Abstract
Integrating nucleic acids (NAs) with nanomaterials has substantially advanced biomedical research, enabling critical applications in biosensing, drug delivery, therapeutics, and the synthesis of nanomaterials. At the core of these advances are the reactions of NAs on nanomaterial surfaces, encompassing conjugation (covalent and non-covalent), detachment (physical and chemical), and signal amplification (enzyme-mediated signal amplification, enzyme-free signal amplification, and DNA Walker). Here, we review the fundamental mechanisms and recent progress in nucleic acid reactions on nanomaterial surfaces, discuss emerging applications for diagnostics, nanomedicine, and gene therapy, and explore persistent challenges in the field. We offer a forward-looking perspective on how future developments could better control, optimize, and harness these reactions for transformative advances in nanomedicine and biomedical engineering.
Graphical abstract
Keywords: Nanomaterials, Interaction, NA reactions, Surface chemistry, Nanomedicine, Gene therapy
Introduction
Nucleic acids (NAs), encompassing oligonucleotides, aptamers, and DNAzymes, are central to numerous cutting-edge breakthroughs in biomedical research [1, 2]. Their extraordinary capacity to specifically recognize and bind a wide variety of biological and chemical targets (e.g., proteins, viruses, small molecules, and cells) underpins critical applications in genomics, bio-detection, and therapeutic development [3, 4]. However, translating these fundamental capabilities into clinically viable tools has long been hampered by challenges related to NA stability, bioavailability, and sensitivity in complex biological environments. A powerful strategy to overcome these limitations is to exploit the interfacial properties of nanomaterials. Owing to their large surface area and tailorable physicochemical features, nanomaterials significantly enhance NA performance by providing robust, multifunctional platforms [5]. Consequently, NA-functionalized nanomaterials have emerged as a promising frontier for applications in biosensing, drug delivery, gene therapy, and medical imaging [6–8]. Yet, the full therapeutic and diagnostic potential of these systems cannot be realized without a deeper understanding of how NAs interact with, attach to, or detach from nanomaterial surfaces.
Recent advances demonstrate that NA attachment, or “conjugation,” onto nanomaterial surfaces often uses either covalent or non-covalent methods, such as thiol-gold linkages, biotin-streptavidin recognition, hydrogen bonding, or electrostatic attraction [9–11]. These techniques reliably anchor NAs to the nanomaterial interface, improving stability and enabling targeted functions. Conjugated complexes have seen particular success in biosensing, where specific NA sequences enable sensitive detection of disease biomarkers, and in targeted drug delivery platforms, where precise NA design directs nanomaterials to specific tissues or cell types. Equally important, though less frequently discussed, is the process of NA detachment. Physical or chemical stimuli can disrupt the connection between NAs and nanomaterials, allowing on-demand release of NA payloads. For instance, adjusting an external magnetic field can cause DNA probes to detach from magnetic nanomaterials, permitting reversible in vitro diagnostics [12]. Meanwhile, photothermal nanomaterials facilitate NA desorption via laser irradiation, enabling controllable gene delivery or smart photothermal-chemotherapeutic strategies [13]. An in-depth understanding of these mechanisms is essential for designing advanced biomedical devices that require precise NA release profiles.
In addition to conjugation and detachment, another dimension of NA–nanomaterial interactions centers on signal amplification. By housing NA signal amplification on nanomaterial surfaces, scientists can dramatically boost the sensitivity of analytical assays [14, 15]. This amplification approach effectively addresses a critical bottleneck in early disease detection: the extremely low abundance of diagnostic molecules in bodily fluids. For example, nanomaterial-based nucleic acid amplification can detect trace pathogens or cancer-specific sequences with minimal false positives, opening new horizons for early-stage intervention. This concept of “NA reactions on nanomaterial surfaces” thus spans a complete spectrum: (i) anchoring NAs (conjugation), (ii) releasing them (detachment), and (iii) leveraging their signal amplification capacity. Despite significant achievements, fundamental questions remain unresolved. How can researchers systematically select the most suitable conjugation strategies for specific biomedical tasks? What factors enable fine control of NA detachment under physiological conditions? And how might signal amplification be further refined to improve throughput, cost-effectiveness, and clinical performance?
Although various reviews address NA-based nanotechnologies, most focus on functional modifications of NAs or the downstream applications of NA-functionalized nanomaterials [16–21]. Relatively few consider the crucial mechanistic underpinnings of NA–nanomaterial surface reactions. As a result, researchers and practitioners may lack vital insights into selecting optimal conjugation or detachment strategies, accurately modeling reaction kinetics, or integrating advanced signal amplification mechanisms into emerging biomedical devices. To bridge this gap, we provide a reaction-oriented perspective, systematically examining the ways in which NAs can be covalently or non-covalently attached, detached via physical or chemical means, and amplified on nanomaterial surfaces. By explicitly discussing these reaction classes, we aim to offer a unified framework that not only consolidates knowledge but also stimulates future research directions in nanomedicine and biomedical engineering.
In this review, we first analyze conjugation reactions, from classical covalent linkages (e.g., amide and disulfide bonds) to more dynamic non-covalent interactions (e.g., hydrogen bonding, π-π stacking, and electrostatic attraction). We then delve into detachment reactions, highlighting both chemical (e.g., competitive displacement and chelating agents) and physical (e.g., thermal and photothermal) methods. Next, we cover signal amplification reactions, encompassing enzyme-mediated NA amplification, enzyme-free signal amplification, and DNA walker, with a focus on leveraging nanomaterial properties to boost sensitivity and specificity. Following this foundational discussion, we spotlight advanced biomedical applications—including nanomaterial synthesis guided by NAs, cutting-edge gene delivery strategies, and next-generation biosensors. Finally, we offer a perspective on current challenges (e.g., reproducibility, in vivo stability, biocompatibility) and speculate on future avenues, emphasizing data-driven and machine learning-based approaches. Our overarching goal is to demonstrate how a deeper, mechanistic understanding of these NA reactions can foster the design of robust, high-impact tools for diagnostics, therapeutics, and beyond, ultimately advancing the frontier of nanomedicine.
Conjugation reaction of nucleic acids on nanomaterial surface
The conjugation of nucleic acids to nanomaterial surfaces constitutes a pivotal process in numerous biomedical applications, as it significantly influences the stability, dispersibility, and biocompatibility of the resultant complexes. By establishing a reliable NA–nanomaterial interface, researchers can enhance the effectiveness of biosensors, drug delivery vehicles, imaging agents, and other clinical technologies [22]. As summarized in Tables 1 and 2, two broad strategies, i.e., covalent and non-covalent approaches, govern NA conjugation. In the following sections, we focus on covalent conjugation reactions, detailing the chemical bonds and reaction mechanisms that confer strong, durable, and tunable interfaces.
Table 1.
Covalent strategies for nucleic acid Conjugation on the surface of nanomaterials
| Conjugation Strategies | Conjugation method | Principle | Bond Disscociation Energy (KJ/mol) | Reaction conditions | Cost | By-products | application | References |
|---|---|---|---|---|---|---|---|---|
| Covalent conjugation | Amide bonds | Dehydration synthesis between amino and carboxyl groups | 421.7 ± 8.4 |
Room temperature EDC: pH 4.5–5 Sulfo-NHS: pH 5–6(react with EDC) Sulfo-NHS: pH 7.2–7.5(react with amine) |
EDC: 1.22 CNY/mg (Thermo Scientific™) Sulfo-NHS: 8.76 CNY/mg (Thermo Scientific™) |
O-Acyl isourea intermediate/R-COOH |
MNPs, Au@graphene NPs, QDs, UCNPs |
[23–27] |
| Thioether bonds | Sulfuralcohol/mercapto group adds across multiple bonds in unsaturated compounds | 713.3 ± 1.2 |
Room temperature pH 6.5–7.5 |
SMCC: 45.94 CNY/mg (Thermo Scientific™) | Maleic acid/Michael addition reaction: Irreversible C-N bond | UCNPs, SiNPs, Ag@SiO2, | [28–31] | |
| Disulfide bonds | The bond between sulfur atoms in the form of -S–S- formed by the oxidation of two -SH groups | 425.3 |
Room temperature pH 7.0–8.0 |
SPDP: 67.78 CNY/mg (Thermo Scientific™) | Hydrolysis of the NHS ester |
AgNPs, AuNPs, QDs |
[32–34] | |
| Au–S/Ag–S/Pt–S | High affinity of sulfhydryl on the surface of noble metal nanomaterials |
Au–S: 418 ± 5 Ag–S: 216.7 ± 4.6 Pt–S: 55 ± 4.8 |
Depends on the conjugation method | 5'-Thiol Modification of Oligonucleotides: 280 CNY/2OD (Sangon Biotech (Shanghai)) | – | AuNPs, AuNP@Pt, AgNPs | [35–37] | |
| Schiff base | Chemical compounds (imines) bearing a hydrocarbyl group on the nitrogen atom R2C = NR′ (R′ ≠ H) | 602 ± 25 |
Room temperature pH 6.0–8.0 |
Glutaraldehyde: 11.31 CNY/ml (Sigma-Aldrich) 5'-Amine Modification of Oligonucleotides: 105 CNY/2OD (Sangon Biotech (Shanghai)) |
Only single component on glutaraldehyde/Michael addition reaction: Irreversible C-N bond |
Fe3O4@PDA NPs, SiNPs |
[38, 39] | |
| Click Chemistry | A highly specific and selective chemical ligation method based on carbon-heteroatom bond (C-X-C) linkage | 944.8 ± 0.1 |
Room temperature pH 7.0–8.5 |
DBCO-PEG4-NHS ester: 720 CNY/mg (MCE) 5'-Azide Modification of Oligonucleotides: 280 CNY/2OD (Sangon Biotech (Shanghai)) 5'-DBCO Modification of Oligonucleotides: 350 CNY/2OD (Sangon Biotech (Shanghai)) |
– |
AuNPs, UCNPs, SiNPs |
[40–42] |
Comprehensive handbook of chemical bond energies provides the bond energy data in the table [43]
Table 2.
