclear Htt IB is devoid of Atg8/LC3 staining (data not shown). To determine whether autophagy is able to facilitate the clearance of nuclear aggregates, Atx1(Q85) aggregation was assessed by filter trap in cells exposed to Atg siRNA (Fig. 3B). A small increase in trapped Atx1(Q85) was observed in cells exposed to Atg5 or Atg8/LC3 siRNA. In contrast, Atg knockdown significantly increased the levels of aggregated cytoplasmic Atx1(Q85)-K772T. Thus, compared with cytoplasmic aggregates, nuclear aggregates are far less accessible to the autophagy machinery and are more refractory to autophagic clearance.
Fig. 3.
Autophagy is ineffective for clearance of nuclear aggregates. (A) Atg8/LC3 colocalizes with cytoplasmic but not nuclear Atx-1(Q85). HeLa cells expressing FLAG-Atx1(Q85) (Top and Middle) or Atx1(Q85) containing a mutant nuclear localization sequence (K772T) (Bottom) were imaged for anti-FLAG immunofluorescence (Left) and Atg8/LC3 immunofluorescence (Center). Nuclei were labeled with bisbenzimide staining (Right). Nuclear (arrowheads) and cytoplasmic (arrows) inclusions are indicated. (Scale bar, 1 μm.) (B) Atg knockdown increases aggregation of cytoplasmic but not nuclear Atx1(Q85). Shown are filter retardation assay of HEK-293 cells transiently expressing the indicated Atx1 constructs and treated with the indicated siRNA. *, P < 0.0001; **, P = 0.0003 vs. Atx1(Q85).
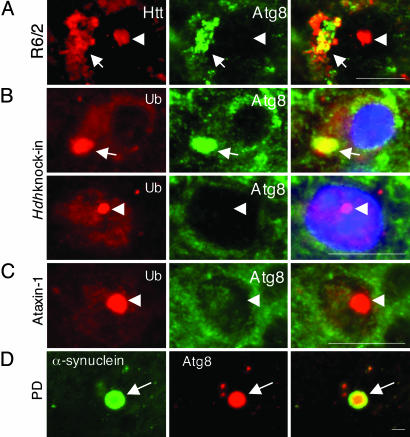
To assess the role of autophagy in the clearance of cytoplasmic and nuclear aggregates in disease pathogenesis, we used immunofluorescence microscopy to examine Atg8/LC3 expression in murine models of polyQ disease (Fig. 4 A–C). R6/2 mice overexpress an N-terminal Htt-Q144 transgene and develop a progressive neurological phenotype with onset at ≈2 months, associated with the presence of ubiquitin-immunoreactive neuronal and cytoplasmic IBs (41). Antibodies to Atg8/LC3 labeled cytoplasmic (arrow) but not nuclear (arrowhead) IBs in cortical sections from R6/2mice (Fig. 4A) were consistent with our observations from studies with cultured cells expressing a similar construct. No significant Atg8/LC3 labeling was detected in sections from age-matched nontransgenic animals or from transgenic animals labeled with preimmune serum (data not shown). To confirm these data in a more physiological model, we examined Atg8/LC3 expression in sections from the brains of homozygous Hdh(CAG)150 knock-in mice (43) (Fig. 4B). These mice express endogenous, full-length murine Q150 Htt (Hdh) under the control of the endogenous Hdh promoter and develop late-onset neurological and neuroanatomical abnormalities consistent with HD, including prominent cortical intranuclear and cytoplasmic IBs (ref. 42 and L.M.E., unpublished data). Ubiquitin-immunoreactive IBs were observed in both the nucleus and cytoplasm in cortical sections from 1-year-old mice homozygous for the Hdh(CAG)150 allele. Strikingly, only the cytoplasmic IBs reacted with antibody to Atg8/LC3. Consistent with this finding, we failed to detect Atg8/LC3 staining in intranuclear IBs within cerebellar Purkinje cells from mice expressing Atx1 containing a pathogenic expanded polyQ tract (43) (Fig. 4C). Finally, we detected robust staining of Atg8/LC3 in cortical α-synuclein-positive Lewy bodies (Fig. 4D) and Lewy neurites (data not shown) from human Parkinson's disease postmortem brain. No significant colocalization was observed with preimmune serum (data not shown).
Fig. 4.
