Abstract
The transcription antiterminator N protein from bacteriophage λ uses its arginine-rich motif to specifically bind a stem-loop RNA hairpin (boxB) as a bent α-helix. A single stacking interaction between a tryptophan (Trp-18) and an adenosine (A7) in the RNA loop is robust and necessary for antitermination activity in vivo. Previously, femtosecond fluorescence up-conversion experiments from this laboratory indicated that the N/boxB complex exists in a dynamical two-state equilibrium between stacked and unstacked conformations and that the extent of stacking depends on the identity of peptide residues 14 and 15. In the present work, we have combined transient absorption and fluorescence up-conversion to determine the nature of interactions responsible for this sequence-dependent behavior. Analysis of mutant complexes supports the idea that the β-carbon of residue 14 enforces the stacked geometry by hydrophobic interaction with the ribose of A7, whereas a positive charge at this residue plays only a secondary role. A positive charge at position 15 substantially disfavors the stacked state but retains much of the binding energy. Remarkably, in vivo antitermination experiments show strong correlation with our femtosecond dynamics, demonstrating how conformational interplay can control the activity of a macromolecular machine.
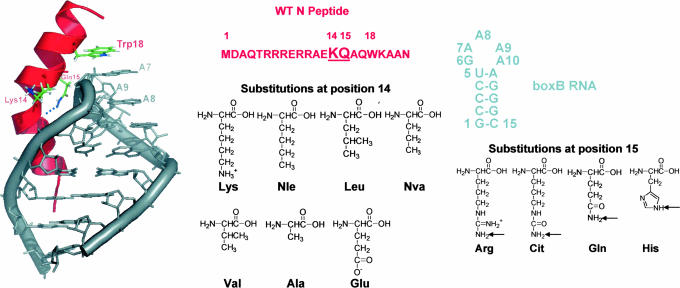
Biological systems are, by their very nature, dynamical. For example, transcription elongation is often regulated by RNA–protein interactions (1) between the mRNA streaming out of the polymerase (at 50 nt/s) and proteins associated with the holoenzyme (2, 3), a so-called cis-acting regulatory mechanism. One of the best-understood cis-acting control systems is antitermination in bacteriophage λ. In this system, the phage N protein enhances transcription elongation, enabling the host RNA polymerase to read through both intrinsic and ρ-dependent terminators (4–6), thereby up-regulating the genes flanking the very early operons (7). This transcription antitermination is initiated by N protein binding to the nascent mRNA stem-loop hairpin termed boxB within the Nut site (N-utilization) (8, 9). The first 22 residues of N contain the boxB RNA-binding domain and feature an Arg-rich motif (10–13). Upon binding, N folds into a bent α-helix and follows the contour of the RNA major groove (11, 14, 15) enforcing the formation of a GNRA tetraloop fold (16, 17) with one base extruded (Fig. 1).
Fig. 1.
Structures and sequences of the boxB RNA, the WT N peptide, and their complex (14, 15). The RNA backbone and bases are gray, and A7, A8, and A9 are labeled. The WT N peptide binds as a bent α-helix, separating the structure into N- and C-terminal helical domains. The peptide chain is represented as red ribbon with the side chains of Lys-14, Gln-15, and Trp-18 shown. The chemical structures of amino acids incorporated into positions 14 and 15 of N peptides are shown at Right. Arrows indicate the protons that are either observed to be involved in H-bonding to carbonyl of Arg-8 in WT complex of box/N (14, 15) or boxB/Nun (28) or hypothesized to participate in similar interactions in the mutant complexes (see text).
In the boxB RNA/N protein complex, the aromatic side chain of Trp-18 stacks on top of the RNA loop and extends the characteristic RNA base π stack by one more step. Structurally, this planar π–π interaction represents an intriguing RNA–protein recognition mechanism. Functionally, early in vitro and in vivo work indicated that this Trp residue was very important for both RNA-binding and processive antitermination (11, 18).
Recently, we used mRNA display experiments to redesign the protein portion of the boxB RNA/N protein interface (12, 13, 19, 20). This work resulted in a series of >100 N peptide variants that bind to the boxB RNA with high affinity and specificity. Interestingly, although functional in vitro, many of these sequences show little or no antitermination activity in vivo. A series of mutants involving peptide positions 14 and 15 were instructive in working to reconcile this apparent inconsistency. NMR, steady-state, and femtosecond fluorescence up-conversion experiments demonstrated that mutations at positions 14 and 15 altered the stacking of the Trp-18/A7 protein–RNA interface. In vivo analysis supported the notion that stacking was essential for functional antitermination (20, 21). These studies revealed that boxB RNA recognition by N proteins is surprisingly robust in that the stacked structures seen in different complexes achieve precisely the same geometry.
Because femtosecond spectroscopy (21) probes the interface for recognition, it was possible to demonstrate that the complexes exist in at least two major types of conformations, one with Trp-18 stacked on the RNA loop as that revealed by NMR structural studies (14, 15) and an ensemble of conformations where the Trp-18 is unstacked from the RNA loop. In that work, we proposed that the two populations are in dynamical exchange (a dynamical two-state model) and that the population distributions are modulated by the residues at positions 14 and 15, one helical turn away from the crucial Trp-18/A7 protein–RNA interface (21). Consistent with this model, we observed that the in vivo function of different complexes correlates with the stacked population but not the peptide binding affinity (20, 21).