Non-Covalent strategies for nucleic acid Conjugation on the surface of nanomaterials
| Conjugation Strategies | Conjugation method | Principle | Characteristics | Application | References | ||
|---|---|---|---|---|---|---|---|
| Non-Covalent conjugation | Electrostatic attraction | Interaction between opposite charges |
(1) Bond Strength: 20 kJ.mol−1 (2) Electrostatic gravity is not directional and the interaction between anions and cations can be in any direction |
Most positively charged nanomaterials | [44–46] | ||
| Hydrogen Bond, π-π interactions and van der Waals force | An attractive interaction between two molecular moieties in which at least one of them contains a hydrogen atom that plays a fundamental role |
(1) Bond Strength: 12–30 kJ.mol−1 (2) Hydrogen bonding is directional, saturated (3) Hydrogen bonds are different from chemical bonds in that they have a small bond energy and a longer bond length (4) π-π interactions are influenced by pH and redox conditions but maintain the chemical structure and biological activity of conjugated NAs |
Carbon-based nanomaterials (graphene oxide, carbon nitride nanosheet), COFs, MOFs |
[47, 48] | |||
| Hydrophobic interaction | The phenomenon of hydrophobic groups aggregating in close proximity to each other to avoid water |
(1) Bond Strength: < 40 kJ.mol−1 (2) The hydrophobic interaction forces are generally stronger than the classical van der Waals forces and exhibit a large range of effects |
PEGylated liposomes, Lipid Nanoparticles, MNPs |
[49–51] | |||
| Biotin-streptavidin | Streptavidin tetramers have a positive effect on biotin Extremely high binding affinity |
(1) One streptavidin protein is able to bind four biotin molecules with high affinity and selectivity (2) The affinity constant of avidin-binding biotin can be millions of times that of antigen–antibody reaction, and the dissociation constant of the complex formed by the combination of the two is very small, irreversible reactivity, and strong stability (3) Thermal instability (> 75 °C) |
Carbon nanoparticles, AuNPs, magnetic nanobeads, SiNPs | [52–55] | |||
| Base affinity | The unique binding force of polynucleotides to certain nanomaterials |
(1) Oligonucleotides do not require additional modifications (2) It belongs to the non-specific adsorption of DNA and is base-dependent (3) It is convenient to control the density of DNA conjugation |
AuNPs, carbon-based nanomaterials, ZnO NPs | [56–58] | |||
Covalent conjugation reactions provide secure and stable NA-nanomaterial assemblies
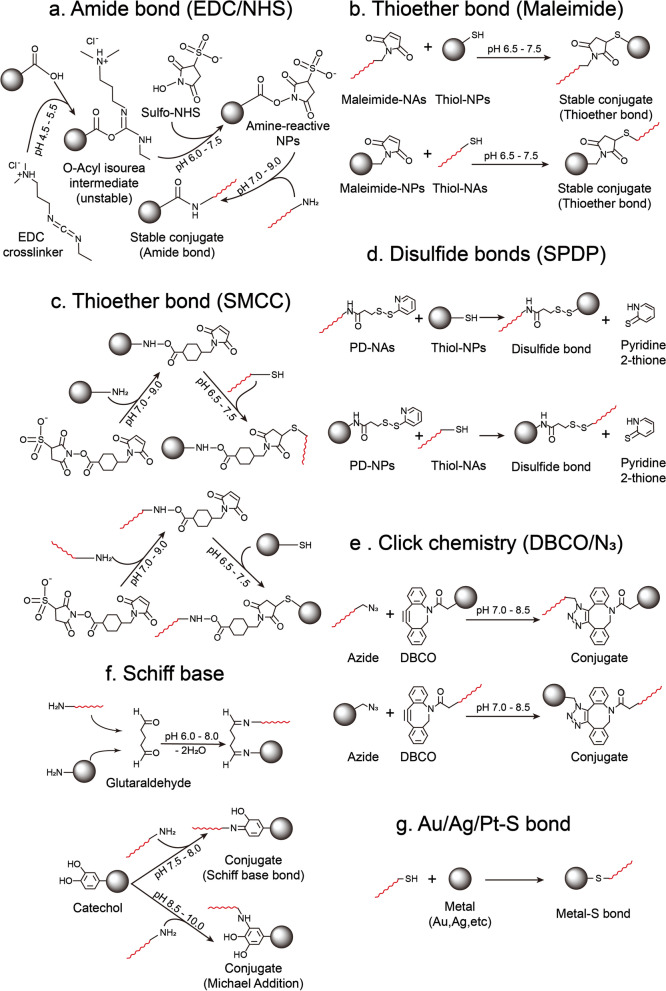
Covalent attachment ensures a robust linkage between NAs and nanomaterial surfaces, safeguarding the functionality of both components in aqueous and physiological environments [59]. This section highlights major covalent chemistries—amide, thioether, disulfide, Au–S bonds, Schiff base formation, and click reactions—that researchers commonly employ to generate stable nanomedicine platforms. Figures 1a and 2 illustrate the diverse covalent bonds that can be harnessed for NA conjugation.
Fig. 1.
Conjugation and detachment reactions of NAs on the surface of nanomaterials. a Covalent conjugation. b Non-covalent conjugation. c Chemical detachment. d Physical detachment. Created by Figdraw
Fig. 2.
Schematic Illustration of Chemical Reactions between Nanomaterials and NAs. a Amide bond (EDC/NHS): Carboxyl-NPs react with crosslinker EDC and sulfo-NHS to generate amine-reactive NPs, then react with amine-NAs to form stable amide bond. b Thioether bond (Maleimide): The formation of thioether bond between maleimide-NAs/NPs and thiol-NPs/NAs. c Thioether bond (SMCC): Amine-NPs/NAs interact with thiol-NAs/NPs to form thioether bond in the existence of SMCC. d Disulfide bonds (SPDP): SPDP active NAs/NPs generate Pyridyldithiol (PD) group, which can interact with thiol-NPs/NAs generate disulfide bond. e Click chemistry (DBCO/N3): Azide-NAs/NPs react with DBCO-NPs/NAs. f Schiff base: Glutaraldehyde react with amine-NPs and amine-NAs generate schiff base. Catechol-NPs react with amine-NAs generate different products in different pH. g Au/Ag/Pt–S bond: Thiol-NAs linked to mental surface through Au/Ag/Pt–S bond
Forming amide bonds: overcoming the challenge of direct condensation
Amide bonds are among the most widely used linkages for creating NA-functionalized nanomaterials. However, direct amide condensation at high temperatures (often above 140 °C) risks denaturing NAs [60]. To circumvent these harsh conditions, researchers typically activate carboxylic acids before coupling them to amines on NAs.
Carbodiimides, particularly water-soluble N-(3-dimethylaminopropyl)-N'-ethylcarbodiimide hydrochloride (EDC·HCl), generate O-acylisourea intermediates that subsequently react with amines to form amide bonds [61, 62]. Adding a nucleophilic additive, such as N-hydroxysuccinimide (NHS), further increases reaction efficiency [63]. This multi-step process is the backbone of EDC/NHS-based functionalization for numerous nanoparticles, including Graphene-encapsulated gold nanoparticles (Au@graphene NPs), Magnetic nanoparticles (MNPs), Quantum dots (QDs), and Upconversion nanoparticles (UCNPs) [23–27]. Conversely, amino-functionalized nanomaterials, such as SiO2-encapsulated magnetic nanoparticles (Fe3O4@SiO2 NPs) or Gold nanoclusters (AuNCs), can be paired with carboxyl-modified oligonucleotides via the same principles [64, 65].
Beyond EDC/NHS coupling, several eco-friendly approaches are emerging. For example, boronic compound-catalyzed dehydration amidation offers low-toxicity conditions, bypassing the need for stoichiometric activating agents [66]. Organosilane chemistry is also promising, with one study employing diphenylsilane and NMPi to directly couple unactivated amines and carboxylic acids in a single step under ambient conditions [67]. Together, these varied amide-bond strategies widen the toolkit for designing stable and biocompatible nanocarriers, biosensors, and diagnostic reagents.
Forming thioether and disulfide bonds: maleimides, crosslinkers, and novel approaches
Sulfhydryl groups on NAs can yield thioether (via Michael-type addition) or disulfide bonds with nanomaterial surfaces. Thioether formation often leverages maleimide-activated nanomaterials, which react specifically with thiols at physiological or near-physiological pH values (6.5–7.5) [28, 68]. For instance, Maleimide (MAL)-treated mSiO2-encapsulated upconversion nanoparticles (UCNP@mSiO2) and Gold nanoparticles (AuNPs) conjugate effectively to thiolated DNA, while MAL-modified oligonucleotides can be bound to thiol-bearing Silica nanoparticles (SiNP) [29, 30, 69].
Disulfide linkages arise from oxidation of free thiols and are generally less stable due to potential thiol–disulfide exchange [70]. Nonetheless, employing cyclic disulfide anchors can confer enhanced stability on silver or gold nanoparticle systems [32–34]. Heterobifunctional crosslinkers such as Sulfo-N-Succinimidyl 4-(Maleimidomethyl)cyclohexane-1-carboxylate, Sodium Salt (Sulfo-SMCC) and 3-(2-pyridyldithio)propionic acid n-hydroxy-succinimide ester (SPDP) greatly expand these functionalities. Sulfo-SMCC combines an NHS ester (for primary amines) with a maleimide group (for thiols), facilitating stable thioether bond formation on a wide range of nanoparticles, including QDs, SiO2-encapsulated silver nanoparticles (Ag@SiO2 NPs), AuNPs, and MNPs [31, 71–75]. SPDP, in contrast, yields reducible disulfide bonds, enabling reversible conjugation but with slightly lower stability at physiological pH [76–79].
Despite their versatility, maleimide-thiol linkages can degrade in thiol-rich or reducing biological environments through undesired side reactions [80]. Contemporary research focuses on stabilizing the maleimide–thiol products or employing next-generation bifunctional reagents [81–84]. While these innovations show promise, classic maleimide chemistries remain widely used, owing to their simplicity and adaptability across most biomedical research scenarios.