Cytoplasmic but not nuclear inclusions from diseased brains recruit Atg8/LC3. (A) Cortex from 16-week-old R6/2 mouse was stained with EM48 anti-Htt antibody (red, Left) and Atg8/LC3 antibodies (green, Center). A representative image of cortical neurons is shown. (B) Cortex from 1-year-old Hdh(CAG)150 knock-in mouse stained with anti-ubiquitin (Ub, red) and Atg8/LC3 (green). Nuclei are stained with bisbenzimide. (C) Cerebellum from Atx1-overexpressing mouse was stained with anti-ubiquitin (Ub, red) and Atg8/LC3 (green). A representative image of Purkinje cells is shown. (D) A midbrain section from a Parkinson's disease patient was stained with LB509 anti-α-synuclein and Atg8/LC3 antibodies as indicated. The image is representative of two sections from four different patients. Arrows indicate cytoplasmic inclusions, and arrowheads indicate nuclear inclusions. (Scale bar, 2 μm.)
The RNAi knockdown data reported here demonstrate that an autophagic process plays an essential role in the clearance of aggregated proteins in cell-culture models of polyQ-mediated neurodegeneration. Immunolocalization of Atgs to cytoplasmic IBs further supports a connection between autophagy and protein aggregation, but elucidation of the precise mechanism by which aggregated proteins are captured by autophagosomes must await further study. Conceivably, conventional autophagosomes could capture small aggregates before their transport to IBs. Alternatively, it is possible that autophagosomes could be recruited to or formed at sites of IB formation. Finally, our data do not preclude the possibility that clearance of aggregated proteins from IBs may involve the formation of morphologically distinct autophagic structures that nonetheless employ the same Atg machinery used in starvation-induced autophagy.
Our data also demonstrate that Atg8/LC3, an autophagic membrane marker, is recruited to sites of cytoplasmic IB formation in Parkinson's disease and in two murine HD models but is absent from nuclear inclusions. How Atg proteins are recruited to IBs is currently unknown, but it is tempting to speculate that microtubules may contribute to this process, by analogy to the delivery of aggregated proteins to aggresomes (39). Autophagy thus appears to be relatively ineffective at clearing protein aggregates that accumulate within the nucleus. The strong correlation between the toxicity of disease-linked misfolded proteins and their import to the nucleus (9, 10) has led to the widely accepted hypothesis that the nucleus is the primary site of action of these proteotoxins. The data presented here suggest an alternative, although not mutually exclusive, interpretation, that the nucleus is a “protected” environment that provides a haven for toxic protein aggregates to escape degradation by autophagy. Thus, the differential toxicity of nuclear and cytoplasmic protein aggregates, which impair the ubiquitin proteasome system throughout the cell, irrespective of the cellular compartment to which they are targeted (44), may reflect the capacity of intrinsic cellular defenses rather than the molecular targets of their action. The observation that Atg knockdown leads to a small (and not statistically significant) increase in accumulation of mutant Atx1 (Fig. 3B) suggests that autophagic processes may be able to contribute to the clearance of nuclear protein aggregates. Perhaps this limited autophagic capacity can explain the observation that mice do recover from proteotoxicity associated with transient expression of mutant Atx1 (16). Clearly, a deeper understanding of the mechanism by which proteins are targeted for autophagy will aid our understanding of pathogenesis and facilitate development of rational treatments for protein conformational diseases.
Acknowledgments
We thank Masaru Kurosawa for technical assistance; Harry Orr and Lisa Duvick for sections from the Atx1-transgenic mouse used in Fig. 4C; Marilyn Farquhar, Neil Bence, Eric Bennett, and members of the Kopito laboratory for insightful discussion; and Karla Kirkegaard for stimulating discussions on autophagy. This work was supported by National Institutes of Health Grants NS042842 (to R.R.K.) and NS40251 (to L.M.E.), grants from the Huntington's Disease Society of America (to R.R.K.), and by grants-in-aid from the Japan Society for the Promotion of Science (to A.I.).
Author contributions: A.I., J.C.C., M.B., and R.R.K. designed research; A.I., J.C.C., M.B., and L.M.E. performed research; J.C.C., M.B., L.M.E., N.N., and L.S.F. contributed new reagents/analytic tools; A.I. and R.R.K. analyzed data; and A.I. and R.R.K. wrote the paper.
Abbreviations: polyQ, polyglutamine; HD, Huntington's disease; Htt, huntingtin; Atx1, ataxin-1; IB, inclusion body; siRNA, short interfering RNA; HEK, human embryonic kidney; TCRα, T cell receptor α.