Our previous studies left unresolved why specific mutations at positions 14 and 15 should alter the stacking of the Trp-18/A7 interface. Further mutational analysis using natural and unnatural amino acids provides a route to uncover the underlying mechanisms. Overall, we want to understand exactly what types of interactions are responsible for the different population distributions and structural specificity in these complexes. Also, because the two populations undergo dynamical exchange, it is likely that unfolding and refolding of the C-terminal helix (residues 12–22) is coupled to the domain motion, leaving the conformation of the unstacked population an open question. Understanding these issues will shed light on the general principles of molecular recognition of RNA and provides a rare opportunity where allosteric control of a macromolecular machine (the 600,000-Da transcription elongation complex) can be understood in molecular detail.
Experimental Procedures
Synthesis and Purification of RNAs and Peptides. The 2-aminopurine (2Ap)-labeled RNA, 5′-GCCCUG(2Ap)AAAAGGGC-3′, was generated by solid-phase synthesis using standard phosphoramidite chemistry, deprotected, and either purified by 20% urea-PAGE or purchased fully purified from Dharmacon (Boulder, CO). Crude peptides were made by automated fluorenylmethoxycarbonyl synthesis using either an Applied Biosystems 432A (R.W.R. laboratory), an Applied Biosystems 433A at the Caltech Biopolymer Synthesis Center, or purchased from AnaSpec (San Jose, CA). The peptides were deprotected by trifluoroacetic acid and purified by reversed-phase C18 HPLC. RNA lacking 2Ap for NMR experiments (5′-GCCCUGAAAAAGGGC3-′) was synthesized by using T7 RNA polymerase and synthetic DNA oligos as described in ref. 22. Concentrations of purified RNA and peptide oligomers were determined by UV absorbance at 260 and 280 nm, respectively (23, 24). For femtosecond time-resolved experiments, RNA (typically 200 μM) was annealed in buffer of 50 mM NaCl and 10 mM phosphate (pH 6). Aliquots of concentrated peptide solutions then were added to the RNA solution to form complexes with 1:1.5 RNA:peptide ratio to ensure complete complexation of the RNA.
Femtosecond Laser Spectroscopy. The experimental set-up used in the fluorescence up-conversion and transient absorption experiments has been published previously (see refs. 25–27). Further details are provided in Supporting Experimental Procedures, which is published as supporting information on the PNAS web site, together with the instruments used for NMR, circular dichroism, and steady-state spectroscopy.
Transcription Antitermination Assay. N and mutant strains were constructed by using the two-plasmid reporter system described in ref. 18. All sequences were verified by DNA sequencing. N mutant strains were plated on tryptone agar supplemented with 0.05 mM isopropyl-β-d-galactopyranoside (IPTG), 0.08 mg/ml X-GAL, and appropriate antibiotics (29). A colorimetric assay based on o-nitrophenyl-β-d-galactopyranoside (ONPG) also was used to quantitate β-gal as a percentage of the wild-type (WT) N reporter construct in the solution (29).
Results and Discussion
Dissecting the Molecular Interactions in boxB/N Complexes. Within the WT boxB/N complex are extensive electrostatic, hydrophobic, and hydrogen-bonding (H-bonding) interactions between the RNA and peptide and within the peptide itself. All of these interactions, particularly electrostatic ones, contribute greatly to binding affinity (11–13, 30), whereas hydrophobic and H-bonding interactions also may contribute to structural specificity in terms of particular binding modes (14, 15). Previously, binding in the N system had been assayed genetically by using in vivo nonsense suppression (18) or in vitro by using Ala scanning mutagenesis (11). These studies approached the system on a residue-by-residue basis and demonstrated the importance of certain side chains at the RNA–protein interface. More recently, we used in vitro selection to redesign the interface at peptide positions 14, 15, and 18 (19, 20). These efforts demonstrated that the sequences at positions 14 and 15 controlled the conformation and structure of the Trp at position 18.
However, these studies did not determine the underlying causes of conformational control. To address this question, we used both femtosecond transient absorption and fluorescence up-conversion to probe the role of these interactions that govern the sequence-dependent behavior of N peptides in recognition of boxB RNA. We constructed a series of N peptides wherein we systematically varied the structure of positions 14 and 15 using both natural and unnatural amino acids (Fig. 1). These amino acids have different electrostatic and hydrophobic characteristics, letting us probe the different interactions independently by measuring (i) the population distribution in these RNA–peptide complexes, (ii) the structural characteristics of the unstacked population, and (iii) the functional consequences of the structural variation in vivo.
