Abstract
The factors that regulate murine ES cell-derived hematopoietic progenitor cell (HPC) commitment to the B lymphocyte lineage remain unclear. Pu.1 plays an essential role in the development of all lymphoid lineages; however, it also regulates commitment to other blood cell lineages. In this study, we found evidence for early B cell lineage commitment as determined by coexpression of CD19 and CD45R (B220) when Pu.1 expression was knocked down in HPC by specific small interfering RNA (siRNA); moreover, the expression of early B cell factor (Ebf) and paired box protein 5 (Pax-5) transcription factors was induced when cells were treated by Pu.1 siRNA, but not by control siRNA. We also found that siRNA-mediated knockdown of Pu.1 expression was more efficient in generating progenitor B cells (pro-B cells) compared with the more common in vitro method of B lymphoid development by means of coculture of CD34+ embryoid body (EB) cells with OP9 stromal cells. To investigate whether this phenomenon also exists in HPC from other sources, we then knocked down Pu.1 gene expression in CD34+ murine bone marrow cells and found a similar effect of increased production of CD19+CD43+CD45R+ progenitor B cells upon the siRNA-mediated decrease in Pu.1 expression. We conclude that, in early B cell development from ES cell-derived HPC, constitutive Pu.1 expression inhibits the earliest B cell development through repressing early B cell factor and paired box protein 5 expression, although lower levels of Pu.1 expression in HPC play a key role in promoting B cell fate determination.
Keywords: RNA interference, differentiation, paired box protein-5, early B cell factor, IL-7
Murine ES cell (mES) differentiation is a robust system to study the regulation of hematopoietic cell development (1, 2). Differentiated mES cells express similar cell surface antigens and molecular expression patterns at the appropriate stages of progenitor cell development as observed in developing murine embryos in vivo. Mature blood cells, such as red blood cells, platelets, neutrophils, eosinophils, mast cells, and dendritic and natural killer cells have been generated from mES cells (3–6). B lymphocyte progenitors have also been generated from ES cell differentiation, through coculture with OP9 stromal cells (7). OP9 cells are a bone marrow stromal cell line that secretes both defined and undefined factors promoting hematopoietic progenitor cell (HPC) commitment to the B cell lineage. However, OP9 cell support for B cell development is not specific because numerous hematopoietic elements emerge from cocultured mES and OP9 cells. Thus, the disadvantage of using OP9 cells is that they introduce greater complexity in understanding the signals involved in hematopoietic progenitor–stromal cell interactions. Therefore, establishment of a nonstromal cell culture system for B cell development from mES cells would provide more precise and direct knowledge of B cell development.
B lymphocyte development from HPC is a multistep process that involves the ordered expression of intracellular and cell surface marker proteins, and stepwise rearrangement of gene segments in the IgH and L chain gene loci (8). Progenitor B (pro-B) cells express both CD19 and CD45R on their surface, and pro-B cells differentiate into precursor B (pre-B) cells when they complete VH to DH-JH recombination and begin to express cytoplasmic IgH protein (8). Previous in vitro studies demonstrate that B cell development from HPC proceeds in several steps requiring specific cofactors: (i) OP9 stromal cell interactions with HPC (no IL-7), (ii) a combination of OP9 stromal cells and IL-7 exposure, and (iii) IL-7 treatment alone. Our theory proposes that IL-7 is only a support factor in early B cell ontogeny and plays no role in B cell fate determination from HPC.
Transcription factors that play an important role in B cell development included Pu.1, Ikaros, E2A, early B cell factor (Ebf), and paired box protein-5 (Pax-5) (9). However, which transcription factors determine the commitment decision in HPC between lymphoid and myeloid fates is not well documented. Dekoter and Singh (10) have established a system for efficient retroviral transduction of murine fetal liver hematopoietic progenitors from Pu.1 heterozygous (Pu.1+/-) and homozygous mutant (Pu.1-/-) mice, and find that different levels of Pu.1 expression in murine fetal liver HPC could affect cell fate determination between the macrophage and B cell lineages; i.e., Pu.1 protein at a low concentration induced a B cell fate, whereas a high concentration promoted macrophage differentiation and blocked B cell development. Pu.1 also inhibits erythroid progenitor development by blocking binding of Gata-1 to DNA in mice (11).
We have used small interfering RNA (siRNA) to knock down Pu.1 expression in CD34+ cells derived from mES cells, and found that Pu.1 is a repressor of Ebf and Pax-5 expression in HPC, and that reduction of Pu.1 expression by siRNA in HPC is sufficient for B cell development. We propose that modulating Pu.1 expression is a powerful method to differentiate B cells from mES cells in vitro and that this system should permit a more detailed analysis of the transcriptional regulation of the emergence of this lineage from HPC.
Materials and Methods
Cells and Reagents. The D3 ES cells were purchased from American Type Culture Collection. The cytokines murine leukemia inhibitory factor (mLIF), murine (m) IL-3, mIL-7, murine granulocyte-macrophage-colony stimulating factor (mGM-CSF), murine stem cell factor (mSCF), and methylcellulose-based ES cell differentiation medium and collagenase were purchased from StemCell Technologies (Vancouver). Rabbit anti-mouse Pu.1 antibody, goat anti-human EBF, and anti-actin antibodies were purchased from Santa Cruz Biotechnology. Rabbit anti-human Pax-5 antibody was purchased from Zymed Laboratories. Oligofectamine was purchased from Invitrogen.