References
- 1.Zoghbi, H. Y. & Orr, H. T. (2000) Annu. Rev. Neurosci. 23, 217–247. [DOI] [PubMed] [Google Scholar]
- 2.Huang, C. C., Faber, P. W., Persichetti, F., Mittal, V., Vonsattel, J. P., MacDonald, M. E. & Gusella, J. F. (1998) Somatic Cell Mol. Genet. 24, 217–233. [DOI] [PubMed] [Google Scholar]
- 3.Scherzinger, E., Lurz, R., Turmaine, M., Mangiarini, L., Hollenbach, B., Hasenbank, R., Bates, G. P., Davies, S. W., Lehrach, H. & Wanker, E. E. (1997) Cell 90, 549–558. [DOI] [PubMed] [Google Scholar]
- 4.Chen, S., Berthelier, V., Yang, W. & Wetzel, R. (2001) J. Mol. Biol. 311, 173–182. [DOI] [PubMed] [Google Scholar]
- 5.Ross, C. A. & Poirier, M. A. (2004) Nat. Med. 10, Suppl., S10–S17. [DOI] [PubMed] [Google Scholar]
- 6.Kakizuka, A. (1998) Trends Genet. 14, 396–402. [DOI] [PubMed] [Google Scholar]
- 7.Taylor, J. P., Tanaka, F., Robitschek, J., Sandoval, C. M., Taye, A., Markovic-Plese, S. & Fischbeck, K. H. (2003) Hum. Mol. Genet. 12, 749–757. [DOI] [PubMed] [Google Scholar]
- 8.Arrasate, M., Mitra, S., Schweitzer, E. S., Segal, M. R. & Finkbeiner, S. (2004) Nature 431, 805–810. [DOI] [PubMed] [Google Scholar]
- 9.Orr, H. T. & Zoghbi, H. Y. (2001) Hum. Mol. Genet. 10, 2307–2311. [DOI] [PubMed] [Google Scholar]
- 10.Ross, C. A., Wood, J. D., Schilling, G., Peters, M. F., Nucifora, F. C., Jr., Cooper, J. K., Sharp, A. H., Margolis, R. L. & Borchelt, D. R. (1999) Philos. Trans. R. Soc. London B 354, 1005–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klement, I. A., Skinner, P. J., Kaytor, M. D., Yi, H., Hersch, S. M., Clark, H. B., Zoghbi, H. Y. & Orr, H. T. (1998) Cell 95, 41–53. [DOI] [PubMed] [Google Scholar]
- 12.Schilling, G., Savonenko, A. V., Klevytska, A., Morton, J. L., Tucker, S. M., Poirier, M., Gale, A., Chan, N., Gonzales, V., Slunt, H. H., et al. (2004) Hum. Mol. Genet. 13, 1599–1610. [DOI] [PubMed] [Google Scholar]
- 13.Peters, M. F., Nucifora, F. C., Jr., Kushi, J., Seaman, H. C., Cooper, J. K., Herring, W. J., Dawson, V. L., Dawson, T. M. & Ross, C. A. (1999) Mol. Cell. Neurosci. 14, 121–128. [DOI] [PubMed] [Google Scholar]
- 14.Hazeki, N., Tukamoto, T., Goto, J. & Kanazawa, I. (2000) Biochem. Biophys. Res. Commun. 277, 386–393. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto, A., Lucas, J. J. & Hen, R. (2000) Cell 101, 57–66. [DOI] [PubMed] [Google Scholar]
- 16.Zu, T., Duvick, L. A., Kaytor, M. D., Berlinger, M. S., Zoghbi, H. Y., Clark, H. B. & Orr, H. T. (2004) J. Neurosci. 24, 8853–8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sapp, E., Schwarz, C., Chase, K., Bhide, P. G., Young, A. B., Penney, J., Vonsattel, J. P., Aronin, N. & DiFiglia, M. (1997) Ann. Neurol. 42, 604–612. [DOI] [PubMed] [Google Scholar]
- 18.Nixon, R. A., Cataldo, A. M. & Mathews, P. M. (2000) Neurochem. Res. 25, 1161–1172. [DOI] [PubMed] [Google Scholar]
- 19.Stefanis, L., Larsen, K. E., Rideout, H. J., Sulzer, D. & Greene, L. A. (2001) J. Neurosci. 21, 9549–9560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kegel, K. B., Kim, M., Sapp, E., McIntyre, C., Castano, J. G., Aronin, N. & DiFiglia, M. (2000) J. Neurosci. 20, 7268–7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petersen, A., Larsen, K. E., Behr, G. G., Romero, N., Przedborski, S., Brundin, P. & Sulzer, D. (2001) Hum. Mol. Genet. 10, 1243–1254. [DOI] [PubMed] [Google Scholar]
- 22.Fortun, J., Dunn, W. A., Jr., Joy, S., Li, J. & Notterpek, L. (2003) J. Neurosci. 23, 10672–10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsen, K. E. & Sulzer, D. (2002) Histol. Histopathol. 17, 897–908. [DOI] [PubMed] [Google Scholar]