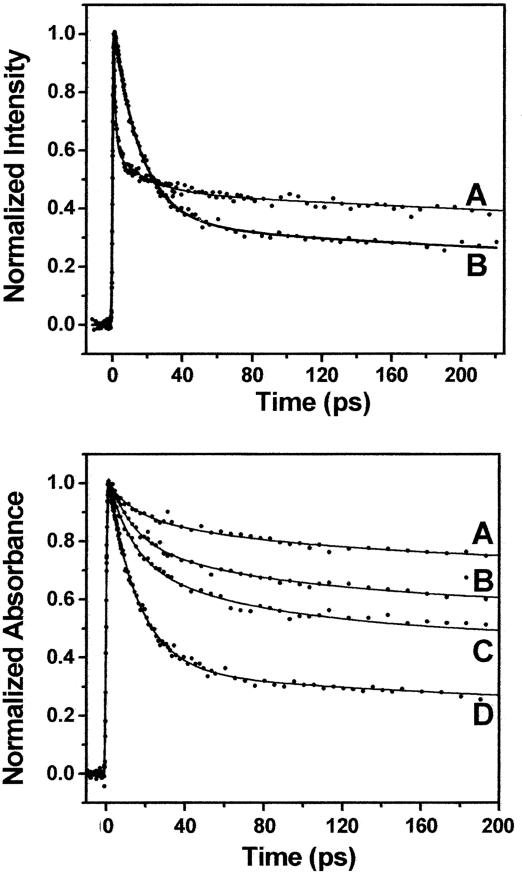
Ultrafast Charge Transfer Dynamics at the RNA/Peptide Interface. We began by exploring ultrafast transient absorption as a tool to probe the stacked vs. unstacked populations in the WT 2Ap-labeled boxB RNA/N peptide complex (Fig. 2 Upper) and compared the results with those of the fluorescence up-conversion approach we had pursued previously (21). In each case, the ultrafast component refers to the fastest decay transient observed in a particular experiment. The stacked WT complex gives rise to a major fluorescence up-conversion transient with τ ≈ 1 ps, whereas the same complex results in a transient absorption signal with τ ≈ 16 ps. A second feature of the transient absorption is that the amplitude of ultrafast component is both larger and less susceptible to initial intensity losses than that of the up-conversion experiments, providing less bias for calculating population distributions.
Fig. 2.
Femtosecond transients. (Upper) Overlay of femtosecond fluorescence up-conversion (trace A) and transient absorption (trace B) for the WT complex with boxB RNA labeled at position A7. (Lower) Both images show an instrument signal rise (t = 0 ps), followed by the measured decay transients (t = 0–200 ps). Femtosecond transient absorption decays measured for the complexes with N variants Glu-14–Arg-15 (trace A); Lys-14–Arg-15 (B); Lys-14–His-15 (C); and Lys-14–Gln-15 (D) (WT). These peptides were previously selected for binding boxB RNA (19).
The photophysics underlying the dynamics of these two methods are very interesting in their own right and are discussed by Wan et al. (31). Briefly, the excitation pulse generates 2Ap*. Before vibrational relaxation, this excited state can undergo direct femtosecond barrierless crossing to a charge-transferred state, a process we call “direct transfer.” On a similar time scale, the direct transfer process competes with vibrational relaxation, which is followed by charge transfer, referred to here as indirect transfer process. The fluorescence up-conversion amplitude is decreased by the direct-transfer process, but the observed dynamics are unchanged, measuring only the loss of the excited state through the fluorescence pathway (τ ≈ 1 ps). Conversely, the amplitude of the transient absorption decay is substantial because this experiment detects 2Ap in both the excited and charge-transfer states. The transient absorbance 16-ps time constant is dominated by the lifetime (recombination) of the charge-transfer state.
For the set of mutant complexes with different amino acids at positions 14 and 15, the primary dynamics of the transient absorption occur on 13–16 ps (Fig. 2 Lower); therefore, the time scale for these complexes is also robust as observed in the up-conversion (21). Moreover, the amplitude of the ultrafast component in the transient absorption follows the same trend as in the up-conversion experiments for the same series of complexes, in the decreasing order of Lys-14–Gln-15 (WT, 66%), Lys-14–His-15 (29%), Lys-14–Arg-15 (22%), and Glu-14–Arg-15 (11%). This result is consistent with the conclusion that the A7/Trp-18 interface stacked population in these complexes decrease in this order.
Fluorescence up-conversion likely does not provide the optimal quantitative means to measure population differences of our RNA–protein complexes. This is because the 2Ap direct-charge transfer pathway can result in substantial fluorescence intensity loss within the time resolution of our experiment (<200 fs). The extent of this loss varies for different RNA/peptide complexes especially compared with free 2Ap (see Fig. 7 Upper, which is published as supporting information on the PNAS web site). The initial loss is greatest for the fully stacked boxB RNA/WT N peptide 22-mer but much smaller for the 11-mer peptide complex that lacks stacking or the boxB RNA alone.
By comparison, the reduction of initial intensity monitored by transient absorbance falls over a much smaller range (Fig. 7 Lower). As before, the largest loss arises from the highly stacked WT complex, whereas the boxB/11mer, boxB alone, and 2Ap samples give similar overall intensities. Mechanistically, the absorbance loss is likely due to the charge transfer state having a smaller cross-section at λ = 600 nm than the 2Ap excited state (31). Overall, these data argue that transient absorbance provides the best ultrafast means to calculate population differences in this system. We note that these population calculations are likely to somewhat underestimate the fraction of stacked molecules due to the relatively small loss in initial intensity in the stacked complexes.