In Vitro Differentiation of ES Cells. ES cell differentiation into embryoid bodies (EBs) was carried out as described in our previous report (12). Briefly, ES cells were added to 0.9% methylcellulose medium (M4100ES, StemCell Technologies, 15% FBS (HCC6900, StemCell Technologies, 100 ng/ml stem cell factor (R & D Systems), and 450 μM monothioglycerol (Sigma) at a cell concentration of ≈5,000–10,000 cells per ml plated in a 33-mm Petri dish. Efficient differentiation of ES cells to EBs occurred after 10 days of culture.
Harvesting EBs and Isolating CD34+ EB Cell Population by Magnetic Associated Cell Sorting (MACS). The EBs were removed from methylcellulose by dilution with Iscove's modified Dulbecco's medium (IMDM). CD34+ EB cell isolation was carried out by using a MACS magnetic separation device as described (12).
Preparation of dsRNA. Both Pu.1 dsRNA and Lamin A dsRNA (Catalog No. D-100-5) were obtained from Dharmacon Research (Lafayette, CO). The 21-nt Pu.1 dsRNA sequence and the protocol for transient transfection of siRNAs have been reported (12).
Transfection of CD34+ EB Cells with Oligos of Pu.1 siRNA. In brief, CD34+ EB cells were diluted with fresh medium without antibiotics and transferred to six-well plates with 1 × 105 cells per well (500 μl per well). Transient transfection of dsRNAs was carried out by using Oligofectamine (Life Technologies, Carlsbad, CA).
Culture Conditions for siRNA-Transfected CD34+ EB Cells. The cells were untreated (control), or treated by Lamin A dsRNA or Pu.1 dsRNA both at 50 nM. In the myeloid differentiation induction group, mGM-CSF and mIL-3 (10 μg/ml) were added to each well of the culture. In a separate maintenance culture, mSCF (10 ng/ml) only was added to each well. After 72 h, cells were harvested, and phenotype analysis was performed by CD19+ cell sorting. In the B lymphoid differentiation culture, mIL-7 (25 ng/ml), mIL-11 (5 ng/ml), and murine Flt3 ligand (mFL) (100 ng/ml) were added in the culture. The cells were stained with FITC-labeled anti-mouse CD19, phycoerythrin (PE)-labeled anti-mouse CD43 and allophycocyanin (APC)-labeled anti-CD45R antibodies. The CD19+CD43+CD45R+ cell population was analyzed by flow cytometry.
Cultured CD34+ EB Cells with OP9 Cells. siRNA-untreated, Lamin A siRNA-transfected, or Pu.1 siRNA-transfected CD34+ EB cells were seeded onto confluent OP9 monolayers in six-well plates at 1 × 104/ml. After 72 h of culture, loosely adherent and nonadherent cells were isolated and stained with FITC-labeled anti-mouse CD19, phycoerythrin-labeled anti-mouse CD43, and allophycocyanin-labeled anti-mouse CD45R antibodies and analyzed by FACS.
CD19+CD43+CD45R+ Cell Culture with mIL-7 and mIL-10 to Develop Pre-B Cell Colonies. Three days after Pu.1 siRNA transfection, cells were harvested for CD19 and CD45R expression analysis by flow cytometry. The CD19+CD43+CD45R+ cells were sorted, and then mixed with Iscove's modified Dulbecco's medium, 20% FCS, 10% BSA (StemCell Technology, Canada), 100 μg/ml bovine transferrin, 10 ng/ml bovine insulin, 0.1 mmol/liter monothioglycerol, 1% methylcellulose, and distributed to four 35-mm bacterial-grade dishes at 1 × 104 to 105 cells per plate. In addition, 25 ng/ml mIL-7 and 10 ng/ml mIL-10 were added, and CFU-pre-B were counted on day 7 of culture as described (13). All of the plates were incubated under 5% O2,5%CO2 at 37°C. The average number of each colony type in each well was adjusted to 1 × 105 seeded cells per well.
Generation of Mature Mitogen-Response Ig-Secreting B Lymphocytes. The individual CFU-pre-B colonies were picked and transferred to six-well plates with mIL-7 and mIL-10 for another 3 weeks and analyzed for CD21 expression by FACS. To demonstrate further the functional capabilities, the in vitro-generated B cells were then treated with LPS for 3 days (3). CD80, a costimulatory molecule that is normally up-regulated on mature B cells after activation (3), was analyzed by flow cytometry following the LPS exposure. Culture supernatants in LPS-treated cultures were collected for ELISA analysis of IgM (Fig. 1).
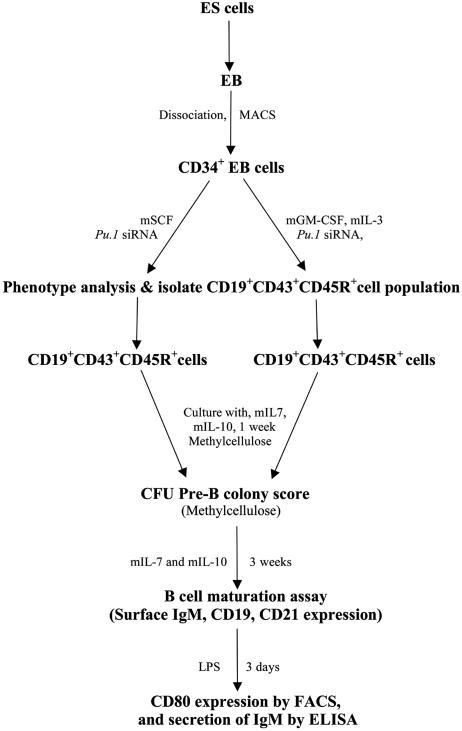
Fig. 1.