- 24.Yuan, J., Lipinski, M. & Degterev, A. (2003) Neuron 40, 401–413. [DOI] [PubMed] [Google Scholar]
- 25.Blommaart, E. F., Krause, U., Schellens, J. P., Vreeling-Sindelarova, H. & Meijer, A. J. (1997) Eur. J. Biochem. 243, 240–246. [DOI] [PubMed] [Google Scholar]
- 26.Qin, Z. H., Wang, Y., Kegel, K. B., Kazantsev, A., Apostol, B. L., Thompson, L. M., Yoder, J., Aronin, N. & DiFiglia, M. (2003) Hum. Mol. Genet. 12, 3231–3244. [DOI] [PubMed] [Google Scholar]
- 27.Ravikumar, B., Duden, R. & Rubinsztein, D. C. (2002) Hum. Mol. Genet. 11, 1107–1117. [DOI] [PubMed] [Google Scholar]
- 28.Klionsky, D. J., Cregg, J. M., Dunn, W. A., Jr., Emr, S. D., Sakai, Y., Sandoval, I. V., Sibirny, A., Subramani, S., Thumm, M., Veenhuis, M. & Ohsumi, Y. (2003) Dev. Cell 5, 539–545. [DOI] [PubMed] [Google Scholar]
- 29.Bence, N., Sampat, R. & Kopito, R. R. (2001) Science 292, 1552–1555. [DOI] [PubMed] [Google Scholar]
- 30.Yu, H. & Kopito, R. R. (1999) J. Biol. Chem. 274, 36852–36858. [DOI] [PubMed] [Google Scholar]
- 31.Kuma, A., Mizushima, N., Ishihara, N. & Ohsumi, Y. (2002) J. Biol. Chem. 277, 18619–18625. [DOI] [PubMed] [Google Scholar]
- 32.Kabeya, Y., Mizushima, N., Ueno, T., Yamamoto, A., Kirisako, T., Noda, T., Kominami, E., Ohsumi, Y. & Yoshimori, T. (2000) EMBO J. 19, 5720–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myers, J. W., Jones, J. T., Meyer, T. & Ferrell, J. E. (2003) Nat. Biotechnol. 21, 324–328. [DOI] [PubMed] [Google Scholar]
- 34.Wanker, E. E., Scherzinger, E., Heiser, V., Sittler, A., Eickhoff, H. & Lehrach, H. (1999) Methods Enzymol. 309, 375–386. [DOI] [PubMed] [Google Scholar]
- 35.Wang, G. H., Mitsui, K., Kotliarova, S., Yamashita, A., Nagao, Y., Tokuhiro, S., Iwatsubo, T., Kanazawa, I. & Nukina, N. (1999) NeuroReport 10, 2435–2438. [DOI] [PubMed] [Google Scholar]
- 36.Tanida, I., Ueno, T. & Kominami, E. (2004) Int. J. Biochem. Cell Biol. 36, 2503–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scalettar, B. A., Swedlow, J. R., Sedat, J. W. & Agard, D. A. (1996) J. Microsc. (Paris) 182, 50–60. [DOI] [PubMed] [Google Scholar]
- 38.Paulson, H. L., Perez, M. K., Trottier, Y., Trojanowski, J. Q., Subramony, S. H., Das, S. S., Vig, P., Mandel, J. L., Fischbeck, K. H. & Pittman, R. N. (1997) Neuron 19, 333–344. [DOI] [PubMed] [Google Scholar]
- 39.Johnston, J. A., Ward, C. L. & Kopito, R. R. (1998) J. Cell Biol. 143, 1883–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rajan, R. S., Illing, M., Bence, N. & Kopito, R. R. (2001) Proc. Natl. Acad. Sci. USA 98, 13060–13065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davies, S. W., Turmaine, M., Cozens, B. A., DiFiglia, M., Sharp, A. H., Ross, C. A., Scherzinger, E., Wanker, E. E., Mangiarini, L. & Bates, G. P. (1997) Cell 90, 537–548. [DOI] [PubMed] [Google Scholar]
- 42.Lin, C. H., Tallaksen-Greene, S., Chien, W. M., Cearley, J. A., Jackson, W. S., Crouse, A. B., Ren, S., Li, X. J., Albin, R. L. & Detloff, P. J. (2001) Hum. Mol. Genet. 10, 137–144. [DOI] [PubMed] [Google Scholar]
- 43.Burright, E. N., Clark, H. B., Servadio, A., Matilla, T., Feddersen, R. M., Yunis, W. S., Duvick, L. A., Zoghbi, H. Y. & Orr, H. T. (1995) Cell 82, 937–948. [DOI] [PubMed] [Google Scholar]
- 44.Bennett, E. J., Bence, N. F., Jayakumar, R. & Kopito, R. R. (2005) Mol. Cell 17, 351–365. [DOI] [PubMed] [Google Scholar]