Formation of Schiff base: leveraging imine chemistry for versatile coupling
Schiff base formation, involving the nucleophilic addition of amines to carbonyl-containing aldehydes or ketones, introduces another robust route to NA-nanomaterial conjugation [85]. For instance, polydopamine (PDA) nanoparticles or PDA-modified nanoparticles leverage their oxidized catechol (o-quinone) species to react with primary amines on NAs, forming stable Schiff base [86]. Similarly, glutaraldehyde (GA), featuring aldehyde moieties at both ends, can serve as a homobifunctional crosslinker to tether amino-modified Fe3O4 NPs or SiNPs with NA strands via a five-carbon bridge [38, 39, 87–90]. These Schiff base chemistries often proceed under mild conditions and can be reversed or modulated by pH, providing a flexible platform for dynamic biosystems or controlled-release applications.
Click chemistry reactions: bioorthogonal, fast, and efficient coupling
Click chemistry offers a set of reactions characterized by high specificity, insensitivity to water or oxygen, and near-quantitative yields [91]. In nucleic acid conjugation, strain-promoted azide-alkyne cycloadditions (SPAAC) and copper-catalyzed azide-alkyne cycloadditions (CuAAC) stand out. By labeling nanomaterials with azide groups and NAs with alkyne groups—or vice versa—researchers can achieve selective linkage under physiological conditions in under two hours [40–42, 92]. Furthermore, Dibenzoazacyclooctyne (DBCO) enables copper-free reactions, removing potential cytotoxicities associated with Cu(I) [93–97]. Commercially available DBCO-DNA or amino-modified DNA with azide-activated nanomaterials expands the scope to metal–organic frameworks, QDs, or polymeric colloids [98, 99].
Formation of Au–S bonds: streamlined pathways to precious metal nanoparticles
Gold, silver, and platinum nanoparticles can directly bind thiolated NAs through covalent-like Au–S, Ag–S, or Pt–S linkages [35–37]. Taking Au–S bonds as a prototypical example, the standard “salt-aging” method incrementally raises ionic strength to stabilize AuNPs and facilitate DNA attachment over one to two days (Fig. 3a) [100, 101]. To accelerate this process, several approaches have emerged. One strategy employs a low-pH environment (pH 3–4) to suppress electrostatic repulsion and rapidly functionalize AuNPs within minutes (Fig. 3b) [102, 103]. Alternatively, freeze–thaw cycles, “solid solution” dehydration in solvents like butanol, and microwave-assisted heating can condense reagents into microdomains, boosting coupling efficiency and minimizing nanoparticle aggregation (Fig. 3c-e) [104–106].
Fig. 3.
Different strategies for conjugating NAs to gold nanosurfaces by Au–S bonds. a The salt-aging method for the NAs conjugation on citrate-stabilized AuNPs. Reprinted with permission from [100]. b The low-pH method for the conjugation of thiol-modified NAs to AuNPs. Reprinted with permission from [103]. c The freeze method AuNPs for the conjugation of thiolated DNA to AuNPs in a few minutes. Reprinted with permission from [104]. d The dehydrated “solid solution” method for the rapid conjugation of thiolated NAs on AuNPs surface. Reprinted with permission from [105]. e The MW-assisted heating-dry method for the conjugation of thiolated NAs to AuNPs. Reprinted with permission from [106]
In parallel with thiolated DNA, phosphorothioate (PS)-modified DNA also attaches to AuNP surfaces by substituting a sulfur atom into the DNA phosphate backbone [56, 107–109]. Although PS-DNA typically demonstrates a slightly weaker affinity for gold, it offers cost advantages and tunable coupling density by varying the polyA tail length. This flexibility makes PS-DNA an appealing alternative for large-scale or cost-sensitive nanomedicine applications.
In summary, covalent conjugation methods, ranging from amide bonds and thioether linkages to click chemistries and Au–S, form the foundation for stable, high-fidelity NA–nanomaterial interfaces in nanomedicine. These strategies facilitate precise reaction control under mild conditions, critical for preserving nucleic acid integrity and nanomaterial functionality. Nonetheless, specific challenges occur with different nanomaterials, requiring careful consideration.
Hydrophobic-to-Hydrophilic Phase Transition: Nanomaterials like UCNPs, QDs, and MNPs are typically synthesized in organic solvents with hydrophobic surface ligands such as oleic acid and Tri-n-octylphosphine/Trioctylphosphine oxide (TOP/TOPO). To enable nucleic acid conjugation, amphiphilic ligand exchange or surface functionalization is essential to render them water-dispersible. Post-modification functional groups (commonly amino, carboxyl, or thiol groups) dictate the choice of covalent conjugation strategy. Reactions must be conducted under optimized pH, low ionic strength, and controlled temperatures to prevent aggregation and maintain colloidal stability.
Ligand Displacement in Gold Nanoparticle Conjugation: For citrate-stabilized gold nanoparticles, thiolated oligonucleotides replace citrate ligands via stronger Au–S bonds. This process requires a substantial molar excess of thiolated oligonucleotides (typically ≥ 100-fold) to drive ligand displacement. In salt-aging protocols, reactions occur under weakly acidic conditions (pH 3–5) to protonate citrate ligands, thereby weakening their binding to gold surfaces. Subsequent gradual addition of NaCl (final concentration 0.1–0.3 M) screens electrostatic repulsion, facilitating oligonucleotide adsorption onto gold surfaces.
Limitations of Click Chemistry: Click chemistry is unsuitable for conjugating nucleic acids to silver nanoparticles or certain metal oxides (e.g., ZnO, CuO). Reaction conditions such as acidic pH or copper ion presence may trigger redox reactions that degrade nanomaterial functionality.
Challenges with Carbon-Based Materials: The chemical inertness of carbon-based materials (e.g., graphene, carbon nanotubes) necessitates harsh oxidation (e.g., carboxylation) or covalent modification to introduce reactive groups (e.g., amino or carboxyl groups). However, such treatments often compromise structural integrity or electrical properties (e.g., reduced conductivity in graphene), making covalent conjugation strategies generally impractical.
As the field expands, it's vital to enhance eco-friendly, safe, and cost-effective solutions. Scaling these chemistries while preserving bioactivity and ensuring compliance is essential for advancing these technologies to biomedicine.
Non-covalent conjugation reactions provide simple and rapid NA-nanomaterial assemblies
Noncovalent conjugation of NAs to nanomaterials relies on intrinsic molecular interactions such as electrostatic attraction, hydrogen bonding, π-π stacking, and hydrophobic forces. The sequence programmability and structural versatility of DNA further enhance noncovalent binding. These methods are cost-effective and preserve NA functionality without requiring chemical modifications but are less stable than covalent methods, being sensitive to environmental factors such as pH and ionic strength [6]. In this section, we discuss common strategies for noncovalent conjugation and their applications (Fig. 1b).
Electrostatic attraction: surface charge modification
Nanomaterials with charged surfaces enable conjugation by attracting oppositely charged NAs. For example, negatively charged nanomaterials bind to positively charged nucleobases in Single-stranded DNA (ssDNA) but repel the negatively charged phosphate backbones of double-stranded DNA (dsDNA) [110]. Surface charge modification can prevent charge repulsion, with molecules like Polyethylenimine (PEI), Cetyltrimethylammonium bromide (CTAB), and Poly(diallyldimethylammonium chloride) (PDDA) commonly used to shift charges from negative to positive [111, 112]. This strategy is widely applied to functionalize AuNPs, carbon dots, and QDs [44, 45, 113]. Additionally, phosphate groups in NAs can adsorb directly onto positively charged nanomaterials, including Gold nanorods (AuNRs) and nanoclusters [46, 114, 115]. These electrostatic interactions improve biosensor performance and facilitate stimuli-responsive release of NAs.
Hydrogen bonding, π-π stacking, and van der Waals forces: Key interactions in DNA conjugation with nanomaterials
The hydrogen bonds between base pairs in DNA form the structural framework of the DNA double helix and ensure its stability. Additionally, π-π stacking and van der Waals forces between adjacent bases in the same strand further enhance the molecule's stability and compactness [116]. These interactions are essential for DNA aggregation, assembly, and conjugation with nanomaterials. For example, NAs bind directly to carbon-based nanomaterials like Graphene oxide (GO), Graphitic carbon nitride (g-C3N4), and Carbon nanotube (CNTs), as well as to organic frameworks such as Metal–organic frameworks (MOFs) and Covalent organic frameworks (COFs) [47, 48, 117]. Similarly, van der Waals forces enable NA conjugation with transition metal oxides and disulfides, including Molybdenum(IV)sulfide (MoS₂), Tungsten disulfide (WS₂), and Manganese dioxide (MnO₂) [118, 119]. These interactions simplify the design of biosensors and nanocarriers by eliminating the need for complex chemical modifications.
Hydrophobic interactions: hydrophobic lipids and host–guest chemistry
Hydrophobic interactions drive the clustering of hydrophobic groups, facilitating processes such as micelle formation, vesicle and bilayer assembly, and protein folding [120]. These interactions are used to conjugate NAs to extracellular vesicles, liposomes, and micelles via hydrophobic lipids such as 2-Distearoyl-sn-Glycero-3-Phosphoethanolamine (DSPE)-PEG2000, diethyl ester-Polyethylene glycol (PEG), and 1,2-Bis(diphenylphosphino)ethane (DPPE) [121–123]. Hydrophobic head groups also enhance conjugation efficiency. For instance, cholesterol-linked ssDNA can attach to exosomal lipid bilayers to create functional exosomes carrying NAs [49]. Similarly, tocopherol-modified DNA can anchor DNA to liposome surfaces by integrating into the phospholipid layer through hydrophobic interactions [50].
Hydrophobic interactions in host–guest chemistry have emerged as an effective strategy for conjugating NAs to nanomaterials. In this approach, NAs modified with host or guest molecules can bind to complementary guest or host molecules on nanomaterial surfaces. Cyclodextrins (CDs), Calixarenes (CAs), and their respective guest molecules are commonly used in these systems. For example, sulfhydrylated CDs on AuNPs enable non-covalent conjugation with azobenzene-modified NAs acting as guest molecules [124]. Similarly, azo-modified DNA can be conjugated to CD-coated MNPs to create an azoreductase-activated imaging probe with"on/off"functionality [51].