General outline of this protocol. ES cells were differentiated into EBs in the absence of murine leukemia inhibitory factor. CD34+ cell populations were isolated from whole EB cell suspension by magnetic associated cell sorting magnetic bead cell separation. Cells were then transfected with Pu.1 siRNA and maintained with mSCF in culture, or secondary differentiation was initiated in myeloid differentiation conditions; i.e., mGM-CSF and mIL-3 were added in the medium. Pro-B cell formation was examined in the CD19+CD43+CD45R+ phenotype. To test whether the B progenitors were functional, cells were matured and examined for CD21 expression, and further examined for their LPS stimulation response, such as CD80 expression and IgM secretion.
Western Blot Analysis. Protein was isolated from cultured cells after 72 h of siRNA treatment. Pu.1 Western blot analysis was performed as described (12). For Ebf and Pax-5 Western blot analysis, 50 μg of protein was separated by SDS/PAGE by using a 10% (wt/vol) polyacrylamide resolving gel and transferred electrophoretically to a nitro-cellulose membrane. The blots were blocked with 5% TBS/T buffer for 1 h. Immunoblotting was performed overnight at 4°C by using the Ebf primary antibody (Santa Cruz Biotechnology) at a 1:700 dilution, Pax-5 antibody at 1:500 dilutions, or actin antibody at 1:1,000 dilutions, and the peroxidase-conjugated secondary antibodies (Amersham Pharmacia). All immunoblots were visualized by ECL reagent according to the manufacturer's instructions (Amersham Pharmacia Biotech).
VDJ Recombination Assay. CD19+CD43+CD45R+ cells were sorted from total cells treated with Pu.1 siRNA for 3 days. The genomic DNA was isolated, and VH7183-DJH4 (VDJ) recombination detection was performed as described (14). The VH7183-specific primer was 5′-TGG TGG AGT CTG GGG GAG GCT TA-3′ and the 3′ JH4 primer was 5′-GGC TCC CTC AGG GAC AAA TAT CCA. The expected size for the VDJH4 product was 0.4 kb.
FACS Sorting of CD34+ Murine Bone Marrow Cells. Bone marrow was collected from adult mice, and the CD34+ cell population was isolated by FACS sorting. Sorted cells were then transfected with Pu.1 siRNA or control siRNA, then placed in culture with mSCF. After 72 h, cells were collected for phenotype assay by using B lymphocyte lineage markers (CD19+CD43+CD45+).
Results
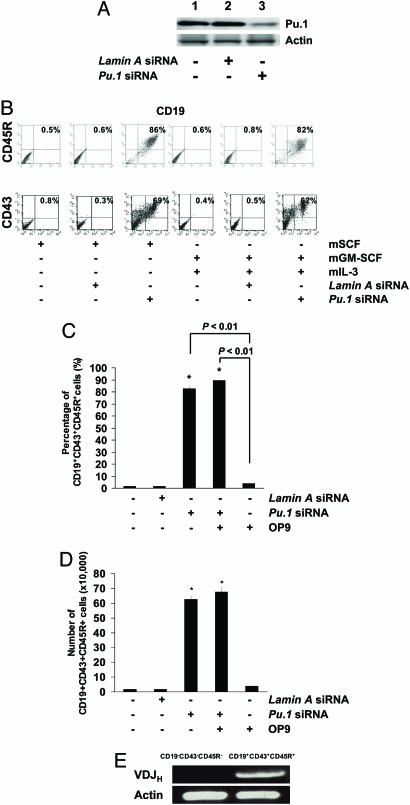
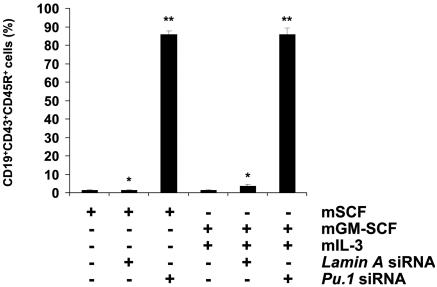
Knockdown of Pu.1 Gene Expression in CD34+ EB Cells Induces the Production of CD19+ Cells. Differentiated ES cells regularly form EBs under specific differentiation conditions. Culture of sorted CD34+ EB cells with mGM-CSF and mIL-3 led to proliferation and differentiation of myeloid cells and inhibition of early B cell development (15). In this study, CD34+ EB cells maintained in culture in the presence of mSCF alone gave rise to a detectable level of CD19 expression in hematopoietic cells when Pu.1 expression was knocked down by Pu.1 siRNA treatment (Fig. 2 A and B). Further experiments demonstrated that CD19 expression was detectable in cells derived from CD34+ EB cells cultured with mGM-CSF and mIL-3 when cells were treated with Pu.1 siRNA, but not with control siRNA (Fig. 2B). These CD19+ cells also coexpressed CD45R and CD43 on their surface (Fig. 2B), but not sIgM (data not shown). Furthermore, siRNA-mediated reduction in Pu.1 expression was associated with a more robust differentiation of CD34+ HPC to the B lymphocyte lineage than CD34+ HPC cocultured with OP9 cells (Fig. 2 C and D). The CD19+ cells did not coexpress progenitor T cell markers, such as CD25 and CD44 (data not shown), indicating that the siRNA effect was restricted to B lymphoid development. In the CD19+CD43+CD45R+cells, VDJ recombination was detected by PCR (Fig. 2E).