Position 14: A β-Carbon Restraint Enforcing the Stacked Conformation. We first looked at the issue of the positive charge of Lys-14 in the WT peptide. Previous work indicated that positive charge at position 14 was necessary for peptide binding with the boxB RNA and that Arg and lysine (Lys) were functionally equivalent at this position (11). Conversely, in vivo experiments showed that some uncharged residues, e.g., leucine (Leu) and valine (Val), were functional at this position (18). We had previously demonstrated that a stacked interface between Trp-18 and A7 was necessary for functional antitermination in vivo (20, 21).
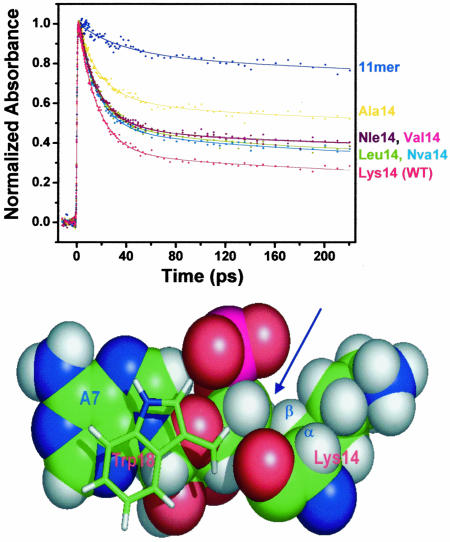
In line with this view, ultrafast transient absorption demonstrates that in the WT complex, the ultrafast component represents ≈66% of the total signal (Fig. 2). Interestingly, various of different uncharged amino acids inserted at position 14 also give rise to a stacked RNA–protein interface. For example, norleucine (Nle) is a residue that has the same aliphatic side chain as Lys but lacks the terminal ε-amino group and positive charge (Fig. 1). The N Nle-14/boxB complex has somewhat smaller amplitude of the ultrafast component than the WT complex (Fig. 3). This finding means that although the Lys-14 positive charge plays some role in binding, it is not required to maintain the stacked interface. We then varied the length and geometry of the aliphatic side chain of position 14 by incorporating other different natural and unnatural amino acids, including Leu, norvaline (Nva), Val, and Ala (Fig. 1). As shown in Fig. 3, Leu-14, Nva-14, and Val-14 complexes have similar stacked populations (≈50%); even the Ala-14 mutant has a significant fraction of stacking (34%) compared with the 11-mer complex, which lacks the stacked C-terminal half of the peptide.
Fig. 3.
Structural effects on dynamics. (Upper) Effects of amino acid residues at position 14 of N peptides on the amplitude of ultrafast component in the transient absorption experiments. The 11-mer peptide/boxB RNA complex provides a no-stacking control because it lacks Trp-18 residue altogether. (Lower) Hydrophobic interactions between the Lys-14 side chain and the boxB A7 ribose observed in the WT complex (indicated by the blue arrow). The aromatic side chain of Trp-18 that stacks on A7 also is shown.
These results suggest that at position 14, the aliphatic side chain, rather than positive charge, determines stacking of the RNA–peptide interface. The most likely structural origin of this conformational bias is that the β-methyl or methylene at position 14 enforces the stacked conformation. In line with this view, the interactions between the methylene groups of Lys-14 with the ribose of A7 were intermolecular interactions observed for the WT system (Fig. 3 Lower) (32). These interactions are presumably important in defining the interface between the base A7 and the Trp-18 residue that is one helical turn from Lys-14.
The best-stacked complexes all have position 14 aliphatic residues with at least three methylene/methyl groups, suggesting that this quantity is the minimal number of hydrophobic contacts sufficient to maintain the interface. In particular, the Leu-14 substitution in N protein, although a moderate affinity boxB binder (Kd = 30 nM vs. Kd = 1 nM in WT) is fully functional in vivo (18). This observation can now be interpreted as being due to the fact that this mutation maintains the stacked interface. Similarly, imino proton NMR experiments demonstrate that the Arg-14–Gln-15 complex has similar structure as the stacked complex (data not shown). The fact that even Ala-14 produces some stacking reveals that even the β-carbon alone provides a sufficient structural restraint to enforce the stacked structure. Conversely, negative charge–charge interactions at position 14 can disfavor the stacked structure, as shown by the fact that the Glu-14 complex (which has β and γ methylene groups) has very little stacked population based on NMR and steady-state fluorescence data (20).
Position 15: Positive Electrostatic Control of Peptide Conformation. We next wanted to understand why the Gln-15 to Arg-15 mutation was seen in many of the tight-binding peptides isolated from mRNA display selection experiments (19). This particular mutation was quite interesting because previous work indicated that it caused the Trp-18/A7 interface to become substantially unstacked [e.g., the Glu-14–Arg-15 complex shows very little stacking (20, 21) (see Fig. 2 Lower)].
We reasoned that the loss of stacking could occur from several mechanisms including (i) negative charge-charge repulsion between adjacent side chains, (ii) inability to pack the large Arg side chain at the RNA–peptide interface, or (iii) positive charge–charge repulsion between the Arg-15 side chain and the cluster of positively charged Arg residues at the RNA–protein interface. In the Glu-14–Arg-15 complex, loss of stacking does not appear to be due to simple charge–charge repulsion from the two consecutive negative charges (Glu-13 and -14), as seen by the fact that neutral substitutions at position 13 (e.g., Gln-13–Glu-14–Arg-15) result in identical up-converion and transient absorption dynamics and a lack of the stacked state (data not shown).