Biotin and streptavidin: high specificity and affinity conjugation
Biotin and streptavidin exhibit exceptional specificity and affinity, forming highly stable complexes [125]. Their rapid binding kinetics and resilience to high pH, temperature fluctuations, and chemical reagents make them widely used in bioconjugation applications. Biotinylated NAs, which are commercially available through nucleic acid synthesis, facilitate straightforward conjugation to streptavidin-functionalized nanomaterials. These include carbon-based, metallic, silicon-based, magnetic, and fluorescent nanomaterials [52–55, 126]. However, a key limitation of the streptavidin–biotin system is the substantial size of the resulting complex. Streptavidin, as a protein, increases the overall dimensions of the construct when biotin is attached to a nanomaterial or biomolecule via spacers (e.g., PEG chains) or cross-linking agents. This added bulk may influence binding kinetics and potentially affect the performance of the final conjugate.
Base affinity: binding patterns of DNA nucleobases with various nanomaterials
DNA can conjugate to nanomaterials through nucleobases, with imine (Metal-N) and ketonic (Metal-O) groups as primary binding sites [127]. The binding affinities of nucleobases vary significantly across different nanomaterials [57]. PolyA induces hydrophobic collapse on the surface of AuNPs, resulting in strong adsorption (Fig. 4a)[128] [129]. The interaction between polyA DNA and AuNPs at high temperatures enables rapid oriented conjugation of non-thiolated DNA (Fig. 4b)[130]. PolyC DNA exhibits stronger affinity for nanocarbons (e.g., GO and single-walled carbon nanotubes), transition-metal disulfides (e.g., WS₂ and MoS₂), and metal oxides (e.g., ZnO and Fe₃O₄ nanoparticles) (Fig. 4c) [58]. On the same nanomaterial, single nucleotides and polynucleotides exhibit different binding patterns. Single nucleotides bind to 5 nm AuNPs in the order dA > dG > dC > dT, while polynucleotides bind in the sequence polyA ≈ polyC ≈ polyT > polyG [131]. The affinity of single nucleotides to GO was dT > rC > rA > rG [132]. The binding affinity of polynucleotides for single-walled carbon nanotubes follows the order polyA > polyG > polyT > polyC, whereas for flat graphite, the affinity order is polyT > polyA > polyG ≈ polyC (Fig. 4d)[133].
Fig. 4.
Base affinity on the surface of different nanomaterials. a Schematic of polyA-DNA adhesion on AuNPs, whose density is dependent on the polyA length. Reprinted with permission from [129]. b Schematic illustrating the thermal drying method for preparing nonthiolated SNAs. DNA with different lengths of the poly-A block had different DNA densities. Reprinted with permission from [130]. c The PolyC-DNA has a much stronger affinity than other DNA homopolymers for nanocarbons, transition-metal dichalcogenides, and metal oxides. Reprinted with permission from [58]. d Pictorial representation of proposed hypotheses to explain the enhanced interaction between ssDNA and SWCNTs, compared to ssDNA with flat graphite. Reprinted with permission from [133]
In summary, non-covalent coupling techniques utilize the inherent properties of NAs and nanomaterials for efficient assembly without chemical modifications, simplifying production. However, the performance of these assemblies varies based on interaction strength, stability, and reversibility, necessitating careful optimization in specific applications.
Weak Interactions and Composite Conjugation Strategies: Non-covalent interactions like hydrogen bonds, π-π stacking, and van der Waals forces are weak, but their stability can be improved with composite conjugation strategies. For example, nucleic acids can be stably attached to graphene oxide surfaces by combining hydrogen bonds, π-π stacking, and base affinity.
Hydrophobic Interactions and Cholesterol-Linked DNA: DNA with two cholesterol linkages aggregates more than single-linked DNA, decreasing conjugation stability. Adding single-stranded DNA overhangs near hydrophobic groups can regulate the hydrophobic interactions of cholesterol-tagged DNA.
Electrostatic Adsorption of DNA on AuNPs: In the DNA and AuNPs electrostatic adsorption system, a NaCl concentration of 0.1–0.3 M reduces charge repulsion and enhances adsorption. Exceeding 0.5 M leads to DNA compaction, decreasing contact area by 30%−40%. Different salts impact adsorption, with monovalent cations like K⁺ being about 1.8 times more effective than Na⁺ due to higher charge density.
Thermal Stability of Streptavidin: Streptavidin loses its biotin-binding ability at high temperatures, with 75 °C causing irreversible inactivation and reduced ssDNA capture. Co-conjugating with PEG can greatly improve its thermal stability and maintain efficient capture of biotinylated ssDNA.
Future development should aim to: (1) create hybrid conjugation systems with multiple interaction modes for improved stability, (2) use computational modeling to design specific binding interfaces, and (3) advance nanomaterial surface engineering for precise control of NA orientation and release. Combining these with stimuli-responsive nanomaterials could lead to advanced smart biosensors and targeted delivery systems with precise spatiotemporal control.
Nucleic acid detachment reaction on the surface of nanomaterials
The detachment of NAs from nanomaterial surfaces is a key focus in numerous studies and applications. Selective detachment and detection of specific DNA or RNA sequences are crucial for diagnosing genetic diseases, identifying pathogens, and monitoring gene expression [134]. In drug delivery, nanomaterials act as carriers for NA-based therapies, enhancing therapeutic payloads while minimizing off-target effects. Controlled release of NAs at target sites improves efficacy and reduces systemic side effects [7]. A thorough understanding of the methods and factors influencing NA detachment is vital for developing advanced nanomaterial-based platforms in biomedical and biotechnological applications. Continued exploration of novel detachment strategies and elucidation of their underlying mechanisms will drive progress in the field, paving the way for innovative applications. This section examines common types of NA detachment reactions on nanomaterial surfaces, highlighting their advantages and limitations. Further details are summarized in Table 3.
Table 3.
Strategies for nucleic acid desorption on the surface of nanomaterials
| Detachment Strategies | Reagents or factors | Mechanism | Application | References |
|---|---|---|---|---|
| Chemical | Urea | Disruption of the hydrogen bonding | Graphene oxide | [135] |
| DMSO | Disruption of hydrophobic interactions | Hydrogel NPs | [136] | |
| Tris–EDTA or Phosphate Buffer | Raise the pH and destroy the high-salt environment | Magnetic NPs | [137] | |
| DTT | Destruction of the Au–S bond or Ag–S bond | AuNPs, AgNPs | [138–140] | |
| KCN | Direct dissolution of nanomaterials | AuNPs | [141] | |
| cDNA or Non-cDNA | Specific hybridization versus non-specific displacement | Graphene oxide, TiO2 NWs, AuNPs, Metal − organic coordination polymers | [142–144] | |
| pH-responsive | Acid-sensitive chemical bonds or protonation of surface groups | AuNPs, inorganic nanoparticles | [145, 146] | |
| UV-responsive | Photolysis of linkers or photoisomerization | AuNPs, UCNPs | [147, 148] | |
| Physical | Heating | Disruption of the Au–S bond | AuNRs, Graphene oxide, AuNPs | [140, 149, 150] |
| Laser | Disruption of the Au–S bond, melting of the dsDNA | gold nanomaterials | [151, 152] |
Chemical methods
Chemical methods for detaching NAs from nanomaterial surfaces typically involve agents that disrupt the interactions between NAs and nanomaterials (Fig. 1c)[135, 138]. For instance, urea or dimethyl sulfoxide (DMSO) can denature NAs by forming hydrogen bonds with nucleotide bases, causing ssDNA to detach from nanomaterials (Fig. 5a)[136, 153]. Magnetic beads modified with silanol or carboxyl groups create"salt"or"electrostatic bridges"with NAs. Tris–EDTA or phosphate buffers disrupt these interactions by lowering the salt concentration or increasing pH, which induces repulsion between the negatively charged NAs and the beads (Fig. 5b)[137, 154, 155]. For metallic nanomaterials, such as AuNPs, chemical destabilizers like dithiothreitol (DTT) can cleave Au–S bonds, effectively detaching conjugated NAs (Fig. 5c)[139]. Additionally, potassium cyanide (KCN) can solubilize metallic nanomaterials directly, facilitating complete release of bound NAs (Fig. 5d)[141].
Fig. 5.
Chemical strategies for the detachment of NAs from the surface of nanomaterials. a Urea and dimethyl sulfoxide inhibit DNA adsorption on the surface of Hydrogel nanoparticle. Reprinted with permission from [136]. b Schematic diagram of MNPs-based NA separation. Reprinted with permission from [154]. c Oligonucleotides were dissociated from the AuNPs surface by using DTT. Reprinted with permission from [139]. d KCN was added to dissolve the AuNPs and fully release the DNA. Reprinted with permission from [141]. e The schematic illustrating the CNPs-based fluorescent nucleic acid detection. Reprinted with permission from [156]. f Detecting ssDNA and dsDNA via fluorescence quenching of fluorophore-labeled DNA probes by AuNPs. Reprinted with permission from [143]. g A few possible mechanisms of cDNA-induced probe DNA detachement from GO surface. Reprinted with permission from [144]. h pH-responsive regulation of the nanoswitch and NAs release. Reprinted with permission from [145]. i A schematic representation depicting the release of DNA from the photocleavable nanoparticles–DNA complex following exposure to UV irradiation. Reprinted with permission from [147]
The use of complementary DNA (cDNA) to detach NAs from nanomaterial surfaces is classified as a chemical method. Since ssDNA and dsDNA have different affinities for nanomaterial surfaces, cDNA-mediated displacement reactions can promote the release of fluorescently labeled NAs [141, 142]. In this process, single-stranded fluorescent probes initially adsorbed onto nanomaterial surfaces are quenched due to their proximity. cDNA, complementary to the single-stranded probes, forms double- or triple-stranded DNA structures away from the surface, restoring fluorescence. Based on this principle, many interesting nanomaterial-based direct nucleic acid detection platforms have been developed (Fig. 5e-f) [143, 156]. Further systematic investigation of cDNA-induced detachment of probe DNA from GO surfaces proposed several mechanisms (Fig. 5g): (1) Langmuir–Hinshelwood model: cDNA adsorbs, diffuses, hybridizes with probe DNA, and desorbs in two phases. (2) Eley–Rideal model: cDNA hybridizes directly with probe DNA without adsorbing. (3) Displacement model: cDNA replaces probe DNA in the solution and hybridizes with another cDNA. (4) Surface Heterogeneity: Probe DNA adsorbed to GO with varying affinities may follow one or both of the above mechanisms [144, 157]. Despite these insights, the mechanisms underlying DNA desorption via strand displacement remain complex and subject to ongoing debate. Experimental evidence suggests that no single model fully accounts for all observed detachment phenomena. Further research is required to quantitatively elucidate these processes and improve our understanding of DNA desorption mechanisms.