Fig. 2.
Knockdown of Pu.1 by siRNA leads to CD19+CD43+CD45R+ pro-B cells. (A) Western blot analysis was performed to confirm the effect of Pu.1 siRNA on the target gene level of expression. As shown, Pu.1 expression was reduced when cells were treated by Pu.1 siRNA (lane 3) compared with siRNA untreated cells (lane 1) or control (Lamin A) siRNA-treated cells for 72 h. Data are representative of one of three individual experiments. (B) Pu.1 siRNA-transfected CD34+ cells maintained with SCF differentiated to CD19+CD43+CD45R+ pro-B cells. (C) Pu.1 siRNA-transfected CD34+ cells cultured with mGM-CSF and mIL-3 differentiated into CD19+CD43+CD45R+ pro-B cells. Data from one representative experiment from three individual experiments is shown. (D) Comparison of pro-B cells generated between siRNA-mediated Pu.1 knockdown of gene expression compared with the OP9 coculture system. Knockdown of Pu.1 expression induced over 80% of the CD34+ cells to become pro-B cells within 72 h. However, simple coculture of CD34+ EB cells with OP9 stromal cells induced only 5% of cells to become pro-B cells in the same time period. Combination of knockdown of Pu.1 expression by means of siRNA treatment with coculture with OP9 cells could not significantly increase the frequency of pro-B cells compared with the siRNA alone group. (E) Generated CD19+CD43+CD45R+ cells express VDJ gene rearrangement examined by RT-PCR.
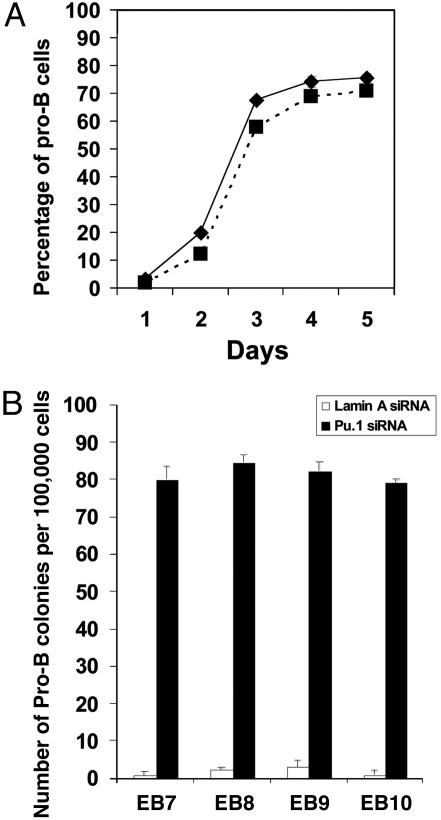
The Kinetics of Pro-B (CD19+CD43+CD45R+) Cell Development in Pu.1 siRNA-Treated CD34+ EB Cells and a Comparison of the Potential for Pro-B Cell Development of These Cells from Different EB Ages. After Pu.1 siRNA treatment, we found that pro-B cell formation was very low (<5%) on day 1; on day 2, ≈20% of cells are pro-B cells; on day 3, the percentage of pro-B cells has been increased to 70%; and it continues to increase slightly till day 5 (Fig. 3A). Our study also reveals that CD34+ EB cells from different age EBs have similar potential for Pro-B cell development under the condition of Pu.1 treatment (Fig. 3B).
Fig. 3.
Kinetics in pro-B cell production from Pu.1 siRNA-treated CD34+ EB cells, and a comparison of the potential for pro-B cell development of these cells from different EB ages. (A) After Pu.1 siRNA treatment, in day 1, <5% of cells are pro-B cells; in day 2, it increased to ≈20%; on day 3, the percentage of pro-B cells has increased to 60–70%; then the percentage of pro-B cells gradually increased until day 5 (solid line, cells cultured with mSCF; dotted line, cells cultured with mGM-CSF and mIL-3). (B) CD34+ EB cells from different age EBs have similar potential in pro-B cell development under the Pu.1 siRNA treatment.
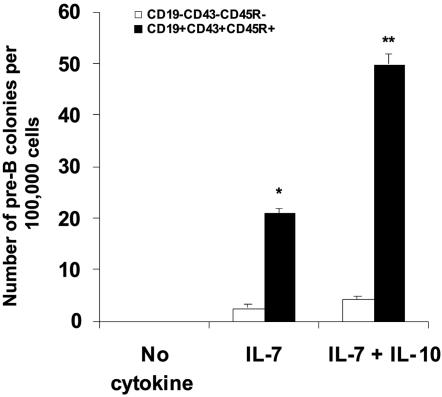
The CD19+CD43+CD45R+ Cell Population Could Develop into CFU-Pre-B Colonies in Vitro in the Presence of mIL-7 and mIL-10. To examine whether the CD19+CD43+CD45R+ cells formed in Pu.1 siRNA-treated CD34+ EB cocultures could form pre-B colonies, we isolated this population by FACS sorting and performed colony-forming assays. CFU-pre-B colonies could be formed with mIL-7 and mIL-10 stimulation in the culture for 7 days (Fig. 4). Without mIL-7 in the culture, there was no colony formation. Moreover, mIL-10 could increase the frequency of CFU-pre-B colonies primed by mIL-7 compared with mIL-7 administration alone (P < 0.05). In contrast, the CD19-CD43-CD45R- cell population had minimal potential to develop to pre-B colonies in the same culture conditions (Fig. 4).