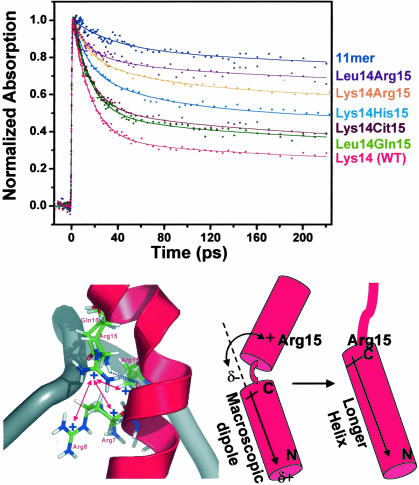
To address the steric and charge effects of Arg-15 on boxB RNA recognition, we compared the dynamics of (i) the WT N peptide (Gln-15), (ii) the Arg-15 mutant, and (iii) a peptide containing the unnatural amino acid citrulline at position 15 (Cit-15) (Fig. 1). We chose Cit because it is a structural mimic of Arg lacking a positive charge, a feature that has been used in analyses of general electrostatic interactions (30). As expected, the Lys-14–Arg-15 RNA/peptide complex significantly reduced stacked population compared with WT Lys-14–Gln-15 (Fig. 4 Upper). Conversely, the Cit-15 mutant shows stacking similar to the WT level. We further narrowed this effect by comparing Gln-15 and Arg-15 in a neutrally charged (Leu-14) background. Leu-14–Arg-15 has significantly lower stacked population compared with WT, whereas the Leu-14–Gln-15 mutant shows near WT levels of stacking (Fig. 4 Upper).
Fig. 4.
Structural effects on dynamics. (Upper) Effects of amino acid residues at position 15 of N peptides on the amplitude of ultrafast component in the transient absorption experiments with position 14 being either Lys or Leu. The decay profile for 11-mer complex is included as a no-ultrafast component control. (Lower Left) Structural model for the Arg cluster interacting with Arg-15 in the complex. Only part of the backbone of RNA is shown; the bases are removed for clarity. Blue dotted lines represent the H-bonds from either Gln-15 in the WT and Arg-15 mutants (modeled based on ref. 37) to the main-chain carbonyl of Arg-8. The positively charged Arg cluster formed by Arg-7, -8, and -11 is labeled. Red double arrows indicate potential repulsive interactions between Arg-15 and the Arg cluster. (Lower Right) Interaction between Arg-15 and the macroscopic dipole moment (indicated by the arrow) in the stacked and helix-extended complexes. The positive charge favors a longer, straighter helix that terminates at Arg-15 (see text).
Together, these data strongly suggest that positive charge of Arg-15 biases the complexes against forming a stacked RNA–peptide interface. Increasing the steric bulk at position 15 (from Gln to Cit) still results in a largely stacked complex. Conversely, introduction of charge (converting Cit to Arg) dramatically destacks the complexes. Consistent with this view, His-15 RNA-peptide complex (Lys-14–His-15) has intermediate stacking, possibly reflecting the intermediate charge state of His (between 0 and +1) around neutral pH.
In the WT complex, there is an important H-bonding interaction between the amide side chain of Gln-15 and the main-chain carbonyl group of Arg-8, which presumably acts to stabilize the peptide structure in the bent-helical form (Figs. 1 and 4 Lower). The fact that Gln, Cit, His, and Arg all can produce some degree of stacking supports the hypothesis that they can make a structurally analogous H-bond by using their amide, guanidinium, or imino protons, respectively (Fig. 1). If this hypothesis is true, then different-length side chains can be accommodated while maintaining this H-bond and preserving the overall bent helical structure in the complex. In this view, the chain length itself is not crucial, and the longer, flexible Arg and Cit side chains can fold and use their appropriate protons (Fig. 1) to form the H-bond with the main-chain carbonyl of Arg-8.
Why does positive charge at position 15 significantly reduce stacking? The most likely culprit can be seen by looking at the WT N peptide/boxB interface (Fig. 4 Lower). In this complex, Arg residues at positions 7, 8, and 11 form a cluster of positive charges that attracts the negatively charged RNA backbone, playing a central role in boxB RNA recognition (14, 15, 18). Previous mutational analysis indicates strong energetic coupling between these charged residues and other portions of the peptide in forming a proper RNA–protein interface (13). Inserting a positive charge at position 15 creates a structural conundrum for the peptide. Attempting to maintain the stacked configuration would likely add favorable peptide–RNA electrostatic interactions. To maintain the H-bond between the side chain of 15 and main-chain carbonyl group, the side chain has to insert into the pocket of positive charges formed mostly by Arg-7, -8, and -11 residues (Fig. 4). However, in the presence of Arg-15, the stacked state interferes with the Arg cluster, which is energetically very unfavorable. This repulsion likely causes the peptide to adopt an alternate conformation with the C-terminal half pushed away from the RNA, thereby disrupting the stacking interaction at the Trp-18/A7 interface.