Stimuli-responsive NA detachment strategies enable controlled release of nucleic acids from nanomaterials under specific external stimuli like pH and UV light, through designed surface chemistry. In pH-responsive systems (Fig. 5h), DNA release is achieved by using pH-sensitive sequences (e.g., i-motif, triplex) [145, 158, 159], or by breaking acid-labile bonds (e.g., hydrazone, ketal)/protonating surface groups (e.g., amines, carboxyls) on nanomaterials [146, 160, 161]. In UV-responsive systems (Fig. 5i), DNA release relies on photolabile linkers or photoisomerization. For example, DNA linked to positively charged AuNPs via photolabile nitrobenzyl ester bonds is released upon UV irradiation (> 350 nm) [147]. Similarly, Ortho-nitrobenzyl (ONB)-functionalized nucleic acids can partially release from nanomaterials under UV light [148]. Additionally, azobenzene-modified DNA on nanoparticles is released when UV converts azobenzene to its cis form, facilitating its detachment from the cyclodextrin cavity [162, 163].
Physical methods
Physical methods for detaching NAs from nanomaterial surfaces primarily involve altering the surface temperature of the nanomaterial to disrupt NA-nanomaterial interactions (Fig. 1d) [140]. Increased temperature enhances nanomaterial motion, weakening interactions with NAs and promoting detachment. It also facilitates the thermal diffusion of NAs, further aiding their release [149]. Common approaches include direct heating and photothermal conversion. Research on these mechanisms has primarily focused on AuNPs. One study examined the kinetics of DNA detachment from AuNP surfaces at temperatures from 40 to 95 °C [164]. Another study investigated the thermal stability of DNA-AuNP conjugates with various organosulfur anchor groups from 25 to 85 °C, finding that bidentate Au–S bonds with cyclic disulfides were less thermally stable than those with thiol or acyclic disulfides (Fig. 6a) [150].
Fig. 6.
Thermal-induced detachment of NAs from the surface of nanomaterials. a Schematic illustrating the fluorescence-based measurement of the released FAM-labeled DNA from DNA-AuNP at different temperatures. Reprinted with permission from [150]. b Near-IR light-induced DNA release. Reprinted with permission from [151]. c DNA release profiles and corresponding fits of the 25, 35, 55, and 70 nm AuNP. Reprinted with permission from [165]. d The siRNA release efficiency of three gold nanoparticles (HGNS, HGNC, and AuNR) at different laser irradiation powers. Reprinted with permission from [152]
The excellent photothermal conversion efficiency of AuNPs has enabled advanced methods for light-stimulated detachment of NAs. Research has shown that near-infrared (NIR) light can detach DNA from AuNRs by generating'hot electrons'that cleave Au–S bonds [166]. Similarly, AuNRs absorb light, causing localized heating that unwinds and releases DNA without significantly raising the solution's temperature, indicating a non-thermodynamic mechanism [167, 168]. Additionally, the effects of continuous wave (CW) and femtosecond pulse lasers on DNA release from gold nanoparticle complexes were different. CW lasers caused photothermal release of dehybridized ssDNA while the complementary strand stayed bound. Femtosecond pulse lasers generated non-equilibrium hot electrons, breaking Au–S bonds and releasing intact dsDNA (Fig. 6b)[151]. Another study with different conclusions noted that silicon core/Au nanoshells irradiated with near-infrared pulsed laser and continuous laser released single- and double-stranded NAs, but pulsed irradiation resulted in higher dsNA release [169]. The size and shape of nanomaterials significantly affect the release process. Smaller AuNPs release ssDNA more rapidly than larger particles (Fig. 6c) [165]. AuNRs demonstrate more efficient siRNA release than Hollow gold nanoshells (HGNS) or Hollow gold nanocages (HGNC) when subjected to 800 nm pulsed laser excitation (Fig. 6d)[152]. In summary, light-stimulated NA detachment using the plasmonic properties of metal nanoparticles offers precise control over release by tuning the wavelength, intensity, and irradiation mode of light. Additionally, the inherent characteristics of nanomaterials and the type of NA conjugation significantly influence detachment outcomes [170].
Detaching NAs from nanomaterial surfaces is complex, influenced by factors like nanomaterial size, shape, surface features, and environmental conditions. Advances in light-responsive and strand displacement nanomaterials enable precise NAs release. Hybrid structures and environmental triggers also enhance responsiveness to biological or external stimuli, boosting nanomaterial applications in biomedicine. Ongoing research aims to create more efficient, targeted, and biocompatible systems for gene therapy and biosensing.
Nucleic acid signal amplification reaction on the Nanomaterials Surface
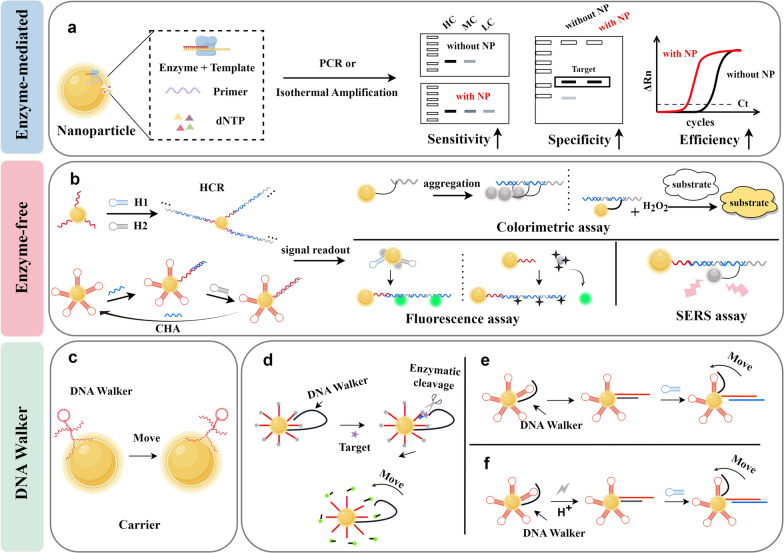
Signal amplification on nanomaterial surfaces holds significant potential for achieving high sensitivity and selectivity in the in situ detection of NAs, thanks to its rapid analytical process and ease of miniaturization [171]. In this section, we divide the NA signal amplification reactions that occur on the surface of nanomaterials into enzyme-mediated signal amplification (nucleic acid amplification) and enzyme-free signal amplification (HCR and CHA), as well as DNA Walker that enables signal amplification in both enzymatic and enzyme-free systems (Fig. 7).
Fig. 7.
Nucleic acid signal amplification reaction on the surface of nanomaterials. a Enzyme-mediated amplification of nucleic acid signals on the surface of nanomaterials. The addition of nanomaterials to the nucleic acid amplification system can improve the sensitivity, specificity, and efficiency b Enzyme-free signal amplification of nanomaterials. Reaction schematics of HCR and CHA on the surface of nanomaterials and their signal readouts include colorimetric, fluorescence, and Raman spectroscopy techniques. c Nanomaterials as carriers provide stability to DNA Walker. DNA Walker can be divided into enzymatic reaction-driven DNA Walker (d), strand displacement reaction-driven DNA Walker (e), and stimulus response-driven DNA Walker (f). Created by Figdraw
Enzyme-mediated nucleic acid amplification reaction
Nucleic acid amplification reactions (NAAR) are enzyme-mediated molecular biotechnologies used to replicate NAs within specific systems [172]. Nanomaterials have been integrated into NAAR development since its early stages, with nanomaterial-assisted PCR (nano-PCR) demonstrating enhanced amplification sensitivity, specificity, and efficiency (Fig. 7a)[173, 174]. Using AuNPs as a representative case, the introduction of optimal AuNP concentrations into NAAR systems induces substantial interfacial interactions between nanomaterial surfaces and key components, including DNA polymerases, double-stranded templates, single-stranded primers, salt ion buffers, and magnesium ions [175]. Lou et al. systematically investigated the interactions between AuNPs and PCR components, revealing dynamic adsorption–desorption processes of these components on AuNP surfaces during thermal cycling. The mechanism is threefold: (1) AuNPs non-specifically adsorb polymerase, binding tightly at lower temperatures to suppress nonspecific amplification; (2) AuNPs facilitate dsDNA product dissociation during denaturation; (3) AuNPs preferentially adsorb ssDNA primers over dsDNA, reducing primer-template mismatch through competitive adsorption [176]. Sedighi et al. demonstrated that AuNPs preferentially bind ssDNA in Helicase-dependent amplification (HDA), enhancing DNA unwinding and subsequently improving amplification speed and sensitivity [177]. Our previous studies corroborate these findings, revealing that AuNPs establish energy barriers to optimize nucleic acid interactions, thereby boosting amplification sensitivity [178]. Although physicochemical properties vary among nanomaterials, their common characteristic of high surface-to-volume ratios universally enhances nucleic acid molecular interactions. Table 4 summarizes the influence of various nanomaterials on NAAR, categorized by temperature requirements into thermal cycling and isothermal amplification [177, 179–193].
Table 4.