Fig. 4.
CFU-pre-B colony development from a CD19+CD43+CD45R+ cell populations (open bars, CD19-CD43-CD45R- cells; filled bars, CD19+CD43+CD45R+ cells). In the control group, without the stimulation by cytokines mIL-7 and mIL-10, there was no pre-B colony formation. When CD19+CD43+CD45R+ cells were treated with mIL-7, some pre-B colonies formed; however, when mIL-7 and mIL-10 were combined, the frequency of CFU-pre-B cells increased dramatically (*, P < 0.05 for CD19+CD43+CD45R+ cells versus CD19-CD43-CD45R- cells in mIL-7 culture; **, P < 0.01 for CD19+CD43+CD45R+ cells versus CD19-CD43-CD45R- cells in mIL-7 plus mIL-10 culture).
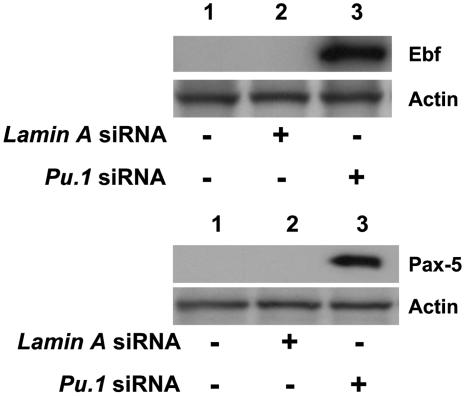
Murine Ebf and Pax-5 Expression Is Induced in CD34+ EB Cell Differentiation When Pu.1 Expression Is Knocked Down by siRNA. To determine whether murine Ebf and Pax-5 genes are involved in the increased emergence of CD19+ cells from CD34+ HPC, whole cell protein was isolated from the cultured cells for Western blot assay. Neither siRNA untreated cells nor cells treated with control siRNA (Lamin A siRNA) expressed Ebf or Pax-5 protein; however, cells treated with Pu.1 siRNA expressed both Ebf and Pax-5 proteins as detected by Western blot analysis (Fig. 5).
Fig. 5.
Knockdown of Pu.1 expression in CD34+ cells induces both Ebf and Pax-5 expression. (Upper) Ebf expression appears in Pu.1 siRNA-treated CD34+ EB cells primed by mGM-CSF and mIL-3 treatment for 72 h. Lane 1, untreated cells; lane 2, cells treated with control Lamin A siRNA; lane 3, cells treated with Pu.1 siRNA. Representative data from one of three individual experiments are shown. (Lower) Pax-5 expression is increased in Pu.1 siRNA-treated CD34+ EB cells. Lane 1, untreated cells; lane 2, cells treated with control Lamin A siRNA; lane 3, cells treated with Pu.1 siRNA. Representative data from one of three individual experiments are shown.
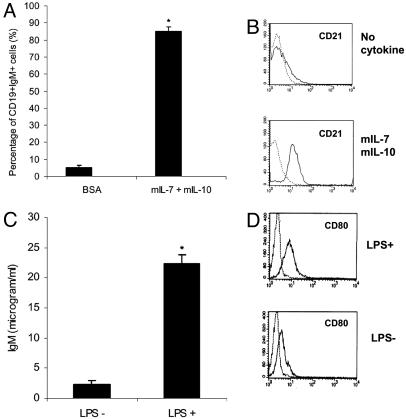
CFU-Pre-B Cells Develop to Mature B Cells. To test whether B precursor cells generated from EB cells are able to develop into mature B cells, single CFU-pre-B colonies were picked and transferred to a six-well plate. After 3 weeks of culture with mIL-7 and mIL-10, the pre-B cells differentiated to mature B cells that coexpress IgM and CD19 (Fig. 6A). These functional B cells also exhibit phenotypic changes, such as up-regulation of CD21 expression, a marker of mature B cells that was highly expressed (Fig. 6B). We then further examined these cells' response to LPS stimulation. CD80 expression was noticeably up-regulated, and secretion of IgM also increased >5-fold (Fig. 6 C and D) when the B cells were cultured with LPS (P < 0.01). Thus, the B lymphoid cells generated from the CD34+ EB cells can fully differentiate into mature B cells in the absence of any supportive stromal elements.
Fig. 6.
Pro-B cell can be further differentiated to mature B cells. (A) Frequency of functional B cell (CD19+IgM+) formation from CD19+CD43+CD45R+ pro-B cells. (B) CD21 expression analysis by flow cytometry. Cytokine stimulation (mIL-7 and mIL-10) is able to up-regulate CD21 expression in these B cells. Representative data from one of three individual experiments are shown. (C) sIgM assay by ELISA on CD19+CD43+CD45R+ cells stimulated by LPS. Data are the mean ± SD of three independent experiments. Lane 1, untreated cells; lane 2, cells treated with LPS. (D) LPS up-regulates CD80 expression in the CD19+CD43+CD45R+ cell population (*, P < 0.01).