Electrostatic interactions between Arg-15 and the helix macroscopic dipole (33, 34) may provide a secondary effect that biases against the stacked conformation and toward an alternate structure (Fig. 4 Lower Right). In the WT structure, residue 15 is adjacent to a partial positive charge created by the start of an α-helix (residues 13–22). Arg-15 would likely have unfavorable charge–charge interactions with this dipole. Conversely, extending the helix by one more turn (residues 1–15) places residue 15 adjacent to the partial negative charge occurring at the α-helix C terminus, a favorable interaction. Preliminary NMR structural analysis of Arg-15 mutants supports this “helix extension” model (R. Austin and R.W.R., unpublished data).
The mutations we examined argue that electrostatic interactions compete with the interactions that bend the helix-hydrophobic contacts between the A7 ribose and the side chain of residue 14 and the stacking interaction between Trp-18 and A7. It is difficult to quantitatively assess the contributions of these interactions experimentally; computational approaches may provide reasonable estimates. Overall, the balance of these opposing interactions determines the population of stacked vs. unstacked conformations. In this context, Arg-15 mutants can be seen as a rare example of a single residue sequence change that maintains one function (high-affinity binding) (19) but substantially alters the backbone conformation. The fact that many Arg-15 mutants lack in vivo antitermination function demonstrates that the elongation complex senses conformation rather than binding affinity (20, 21).
Pro-13: A Stacking Enhancer. Our previous studies had shown that some mutations (such as Arg-15) dramatically disfavor the stacked state. However, we were curious as to whether any mutations could be designed that enhance the formation of stacked complexes. The N peptide/boxB complex normally contains a bend at position 11 (14). Generally, a Pro substitution into the middle of the helix can induce a kink naturally (34, 35). Pro substitution at position 13 (Pro-13) has been shown to have WT properties in vitro and in vivo but led to loss of function at other positions (11, 18, 36).
Given this work, we decided to test the effect of Pro-13 insertion on the formation of the stacked state. Insertion of Pro-13 into unstacked complexes (e.g., Lys-14–Arg-15 and Glu-14–Arg-15) results in a dramatic increase in the stacked state (see arrows in Fig. 8 Top and Middle, which is published as supporting information on the PNAS web site). Pro-13 insertion into the WT background (Lys-14–Gln-15) has little effect on the amount of stacked complex. These findings are consistent with NMR analysis of the complexes (data not shown). Interestingly, the functionally inactive Glu-14–Arg-15 mutant can be partially rescued in vivo by proline substitution due to the enhancement of stacking population (Fig. 8 Bottom). These data further support the notion that the stacked state is required for functional antitermination (20, 21).
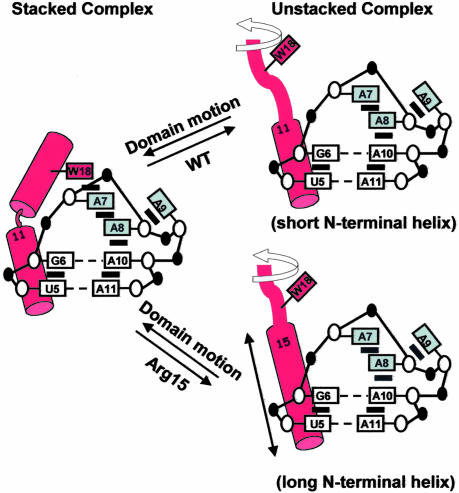
Dynamic Two-State Model: “Helix Extension.” In light of these new studies, we now propose a detailed dynamic two-state model for the boxB/N complexes. In this model, the WT and Arg-15 mutants are treated slightly differently (see Fig. 5). All complexes share the same stacked conformation whose structure is consistent with the NMR model (Fig. 1). Our observation that there are also much slower components in dynamics measurements indicates the existence of other conformations where quenching and charge transfer pathways have been shut off. All of the complexes are structurally heterogeneous, demonstrating a dynamic interchange between states. In the WT complex, the residues 1–11 in the unstacked population are helical and anchor the entire peptide on the RNA as in the stacked population, but the rest of the peptide (residues 12–22) has a random coil conformation. The unstacked population in Arg-15 mutants is different from the WT complex. There, the N-terminal residues undergo helix extension up to residue 15 and become a random coil beyond that (residues 16–22). In addition to the data presented here, formation of this longer, straighter helix is also consistent with CD (20) and NMR 13Cα chemical-shift perturbation studies conducted in our laboratory (R. Austin and R.W.R., unpublished observations).
Fig. 5.
Dynamic two-state models of conformational equilibrium distribution in the WT and Arg-15 mutants RNA-peptide complexes. Arg-15 mutants undergo helix extension when forming the unstacked complex.