Nucleic acid amplification reaction on the surface of nanomaterials
| Reaction type | Methods | Nanomaterial | Effect | Ref |
|---|---|---|---|---|
| Thermal cycling | PCR |
Metal nanomaterials (AuNPs, AgNPs, PtNPs) Carbon-based nanomaterials (SWCNTs, GO, MWCNTs, GNFs, CNPs) Oxide nanomaterials (TiO2, Fe3O4, ZnO, SiO2, MgO) Fluorescent nanoparticles (QDs, UCNPs) |
(1) 10–104 times more sensitive (2) Improved the specificity (3) Enhanced the efficiency (Increased the yield) |
[180, 183, 184] |
| qPCR | AuNPs, QDs | Improved the specificity and sensitivity | [185, 186] | |
| Two-round PCR | AuNPs, MWCNTs | Improved specificity and efficiency | [174, 187] | |
| Multiplex PCR | QDs, AuNPs | Improved the specificity | [189, 191] | |
| Long PCR | CNTs | Significantly improved the amplification efficiency | [190] | |
| AS-PCR | AuNPs | AuNPs can selectively inhibit the amplification of mismatched primer–template pairs and enhance the specificity of AS-PCR | [191] | |
| Isothermal | LAMP | AuNPs | False positive decreased from 76 to 0% | [179] |
| RPA | TiO2 NPs | Shortened the reaction time and the nonspecific amplification of the RPA reaction with TiO2 nanoparticles was reduced by 39 − 87% | [192] | |
| SDA | AuNPs | The sensitivity of the detection of telomerase activity in complex samples is improved five-fold compared with the traditional assay | [175] | |
| SDA | GNFs | tenfold enhancement in the PCR yield | [193] | |
| HDA | AuNPs | Improved sensitivity and specificity | [177] |
In addition, the rational design of nanomaterial-mediated nucleic acid (NA) amplification systems is essential for practical implementation. For instance, excessive gold nanoparticle (AuNP) concentrations (> 1 nM) suppress NA amplification, whereas optimal concentrations (0.4 nM) significantly enhance amplification efficiency [174]. Larger AuNPs are more inhibitory to PCR than smaller ones at the same concentration [194]. Primers containing palindrome sequences (GGATCC or ACCGGT), when combined with 60 nm AuNPs, demonstrate remarkable enhancement in quantitative real-time PCR amplification performance [195]. Polyvinylpyrrolidone (PVP)-modified and methoxy polyethylene glycol thiol (mPEG-SH)-modified AuNPs exhibit no interference with PCR, whereas PDDA- and CTAB-functionalized particles may demonstrate inhibitory effects on the reaction [196]. Ongoing research focused on optimizing nanomaterial physicochemical properties and surface functionalization strategies will drive technological innovations, thereby providing more efficient and precise solutions for nucleic acid amplification and detection.
Enzyme-free signal amplification on the surface of nanomaterials
Unlike enzyme-mediated NAAR on nanomaterials, Hybridization Chain Reaction (HCR) and Catalytic Hairpin Assembly (CHA) represent enzyme-free amplification systems triggered by short DNA strands [197]. This process initiates a cascade reaction that unfolds hairpin probes, ultimately forming dsDNA polymers. The fundamental distinction between CHA and HCR lies in their reaction mechanisms: In CHA, the target molecule is regenerated into the system after initiating cascade amplification to act as a"catalyst"for subsequent reaction cycles, rather than being incorporated into the final product. Owing to their unique optical properties and high specific surface area, nanomaterials not only transduce molecular signals from HCR/CHA into detectable physical signals, but also enable the loading of functional biomolecules, thereby significantly enhancing detection sensitivity [198]. Figure 7b classifies nanomaterial-based HCR/CHA systems according to optical detection principles, including colorimetric, fluorescent, and surface-enhanced Raman spectroscopic methods.
Colorimetric detection in HCR/CHA systems can be divided into two methods. The first involves attaching DNA initiators to nanomaterials, triggering HCR/CHA and causing nanomaterial aggregation, which changes color visibly [199]. For instance, when target DNA opens a hairpin, it hybridizes with a complementary sequence on AuNPs, causing aggregation and a color shift [200]. This method was used for detecting miRNA-21, ATP, thrombin, and acetylcholinesterase activity [201, 202]. The second method uses DNA polymers from HCR/CHA as conjugates with nanomaterials that mimic peroxidase activity, catalyzing substrates like 3,3',5,5'-Tetramethylbenzidine (TMB) or 2, 2'-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) with H₂O₂ to produce a color change [203, 204].
Fluorescence detection in HCR/CHA systems mainly follows two approaches. The first involves using nanomaterials to quench fluorescence by adsorbing labeled hairpins. When a target molecule triggers HCR/CHA, the hairpins open, restoring fluorescence [205, 206]. For instance, Miao et al. used a GO/Au nanocluster system to detect Aflatoxin B1 (AFB1), where AFB1 weakened hairpin adsorption on graphene oxide, allowing fluorescence recovery for detection [207]. Similarly, Li et al. detected microRNAs (miRNAs) by fluorescence generated by a local CHA on AuNPs [208]. The second approach uses nanomaterials to alter fluorescence by interacting with HCR/CHA products [209]. He et al. developed a method to detect the tumor biomarker transmembrane glycoprotein mucin 1(MUC1) using HCR with luminescent ruthenium (II) complexes and CdZnTeS quantum dots. MUC1 initiated HCR to form long dsDNA attached to MNPs. Ruthenium (II) complexes embedded in the dsDNA were magnetically separated, leaving minimal complexes in the supernatant and causing negligible fluorescence quenching of the QDs. Significant quenching occurred when complexes remained [210]. Similarly, Zhang et al. developed a miRNA-10b detection sensor based on target-triggered CHA signal amplification and Luminescence resonance energy transfer (LRET) between UCNPs and AuNPs [211].
In SERS detection, nanomaterials like AuNPs and Silver nanoparticles (AgNPs) are linked with organic dyes through biotin-avidin interactions or nucleic acid hybridization to serve as Raman signal probes for quantifying target substances [212, 213]. Systems using nanoparticles with HCR/CHA achieve high sensitivity, detecting as low as femtomolar levels and even single cells. For instance, a SERS biosensor combining gold Nanowires (AuNWs), silver staining, and HCR was developed for ultra-sensitive miRNA detection, achieving a detection limit of 0.03 fM [214]. A CHA and SERS-based dual signal amplification method was developed for the simultaneous detection of miRNA-21 and miRNA-155 with detection limits of 77 aM and 93 aM, respectively [215].
DNA walker on the surface of nanomaterials
DNA walkers are nanomachines made of DNA that move autonomously along a DNA track using a driving force, a walking chain, and a track. Activation of the driving force disrupts equilibrium, converting energy into motion and propelling the walker. Consuming fuel molecules restores equilibrium and generates a signal, which can be amplified through repeated cycles [216, 217]. This unique movement and signal amplification make DNA walkers valuable for target detection and biological analysis, often enhanced by nanomaterials like AuNPs and MNPs that stabilize and facilitate their movement (Fig. 7c) [218–221]. DNA walkers can be categorized by their driving force into enzymatic and non-enzymatic reactions.
Enzymatic reactions harness restriction endonucleases, exonucleases, and DNAzymes to act on the DNA phosphate backbone, using the energy from covalent bond cleavage to drive DNA walker movement [222]. As shown in Fig. 7d, the process includes: (1) DNA walker attachment to the track via complementary pairing; (2) enzyme recognition of a specific site on the DNA walker; (3) enzyme-mediated cutting or extension of the DNA chain to release the walker; and (4) walker movement to an adjacent site based on enzyme action direction. These reactions offer high catalytic efficiency and specificity, facilitating rapid and precise DNA walker movement. For instance, Cheng et al. developed a Flap endonuclease 1 (FEN 1)-powered DNA walker for mutant DNA detection. The target DNA binds to the DNA track on the AuNP surface, enabling FEN 1 to cleave the overlapping DNA track chain, generating a fluorescent signal and enhancing signal amplification [223].
Non-enzymatic reactions include strand displacement reactions (Fig. 7e) and environmentally responsive reactions (Fig. 7f). Strand displacement reactions are based on the principle of strict complementary pairing in DNA double strands, enabling DNA strand displacement and walker movement through hybridization and de-hybridization between the walking strand and the substrate or fuel strand [224]. Jiang et al. designed an enzyme-free three-dimensional DNA walker that uses hairpin catalytic assembly to facilitate its movement [225]. Environmentally responsive reactions are driven by chemical agents (e.g., H+/OH−, Hg2+/cysteine) or light stimuli, which interact with the DNA walker or track to propel its movement [226]. Xian et al. developed a NIR light-regulated DNA walker system that uses UCNPs to convert NIR light into ultraviolet light, initiating DNA walker movement for precise biological imaging [227].
In conclusion, Enzymatic reaction-based DNA Walker system utilize a"biocatalytic drive"to prioritize efficiency over stability, whereas enzyme-free systems employ a"structural programming drive"to prioritize controllability over speed. The selection of different systems should be tailored to specific application scenarios. For instance, enzymatic reaction systems are typically preferred for in vitro diagnostics, while enzyme-free systems are more suitable for in vivo applications.
Application of nucleic acid reactions on the nanomaterials surface for biomedicine
The seamless integration of NAs with nanomaterials has unlocked numerous applications in the biomedical field, attributed to their exceptional biocompatibility and stability. In this section, we explore several key application areas arising from the interactions of NAs on nanomaterial surfaces and highlight representative examples.