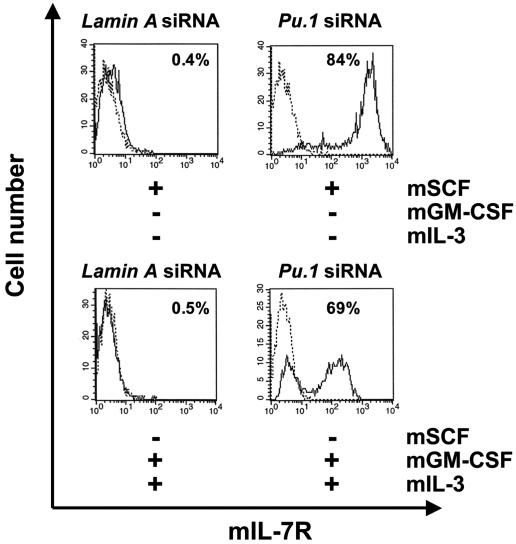
mIL-7 Receptor (mIL-7R) Expression Was Induced When Pu.1 Gene Was Knocked Down by siRNA. In this study, we found that neither CD34+ HPC in mouse bone marrow or CD34+ EB cells express the mIL-7R (data not shown). Furthermore, mIL-7R was not expressed in CD34+ EB cells after 3 days of culture in which the cells were maintained in the presence of SCF alone. However, mIL-7R expression was induced when Pu.1 gene expression was knocked down by siRNA treatment of the CD34+ EB cells. Further experiments revealed that mIL-7R was also induced in the progenitor cell differentiation culture of CD34+ EB cells driven by mGM-CSF and mIL-3 when Pu.1 expression was knocked down (Fig. 7).
Fig. 7.
Knockdown of Pu.1 expression results in mIL-7R expression on CD34+ EB cells cultured either in maintenance conditions (mSCF alone) or myeloid differentiation conditions (mGM-CSF and mIL-3). (Upper) CD34+ EB cells cultured with mSCF did not express mIL-7R when treated with control siRNA; however, upon treatment with Pu.1 siRNA, mIL-7R expression was induced and detected by flow cytometry. Representative data of one of three individual experiments are shown. (Lower) In CD34+ EB cells cultured with mGM-CSF and mIL-3, mIL-7R expression was also induced when cells were treated by Pu.1 siRNA, but not control siRNA. Representative data from one of three individual experiments are shown.
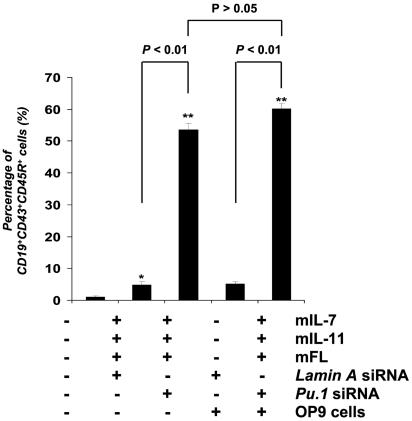
Knockdown of Pu.1-Enhanced Pro-B Cell Development Mediated by Cytokines mIL-7, mIL-11, and Murine Flt3-Ligand (mFL). Early B cell development from HPC can be induced in vitro by several cytokines without stromal cell support. Combinations of mIL-7, mIL-11, and mFL have been reported to induce early B cell development from HPC isolated from mouse fetal liver after 5–10 days (16). We added these cytokines to cultured CD34+ cells derived from mES cells. We found that, after 72 h, <5% of cells coexpressed both CD19, CD43, and CD45R; however, knockdown of Pu.1 in CD34+ cells by means of siRNA treatment enhanced the production of CD19+CD43+CD45R+ pro-B cells by 10-fold. Moreover, coculture of CD34+ cells with OP9 cells for 72 h produces fewer than 5% pro-B cell formation; however, knockdown of Pu.1 on CD34+ cells plus coculture with OP9 cells resulted in significantly increased in pro-B cell formation compared with the CD34+ cells cocultured only with OP9 cells. There is no significant difference in pro-B cell formation between CD34+ cells treated with Pu.1 siRNA compared with CD34+ cells treated with Pu.1 siRNA plus coculture with OP9 cells (P > 0.05) (Fig. 8).
Fig. 8.
Knockdown of Pu.1 expression on CD34+ EB cells enhances the frequency of CD19+CD43+CD45R+ cell production mediated by mIL-7, mIL-11, and mFL in vitro. CD34+ EB cells were transfected with either Lamin A siRNA (control) or Pu.1 siRNA (50 nM), and cultured with or without OP9 cells and with or without cytokines mIL-7, mIL-11, and mFL for 72 h. Cells were then collected and analyzed by flow cytometry for cell surface marker expression. Data are the mean ± SD of three independent experiments. Pu.1 siRNA-treated CD34+ cells showed a 10-fold increase in the production of CD19+CD43+CD45R+ cells compared with the cells treated with control siRNA (*, P < 0.01).
Knockdown of Pu.1 in CD34+ Bone Marrow Cells also Promotes Pro-B Cell Development. To clarify whether HPC differentiation into B cells is enhanced by knockdown of Pu.1 expression in primary CD34+ mouse bone marrow cells, we sorted CD34+ hematopoietic cells from mouse bone marrow and transfected with Lamin A siRNA (as a control) or Pu.1 siRNA, then cultured cells with the cytokines mSCF or mGM-CSF plus mIL-3. As shown in Fig. 9, we determined that knockdown of Pu.1 expression in primary CD34+ mouse bone marrow cells by means of siRNA treatment also promotes pro-B cell development.
Fig. 9.
Knockdown of Pu.1 expression in CD34+ murine bone marrow cells enhances cell fate determination to pro-B cells. CD34+ marrow cells were transfected with control siRNA or Pu.1 siRNA and then cultured with mSCF or mGM-CSF plus mIL-3 for 72 h, before harvesting for FACS analysis. CD19, CD43, and CD45R expression was significantly induced in these bone marrow hematopoietic cells when they were treated by Pu.1 siRNA for 72 h. Data are the mean ± SD of three independent experiments.