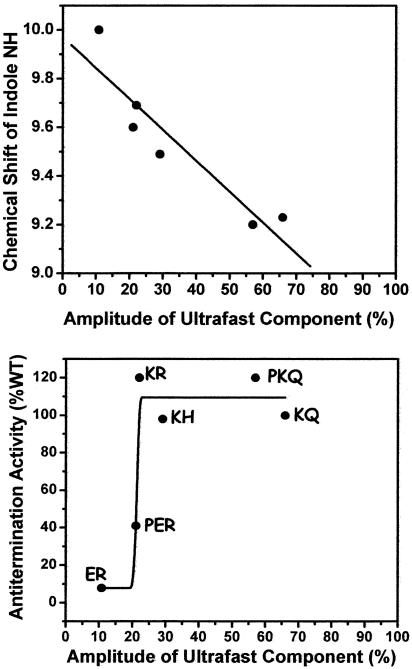
Our ultrafast dynamics shows good correlation between NMR chemical-shift perturbations and dynamics (Fig. 6). Together, these two types of data provide different but complementary information on both the ensemble average (NMR) and population-resolved (dynamics) properties of the system. Combining these spectroscopies with functional studies results in a much richer, more detailed understanding of the structure and dynamics that regulate the transcription elongation complex. Here, we have shown that the ultrafast dynamics amplitude correlates with in vivo function. The N/boxB system now represents a rare example where conformational biases at the single-residue level can be connected with function of a central macromolecular machine within the cell (Fig. 6).
Fig. 6.
Function–dynamics correlation. (Upper) Correlation between chemical shift of indole NH protein of Trp-18 in different complexes (20) and the amplitude of ultrafast component from transient absorption experiments. (Lower) Correlation between transcription antitermination level and the amplitude of ultrafast component from transient absorption experiments. Solid curve is for guiding the eyes only.
Structural Implications of Signal Robustness. The ultrafast transient absorption dynamics in the RNA–protein complexes are the same (τ = 13–16 ps) and also are the same as we observe for 2Ap and Trp mixtures in free solution (31). The fact that the time constants are the same (i.e., robust) indicates that 2Ap and Trp interact in a precise geometry in solution and that the fully stacked RNA–protein complexes achieve this same underlying structure. Together, the fluorescence and absorption dynamics experiments support a model wherein Trp and adenosine interact in only one preferred geometry rather than a series of slipped structures. These data lead us to conclude that the evolution of these macromolecular complexes has created an RNA–peptide interface that mimics the optimal robust structure seen for individual monomers in free solution.
Conclusion
Aside from achieving the design goal of creating functional ligands, the power of combinatorial design (e.g., using mRNA display) lies in the discovery of unexpected and interesting solutions to a recognition problem. Understanding how the selected winners bind the target differently and the physical nature of these differences has two major benefits demonstrated in this work. First, detailed analysis of these complexes improves our fundamental understanding of the principles of molecular recognition. Here, we have identified the different roles of electrostatic and hydrophobic interactions in conferring a particular binding mode, beyond their general contributions to binding affinity. Our work emphasizes that “binding affinity” and “structural specificity” can be achieved with different mechanisms. Second, combinatorial redesign can lead to unexpected insights into biological function. It is remarkable that a single stacking interaction can control the function of a macromolecular machine.
We chose this particular RNA/peptide complex as a model system because it provides experimental access to studies of structure, energetic, dynamic, and biological function. The unique aspect of the ultrafast dynamics is in showing that macromolecular structures do not exist in a rigid, single conformation but that structural heterogeneity represents an essential feature in controlling function. As such, femtosecond resolution of dynamics provides crucial information on the functional population distribution that other structural tools cannot because of the time scale and should be a powerful methodology in the future to address the significant transition from structural biology to function.
Supplementary Material
Acknowledgments
We thank Prof. Naomi Franklin (University of Utah, Salt Lake City) for the two-plasmid N expressor-β-gal reporter constructs; Prof. Douglas Turner for helpful comments, Dr. Adam Frankel (now at University of British Columbia, Vancouver) for help with the transcription antitermination assay, Dr. Suzanna Horvath for purification of some peptides used in this work, and Ms. Nancy Guillen for help with measurements of binding constants. This work was supported by the National Science Foundation through the Laboratory of Molecular Sciences, making it possible to integrate expertise in chemical physics and molecular biology.
Author contributions: R.W.R. and A.H.Z. designed research; T.X. and C.W. performed research; and T.X., C.W., R.W.R., and A.H.Z. wrote the paper.
Abbreviations: 2Ap, 2-aminopurine; Cit, citrulline.
References
- 1.Nagai, K. & Mattaj, I. W. (1994) RNA–Protein Interactions (Oxford Univ. Press, New York).
- 2.Das, A. (1993) Annu. Rev. Biochem. 62, 893–930. [DOI] [PubMed] [Google Scholar]
- 3.Uptain, S. M., Kane, C. M. & Chamberlin, M. J. (1997) Annu. Rev. Biochem. 66, 117–172. [DOI] [PubMed] [Google Scholar]
- 4.Greenblatt, J., Nodwell, J. R. & Mason, S. W. (1993) Nature 364, 401–406. [DOI] [PubMed] [Google Scholar]
- 5.Friedman, D. I. & Court, D. L. (1995) Mol. Microbiol. 18, 191–200. [DOI] [PubMed] [Google Scholar]
- 6.Weisberg, R. A. & Gottesman, M. E. (1999) J. Bacteriol. 181, 359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ptashne, M. (2004) A Genetic Switch: Phage Lambda Revisited (Cold Spring Harbor Lab. Press, Woodbury, New York).