NAs-guided nanomaterial synthesis and assembly
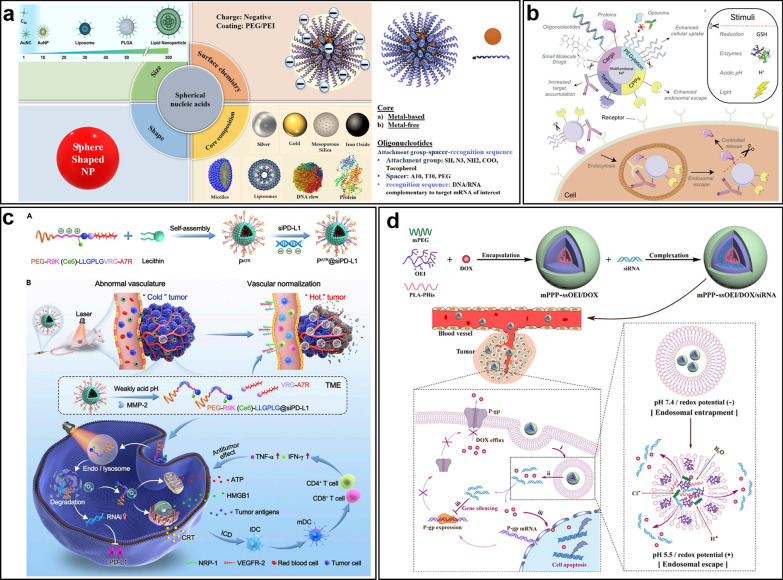
Nucleic acid-based conjugation with nanomaterials has greatly advanced precision assembly of nanomaterials. NAs act as templates for controlled synthesis, allowing precise regulation of nanoparticle size and shape, and imparting unique properties. Thiol-modified ssDNA or poly A/C/G/T chains have been used to shape metal nanocrystals. Specifically, ssDNA with 30 cytosine (C30) or adenine (A30) units turned spherical AuNP seeds into stable, spiky nanoflowers, while thymine-based DNA (T30) formed spherical nanoparticles (Fig. 8a)[228]. Similarly, Different DNA sequences affect the shape and fluorescence of AgNPs made from nanoseeds. Poly-oligo-A10 and -T10 convert nanocubic seeds into stellate octahedral AgNPs with different truncations. Poly-oligo-C10 produces truncated tetrahedral AgNPs, while oligo-G10 maintains the cubic shape and size of the AgNPs (Fig. 8b)[229].
Fig. 8.
NAs-guided nanomaterial synthesis and assemble. a The effects of different DNA molecules of the same length on the morphology of gold nanoparticles during synthesis. Reprinted with permission from [228]. b The effects of different DNA sequences on the morphologies of AgNPs grown from Ag nanocube seeds. Reprinted with permission from [229]. c Parallel CNT arrays were constructed by arranging DNA-coated CNTs via DNA hybridization using DNA brick crystal-based nanogrooves. Reprinted with permission from [230]. d Schematic illustration of colloidal crystals assembled from cube NCs functionalized with different lengths of DNA. Reprinted with permission from [231]
Beyond synthesis, NAs serve as"glue"for organizing nanoparticles, allowing imperfect shapes to form ordered structures. Flexible DNA enhances assembly, enabling complex crystals with improved symmetry. For instance, ssDNA can wrap CNTs and integrate them with DNA origami via hybridization, resulting in parallel CNT arrays with uniform spacing of 10.4 nm (Fig. 8c)[230]. Mirkin and his team have made significant contributions in this field [232–234]. They developed space-filling nanocrystals by pairing DNA-guided, shape-complementary polyhedra, expanding the possibilities for designing metamaterials (Fig. 8d)[231].
Nanoplatforms of nucleic acid therapeutics
The conjugation and detachment of nucleic acids on nanomaterials are crucial processes in nucleic acid therapeutics. Nucleic acid drugs, including DNA, miRNA, and siRNA, must enter cells to function effectively. However, their large molecular size and negative charge lead to repulsion against the cell membrane's lipid bilayer, hindering their entry into cells [235]. Spherical nucleic acids (SNAs), synthesized by conjugating oligonucleotides to nanoparticles, represent a promising platform for nucleic acid delivery, attributed to their distinctive three-dimensional architecture [236, 237]. SNAs are composed of densely functionalized and highly oriented nucleic acids on the surface of a nanoparticle. Since the properties of SNAs originate from their nucleic acid shell, diverse core materials can be employed, including metal nanoparticles (e.g., Au, Pt), liposomes, and polymers (Fig. 9a)[238]. SNAs exhibit low immunogenicity, enable reagent-free transfection, and possess the capability to cross biological barriers, such as the blood–brain barrier [238]. Covalent coupling stands as an effective strategy for immobilizing nucleic acids on the surface of nanomaterials, such as conjugating nucleic acids to gold nanoparticles via thiol groups [239]. However, given the negative charge of siRNA, non-covalent electrostatic interactions are frequently employed as an alternative. For instance, nitrogen-doped graphene quantum dots can directly bind to siRNA [240]. Typically, nanomaterial surfaces are modified with molecules like PEI and PDDA to alter their surface charge from negative to positive [241, 242]. Nevertheless, these modifications may compromise biocompatibility, thereby necessitating the development of improved strategies. For instance, tyrosine-modified PEI and PEGylated PEI coatings can enhance both efficiency and biocompatibility [243, 244].
Fig. 9.
Nanoplatforms of nucleic acid therapeutics. a Schematic display of Spherical Nucleic Acid (SNA) nanoconjugate. Reprinted with permission from [238]. b External and internal stimuli for controlled release of indicated functional groups. Reprinted with permission from [245]. c Illustration of MMP-2-responsive, peptide-assembled micelleplexes for enhanced photoimmunotherapy. Reprinted with permission from [246]. Schematic illustration of the pH/redox dual-responsive codelivery polyplex with effective endo-lysosomal escape. d Reprinted with permission from [247]
Once stable SNAs have successfully translocated across the target cell membrane, the subsequent detachment of nucleic acids from the nanomaterial surface becomes a critical factor for achieving effective nucleic acid therapy. Commonly employed nucleic acid detachment mechanisms encompass reduction, enzymatic cleavage, pH sensitivity, and photo-stimulation (Fig. 9b)[245]. The choice of an appropriate release strategy depends on the stimulus-responsive design of the delivery system. For instance, disulfide bonds enable reduction-sensitive nanocarriers to release nucleic acid drugs in response to cellular reductants [248]. Polycationic micellar nucleic acid carriers, which rely on electrostatic interactions and are coated with thiolated hyaluronic acid, facilitate the delivery of plasmids in the presence of hyaluronidase-mediated degradation [249]. Imine bonds and pH-sensitive coatings introduced into PEGylated liposomes enable targeted delivery of chemotherapeutic and gene therapy agents to tumors [250]. pH-sensitive nanomaterials, including metal–organic frameworks, quantum dots, and micelles, are capable of directly releasing gene drugs [251]. The exceptional photothermal properties of AuNPs facilitate the light-induced release of nucleic acids, thereby promoting significant restoration of target gene expression [147]. Moreover, the system design permits the integrated control of siRNA release through multiple mechanisms. Yi et al. developed a Matrix metalloproteinase-2 (MMP-2)-responsive immunotherapy that releases siRNA via reduction and enzymatic cleavage-stimulation (Fig. 9c)[246]. Gao et al. created a pH/reduction-responsive polycation for delivering Multidrug resistance gene 1 (MDR1) siRNA and doxorubicin to combat multidrug resistance (Fig. 9d)[247]. While multi-stimuli-responsive nanocarriers are promising, it's unclear if their benefits outweigh the associated complexity. Through comprehensive investigation of nucleic acid conjugation and detachment reactions on nanomaterial surfaces, it is anticipated that more efficient and intelligent stimulus-responsive nanoparticles will soon demonstrate their clinical utility.
Biosensor
NA reactions on nanomaterial surfaces, including conjugation, detachment, and signal amplification, have revolutionized biosensors by improving detection sensitivity, specificity, speed, and functionality. These advancements overcome traditional biosensor limitations, enabling trace detection, dynamic monitoring, and adaptability to complex samples, thereby advancing biosensing technology from lab research to clinical diagnosis.
NA conjugation reactions form the foundation for constructing robust and highly specific biosensors. NA probes are securely anchored to the surface of nanomaterials through covalent and non-covalent coupling strategies. This anchoring prevents probe detachment during detection and enhances biosensor stability. The high specific surface area of nanomaterials significantly increases probe loading density, thereby ensuring efficient target molecule capture by each detection unit and enhancing the biosensor's directional identification capability. Moreover, the surface modification of nanomaterials participating in the conjugation reaction, such as PEG and Polyacrylic acid (PAA), reduces non-specific adsorption in complex biological samples, thereby enhancing the detection accuracy of biosensors [252]. NA detachment reactions are essential for developing dynamic biosensors. By utilizing competitive and stimulus-responsive detachment processes, targets can dynamically modulate the binding and dissociation equilibrium between nucleic acids and nanomaterials. This modulation can induce rapid alterations in the optical and electrical properties of nanomaterials. For instance, when a target binds more strongly to the nanomaterial's surface nucleic acid than to the nanomaterial itself, the nucleic acid detaches, rapidly changing the solution's color (Fig. 10a)[253]. This dynamic reversibility also enables sensor reuse, significantly reducing inspection costs [254]. NA signal amplification reactions are pivotal in transcending the detection sensitivity thresholds of biosensors. Leveraging enzymatic amplification techniques (e.g., PCR and isothermal nucleic acid amplification), enzyme-free self-assembly methods (e.g., HCR and CHA), and the synergistic enhancement provided by nanomaterials within cascade amplification effects, a singular target binding event can be converted into a detectably amplified signal of exponential magnitude [255]. The combined efforts of the three have accelerated the development of many cutting-edge biosensors in the field of biomedicine. For instance, surface coupling of magnetic nanoparticles with capture nucleic acids for target enrichment enables the construction of electrochemical biosensors for miRNA detection following magnetic dissociation or signal amplification (Fig. 10b)[256]. Quenched fluorescent probes non-covalently adsorbed on graphene surfaces can be restored to fluorescence by the target's rolling circle amplification products, enabling single-cell imaging (Fig. 10c)[257]. AgNPs coated with biotin-streptavidin coupled single guide RNA (sgRNA) can trigger the CRISPR/dCas9 signal amplification system, constructing a surface-enhanced Raman scattering biosensor for gene mutation detection (Fig. 10d)[258]. Similar strategies have also been employed to construct biosensors for detecting bacterial pathogens and cancer biomarkers [259, 260]. As materials science and molecular biology become increasingly integrated, biosensor development is expected to accelerate toward enhanced portability, intelligence, and multifunctionality.
Fig. 10.