Discussion
HPC target gene expression can be efficiently modulated by siRNA treatment in vitro (for a review, see ref. 17). In this article, we described our observation that B cell development was induced when Pu.1 gene expression was knocked down by siRNA treatment in CD34+ EB cells derived from mES cells. Pu.1 levels played a key role in cell fate determination from HPC to pro-B cell differentiation. Up to 90% of cells expressed CD19, CD43, and CD45R when CD34+ cells were treated with Pu.1 siRNA (Fig. 2). Further examination with PCR revealed that these CD19+CD43+CD45R+ cells exhibited VDJ rearrangement, which is a critical marker of pro-B cells. These cells were negative for progenitor T cell markers, such as CD25 and CD44 (data not shown), suggesting that the siRNA effect was specific to B lymphoid development.
It is generally agreed that acquisition of cell surface expression of CD19, CD43, and CD45R signifies commitment to the B cell lineage (8). To exclude the possibility that isolated CD19+ cells were follicular dendritic cells (FDC) (which also express this marker), flow cytometry was performed to examine CD35 expression, which is a specific marker on FDC. CD35 was not expressed on these CD19-expressing cells (data not shown). Thus, these CD19+ cells are B precursors.
mIL-7R expression is one of the earliest detectable events in B lymphoid development from hematopoietic precursors. In our study, mIL-7R expression could be detected when CD34+ cells were also induced to express CD19 after Pu.1 siRNA treatment (Fig. 7). Blocking of mIL-7R expression induced by Pu.1 siRNA treatment by anti-mIL-7R antibody could inhibit the CD19 expression in CD34+-derived progeny (data not shown). Thus, CD19 expression seems to be triggered by endogenous mIL-7/mIL-7R interactions as B lymphoid precursors differentiate from CD34+ HPC. Because the reduction of Pu.1 in CD34+ cells promotes mIL-7R expression in this study, we speculate that constitutive Pu.1 expression in the CD34+ HPC may prevent mIL-7R expression and thus block B cell development. This finding is consistent with other observations in which mIL-7R–mediated signaling is necessary for the development of the earliest B cells (18).
These CD19+ cell populations generated from the CD34+ HPC matured in the presence of LPS and secreted IgM, indicative of B cell maturation. Therefore, our data demonstrate that differentiation of mES cell-derived CD34+ HPC to an early B lymphoid progenitor stage is highly influenced by the level of Pu.1 expression (Figs. 8 and 9). In separate experiments, we also found the same phenomenon using adult murine bone marrow hematopoietic cells. Knockdown of Pu.1 expression in bone marrow CD34+ cells also causes cell commitment to pro-B cells as judged by coexpression of CD19, CD43, and CD45R in cells cultured with either mSCF alone or mGM-CSF plus mIL-3 (data not shown). These data support an interpretation that a low level of Pu.1 expression is a critical factor in the transition of a HPC to a pro-B cell.
Transcription factors play a key role in control of the commitment of progenitors along different lineages. DeKoter and Singh (9) described that low concentrations of Pu.1 protein induced a B cell fate, whereas a high concentration promoted macrophage differentiation and blocked B cell development in fetal liver HPC. Recently, they further reported (19) that transduction of exogenous Ebf into Pu.1-/- murine fetal liver progenitors restored early B cell development. The transcription factors Ebf and Pax-5 are essential regulators in early B cell development (20). Ebf can restrict lymphopoiesis to the B cell lineage by blocking development of other lymphoid-derived cell pathways (21). Ebf is an upstream regulator of Pax-5 gene transcription in B cells (20). Pax-5 acts as a traffic signal in the commitment of progenitors. When it is on, hematopoiesis will proceed to the B-lineage; when it is off, the cell commitment is routed to the myeloid and T-lymphoid lineages (22). We propose that induction of both Ebf and Pax-5 gene expression in CD34+ cells treated with Pu.1 siRNA leads to B cell differentiation. Conversely, constitutive Pu.1 expression leads to the suppression of Ebf and Pax-5 expression, thus preventing HPC commitment to the B cell lineage.
Stromal cells OP9 are frequently required to study B cell ontogeny from lineage uncommitted hematopoietic stem cells or from ES-derived EB cells (23, 24). The disadvantage of this strategy is that the signals required inducing B cell lineage commitment from the stem cells or ES-derived EB cells are unknown. In the present work, we have determined that decreasing the level of Pu.1 expression in combination with mGM-CSF, mIL-3, or mSCF stimulation is sufficient to induce B cell lineage commitment from EB-derived CD34+ cells. Knockdown of Pu.1 is more efficient in inducing B cell lineage commitment than the OP9 coculture system. We propose that this method of siRNA-mediated regulation of transcription factor expression will further promote discovery of the molecular signals regulating B cell lineage development in ES-derived cells and may be applicable to human ES cells, where B cells have been recently reported to emerge from CD34+ hEB cells in OP9 coculture (25).
Acknowledgments
We thank Becky Chan for reading the manuscript. We thank Peter Zandrstra and Gordon Keller for critical review of our manuscript. This research was supported by National Institutes of Health Grant CA 84405 (to J.D.R.) and by the Spastic Paralysis Foundation of the Illinois–Eastern Iowa Division of Kiwanis International (J.D.R.).