- 8.Mogridge, J., Mah, T. F. & Greenblatt, J. (1995) Genes Dev. 9, 2831–2845. [DOI] [PubMed] [Google Scholar]
- 9.Zhou, Y., Mah, T. F., Yu, Y. T., Mogridge, J., Olson, E. R., Greenblatt, J. & Friedman, D. I. (2001) J. Mol. Biol. 310, 33–49. [DOI] [PubMed] [Google Scholar]
- 10.Cilley, C. D. & Williamson, J. R. (1997) RNA 3, 57–67. [PMC free article] [PubMed] [Google Scholar]
- 11.Su, L., Radek, J. T., Hallenga, K., Hermanto, P., Chan, G., Labeots, L. A. & Weiss, M. A. (1997) Biochemistry 36, 12722–12732. [DOI] [PubMed] [Google Scholar]
- 12.Austin, R. J., Xia, T., Ren, J., Takahashi, T. T. & Roberts, R. W. (2002) J. Am. Chem. Soc. 124, 10966–10967. [DOI] [PubMed] [Google Scholar]
- 13.Austin, R. J., Xia, T., Ren, J., Takahashi, T. T. & Roberts, R. W. (2003) Biochemistry 42, 14957–14967. [DOI] [PubMed] [Google Scholar]
- 14.Legault, P., Li, J., Mogridge, J., Kay, L. E. & Greenblatt, J. (1998) Cell 93, 289–299. [DOI] [PubMed] [Google Scholar]
- 15.Scharpf, M., Sticht, H., Schweimer, K., Boehm, M., Hoffmann, S. & Rosch, P. (2000) Eur. J. Biochem. 267, 2397–2408. [DOI] [PubMed] [Google Scholar]
- 16.Jucker, F. M., Heus, H. A., Yip, P. F., Moors, E. H. & Pardi, A. (1996) J. Mol. Biol. 264, 968–980. [DOI] [PubMed] [Google Scholar]
- 17.Heus, H. A. & Pardi, A. (1991) Science 253, 191–194. [DOI] [PubMed] [Google Scholar]
- 18.Franklin, N. C. (1993) J. Mol. Biol. 231, 343–360. [DOI] [PubMed] [Google Scholar]
- 19.Barrick, J. E., Takahashi, T. T., Ren, J., Xia, T. & Roberts, R. W. (2001) Proc. Natl. Acad. Sci. USA 98, 12374–12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia, T., Frankel, A., Takahashi, T. T., Ren, J. & Roberts, R. W. (2003) Nat. Struct. Biol 10, 812–819. [DOI] [PubMed] [Google Scholar]
- 21.Xia, T., Becker, H. C., Wan, C., Frankel, A., Roberts, R. W. & Zewail, A. H. (2003) Proc. Natl. Acad. Sci. USA 100, 8119–8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milligan, J. F., Groebe, D. R., Witherell, G. W. & Uhlenbeck, O. C. (1987) Nucleic Acids Res. 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fasman, G. D. (1975) Handbook of Biochemistry and Molecular Biology: Nucleic Acids (CRC Press, Cleveland), Vol. 1.
- 24.Sambrook, J. & Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Woodbury, NY), Vol. 3.
- 25.Fiebig, T., Wan, C. & Zewail, A. H. (2002) Chem. Phys. Chem. 3, 781–788. [DOI] [PubMed] [Google Scholar]
- 26.Wan, C., Fiebig, T., Schiemann, O., Barton, J. K. & Zewail, A. H. (2000) Proc. Natl. Acad. Sci. USA 97, 14052–14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pal, S. K., Peon, J. & Zewail, A. H. (2002) Proc. Natl. Acad. Sci. USA 99, 1763–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faber, C., Scharpf, M., Becker, T., Sticht, H. & Rosch, P. (2001) J. Biol. Chem. 276, 32064–32070. [DOI] [PubMed] [Google Scholar]
- 29.Peled-Zehavi, H., Smith, C. A., Harada, K. & Frankel, A. D. (2000) Methods Enzymol. 318, 297–308. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Garcia, C. & Draper, D. E. (2003) J. Mol. Biol. 331, 75–88. [DOI] [PubMed] [Google Scholar]
- 31.Wan, C., Xia, T., Becker, H.-C. & Zewail, A. H. (2005) Chem. Phys. Lett. 412, 158–163. [Google Scholar]
- 32.Zwahlen, C., Legault, P., Vincent, S. J. F., Greenblatt, J., Konrat, R. & Kay, L. E. (1997) J. Am. Chem. Soc. 119, 6711–6721. [Google Scholar]
- 33.Shoemaker, K. R., Kim, P. S., York, E. J., Stewart, J. M. & Baldwin, R. L. (1987) Nature 326, 563–567. [DOI] [PubMed] [Google Scholar]
- 34.Richardson, J. S. & Richardson, D. C. (1988) Science 240, 1648–1652. [DOI] [PubMed] [Google Scholar]
- 35.O'Neil, K. T. & DeGrado, W. F. (1990) Science 250, 646–651. [DOI] [PubMed] [Google Scholar]
- 36.Tan, R. & Frankel, A. D. (1995) Proc. Natl. Acad. Sci. USA 92, 5282–5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.