Author contributions: G.-M.Z. designed research; G.-M.Z., J.-J.C., and W.W. performed research; G.-M.Z., M.C.Y., and J.D.R. analyzed data; M.C.Y. and J.D.R. contributed new reagents/analytic tools; and G.-M.Z. wrote the paper.
Abbreviations: EB, embryoid body; siRNA, small interfering RNA; pro-B cells, progenitor B cells; pre-B cells, precursor B cells; Ebf, early B cell factor; Pax-5, paired box protein-5; HPC, hematopoietic progenitor cells; m, murine; mES, murine ES; mSCF, murine stem cell factor; mGM-CSF, murine granulocyte-monocyte-colony stimulating factor; mFL, murine Flt3 ligand.
References
- 1.Keller, G., Lacaud, G. & Robertson, S. (1999) Exp. Hematol. 27, 777-787. [DOI] [PubMed] [Google Scholar]
- 2.Daley, G. Q. (2003) Ann. N.Y. Acad. Sci. 996, 122-133. [DOI] [PubMed] [Google Scholar]
- 3.Hamaguchi-Tsuru, E., Nobumoto, A., Hirose, N., Kataoka, S., Fujikawa-Adachi, K., Furuya, M. & Tominaga, A. (2004) Br. J. Haematol. 124, 819-827. [DOI] [PubMed] [Google Scholar]
- 4.Lieber, J. G., Webb, S., Suratt, B. T., Young, S. K., Johnson, G. L., Keller, G. M. & Worthen, G. S. (2004) Blood 103, 852-859. [DOI] [PubMed] [Google Scholar]
- 5.Fujimoto, T. T., Kohata, S., Suzuki, H., Miyazaki, H. & Fujimura, K. (2003) Blood 102, 4044-4051. [DOI] [PubMed] [Google Scholar]
- 6.Nakayama, N., Fang, I. & Elliott, G. (1998) Blood 91, 2283-2295. [PubMed] [Google Scholar]
- 7.Cho, S. K., Webber, T. D., Carlyle, J. R., Nakano, T., Lewis, S. M. & Zuniga-Pflucker, J. C. (1999) Proc. Natl. Acad. Sci. USA. 96, 9797-9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schweitzer, B. L. & DeKoter, R. P. (2004) J. Immunol. 172, 144-154. [DOI] [PubMed] [Google Scholar]
- 9.Kee, B. L. & Murre. C. (2001) Curr. Opin. Immunol. 13, 180-185. [DOI] [PubMed] [Google Scholar]
- 10.DeKoter, R. P. & Singh. H. (2000) Science 288, 1439-1441. [DOI] [PubMed] [Google Scholar]
- 11.Dahl, R., Walsh, J. C., Lancki, D., Laslo, P., Iyer, S. R., Singh, H. & Simon, M. C. (2003) Nat. Immunol. 4, 1029-1036. [DOI] [PubMed] [Google Scholar]
- 12.Zou, G. M., Wu, W., Chen, J. & Rowley, J. D. (2003) Biol. Cell 95, 365-371. [DOI] [PubMed] [Google Scholar]
- 13.Fine, J. S., Macosko, H. D., Grace, M. J. & Narula, S. K. (1994) Exp. Hematol. 22, 1188-1196. [PubMed] [Google Scholar]
- 14.Carlsson, L., Candeias, S., Staerz, U. & Keller, G. (1995) Eur. J. Immunol. 25, 2308-2317. [DOI] [PubMed] [Google Scholar]
- 15.Fairchild, P. J., Brook, F. A., Gardner, R. L., Graca, L., Strong, V., Tone, Y., Tone, M., Nolan, K. F. & Waldmann, H. (2000) Curr. Biol. 10, 1515-1518. [DOI] [PubMed] [Google Scholar]
- 16.Ray, R. J., Paige, C. J., Furlonger, C., Lyman, S. D. & Rottapel. R. (1996) Eur. J. Immunol. 26, 1504-1510. [DOI] [PubMed] [Google Scholar]
- 17.Zou, G. M. & Yoder, M. C. (2005) Biol. Cell 97, 211-219. [DOI] [PubMed] [Google Scholar]
- 18.Purohit, S. J., Stephan, R. P., Kim, H. G., Herrin, B. R., Gartland, L. & Klug, C. A. (2003) EMBO J. 22, 5511-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medina, K. L., Pongubala, J. M., Reddy, K. L., Lancki, D. W., Dekoter, R., Kieslinger, M., Grosschedl, R. & Singh H. (2004) Dev. Cell 7, 607-617. [DOI] [PubMed] [Google Scholar]
- 20.Maier, H. & Hagman. J. (2002) Semin. Immunol. 14, 415-422. [DOI] [PubMed] [Google Scholar]
- 21.Zhang, Z., Cotta, C. V., Stephan, R. P., deGuzman, C. G. & Klug. C. A. (2003) EMBO J. 22, 4759-4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Busslinger M. (2004) Annu. Rev. Immunol. 22, 55-79. [DOI] [PubMed] [Google Scholar]
- 23.Vieira, P. & Cumano. A. (2004) Methods Mol. Biol. 271, 67-76. [DOI] [PubMed] [Google Scholar]
- 24.Cho, S. K. & Zuniga-Pflucker, J. C. (2003) Methods Enzymol. 365, 158-169. [DOI] [PubMed] [Google Scholar]
- 25.Vodyanik, M. A., Bork, J. A., Thomson, J. A. & Slukvin II (2005) Blood 105, 617-626. [DOI] [PubMed] [Google Scholar]