Nucleic acid reactions on the surface of nanomaterials facilitate the development of biosensors. a Schematic diagram of the application of nucleic acid desorption reaction on the surface of AuNPs in the colorimetric detection of bacteria. Reprinted with permission from [253]. b Illustration of electrochemical biosensor based on the NA dissociation reaction and signal amplification reaction on MNP surface. Reprinted with permission from [256]. c Schematic representation of biosensor for single-cell imaging based on the integration of RCA with the quenching function of GO. Reprinted with permission from [257]. d Schematic representation of a double-clamp structure constructed using the CRISPR/dCas9 system in combination with a SERS nanotag to achieve specific recognition of KRAS gene mutations. Reprinted with permission from [258]
Challenges and perspectives
Despite extensive research into nucleic acid interactions with nanomaterial surfaces, significant challenges remain in the field. Variations in the size, shape, and surface properties of nanomaterials introduce heterogeneity, complicating the assurance of reaction specificity and reproducibility. Structural alterations of NAs on nanomaterial surfaces—such as folding, aggregation, or fragmentation—can impair target binding and reduce reaction efficiency. Optimizing coupling and dissociation reactions requires precise control of environmental factors, including pH, temperature, and ionic strength.
Biocompatibility is another critical concern, as nanomaterial-induced toxicity or adverse biological effects must be minimized. While current research often emphasizes the functional capabilities of NAs post-coupling with nanomaterials, less attention is given to the stability of these conjugates during storage and application. Additionally, the reactions of NAs on nanomaterial surfaces generate complex datasets, encompassing thermodynamic and kinetic parameters. This complexity demands multidimensional analysis and diverse analytical techniques, necessitating interdisciplinary expertise and effective communication among researchers.
Fortunately, advances in machine learning are poised to transform the study of nucleic acid reactions on nanomaterial surfaces. By analyzing extensive experimental datasets, machine learning can uncover patterns and underlying principles governing these reactions, aiding researchers in elucidating their mechanisms. Furthermore, machine learning models can predict the outcomes of nucleic acid reactions under specific conditions, reducing reliance on trial-and-error experimentation. Currently, machine learning is making substantial strides in understanding nanomaterial-protein interactions, generating innovative theories and technologies. It is only a matter of time before these insights are applied to nucleic acid research, driving further advancements in the field.
Conclusions
In summary, this review explores the prevailing methodologies for conjugating NAs to nanomaterial surfaces and their subsequent detachment in response to external stimuli. It also provides an in-depth analysis of nucleic acid signal amplification mechanisms on nanomaterial surfaces, including enzyme-mediated signal amplification, enzyme-free signal amplification, and DNA Walker. The specific recognition properties of NAs, when synergistically integrated with the versatile and robust signal transduction capabilities of nanomaterials, make surface-functionalized nanomaterials highly suitable for both in vitro and in vivo nanomedicine applications.
NAs conjugation reactions on nanomaterial surfaces can be broadly classified into covalent and non-covalent interactions. Covalent conjugation relies on pre-modified NAs or nanomaterials linked via chemical cross-linking agents, forming stable complexes ideal for long-term storage and in vivo delivery. Non-covalent conjugation, by contrast, uses weaker intermolecular forces—such as hydrogen bonding, van der Waals forces, and electrostatic interactions—offering enhanced biocompatibility and safety, though with reduced stability. NAs detachment from nanomaterial surfaces can be achieved through physical or chemical methods. Physical methods, such as heating and laser techniques, desorb NAs by altering the surface energy state or nanomaterial properties. Chemical methods employ replacement chains or specific chemicals to modulate adsorption equilibrium, facilitating the detachment process. In amplification reactions, particular emphasis is placed on the adsorption of amplification components onto nanomaterial surfaces. This creates a concentrated microenvironment that enhances specificity, sensitivity, and efficiency. In HCR/CHA, nanomaterials primarily act as signal amplifiers, categorized into colorimetric, fluorescence, and SERS detection methods. In DNA walkers, nanomaterials serve critical roles as carriers, signal amplifiers, and facilitators of imaging processes. These applications introduce innovative approaches for biomedicine.
Collectively, these NAs reactions on nanomaterial surfaces represent a multidisciplinary research field bridging materials science, chemistry, and biology. Detailed study of these interactions unveils new insights into the complex behaviors of NAs on nanomaterials, offering a theoretical foundation and technical support for advancing bio-detection technologies, medical diagnostics, and therapeutic treatments.
Abbreviations
- ABTS
2, 2'-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)
- AFB1
Aflatoxin B1
- Ag@SiO2 NPs
SiO2-encapsulated silver nanoparticles
- AgNPs
Silver nanoparticles
- AS-PCR
Allele specific PCR
- Au@graphene
Graphene-encapsulated gold nanoparticles
- AuNCs
Gold nanoclusters
- AuNPs
Gold nanoparticles
- AuNRs
Gold nanorods
- AuNWs
Gold Nanowires
- CAs
Calixarenes
- cDNA
Complementary DNA
- CDs
Cyclodextrins
- CHA
Catalytic hairpin assembly
- CNPs
Carbon nanoparticles
- CNTs
Carbon nanotube
- CNY
Chinese Yuan
- COFs
Covalent organic frameworks
- CTAB
Cetyltrimethylammonium bromide
- CuAAC
Copper-catalyzed azide-alkyne cycloadditions
- CW
Continuous wave
- DBCO
Dibenzoazacyclooctyne
- DMSO
Dimethyl sulfoxide
- DPPE
1,2-Bis(diphenylphosphino)ethane
- dsDNA
Double-stranded DNA
- DSPE
2-Distearoyl-sn-glycero-3-phosphoethanolamine
- DTT
Dithiothreitol
- EDC·HCl
N-(3-dimethylaminopropyl)-N'-ethylcarbodiimide hydrochloride
- Fe3O4@PDA
PDA-encapsulated magnetic nanoparticles
- Fe3O4@SiO2 NPs
SiO2-encapsulated magnetic nanoparticles
- FEN 1
Flap endonuclease 1
- GA
Glutaraldehyde
- g-C3N4
Graphitic carbon nitride
- GNFs
Graphene nano flakes
- GO
Graphene oxide
- HCR
Hybridization chain reaction
- HDA
Helicase-dependent amplification
- HGNC
Hollow gold nanocages
- HGNS
Hollow gold nanoshells
- KCN
Potassium cyanide
- LAMP
Loop-mediated isothermal amplification
- LRET
Luminescence resonance energy transfer
- MAL
Maleimide
- MDR1
Multidrug resistance gene 1
- MMP-2
Matrix metalloproteinase-2
- MnO₂
Manganese dioxide
- MNPs
Magnetic nanoparticles
- MOFs
Metal–organic frameworks
- MoS₂
Molybdenum(IV)sulfide
- MUC1
The transmembrane glycoprotein mucin 1
- MWCNTs
Multi-walled carbon nanotubes
- NHS
N-hydroxysuccinimide
- NIR
Near-infrared
- NMPi
N-methylpyrrolidinone
- ONB
Ortho-nitrobenzyl
- PAA
Polyacrylic acid
- PCR
Polymerase chain reaction
- PD
Pyridyldithiol
- PDA
Polydopamine
- PDDA
Poly(diallyldimethylammonium chloride)
- PEG
Polyethylene glycol
- PEI
Polyethylenimine
- PtNPs
Platinum nanoparticles
- PVP
Polyvinylpyrrolidone
- QDs
Quantum dots
- qPCR
Quantitative real-time PCR
- RPA
Recombinase polymerase amplification
- SDA
Strand displacement amplification
- sgRNA
Single guide RNA
- SiNP
Silica nanoparticles
- SNAs
Spherical nucleic acids
- SPAAC
Strain-promoted azide-alkyne cycloadditions
- SPDP
3-(2-Pyridyldithio)propionic acid n-hydroxy-succinimide ester
- ssDNA
Single-stranded DNA
- Sulfo-SMCC
Sulfo-N-succinimidyl 4-(maleimidomethyl)cyclohexane-1-carboxylate, Sodium Salt
- SWCNTs
Single-walled carbon nanotubes
- TiO2 NPs
Titanium dioxide nanoparticles
- TiO2 NWs
TiO2 nanowires
- TMB
3,3',5,5'-Tetramethylbenzidine
- TOP/TOPO
Tri-n-octylphosphine/trioctylphosphine oxide
- UCNPs
Upconversion nanoparticles
- WS₂
Tungsten disulfide
Author contributions
Zhenrui Xue: Writing-original draft, Investigation, Data curation, Visualization, Conceptualization. Lu Wang: Investigation, Data curation, Visualization. Shengnan Pan: Investigation, Data curation. Jie Yan: Investigation, Data curation. Minli You: Writing-review & editing, Supervision, Investigation, Funding acquisition, Conceptualization. Chunyan Yao: Writing-review & editing, Supervision, Investigation, Funding acquisition, Formal analysis, Conceptualization.
Funding
This work was supported by the National Natural Science Foundation of China (22104119, 82470235) and the National Natural Science Foundation Key International Cooperative Research Project (W2411086). Chongqing Municipal Science and Health Joint Medical Research Project (2023MSXM100).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors in the paper agree to be published.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhenrui Xue is listed first as the first author, and Lu Wang is listed second as the co-first author.
Contributor Information
Minli You, Email: youminli@xjtu.edu.cn.
Chunyan Yao, Email: yaochunyan@tmmu.edu.cn.
References
- 1.Banoub JH, Newton RP, Esmans E, Ewing DF, Mackenzie G. Recent developments in mass spectrometry for the characterization of nucleosides, nucleotides, oligonucleotides, and nucleic acids. Chem Rev. 2005;105:1869–916. [DOI] [PubMed] [Google Scholar]
- 2.Gao F, Shen XZ, Jiang F, Wu Y, Han C. DNA-guided genome editing using the Natronobacterium gregoryi Argonaute. Nat Biotechnol. 2016;34:768–73. [DOI] [PubMed] [Google Scholar]
- 3.Yang K, Mitchell NM, Banerjee S, Cheng ZZ, Taylor S, Kostic AM, Wong ISB, Sajjath S, Zhang YM, Stevens J, et al. A functional group–guided approach to aptamers for small molecules. Science. 2023;380:942–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McConnell EM, Cozma I, Mou QB, Brennan JD, Lu Y, Li YF. Biosensing with dnazymes. Chem Soc Rev. 2021;50:8954–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu M, Wang S. Functional nucleic-acid-decorated spherical nanoparticles: preparation strategies and current applications in cancer therapy. Small Sci. 2021;1:2000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi S, Chen J, Wang X, Xiao M, Chandrasekaran AR, Li L, Yi C, Pei H. Biointerface engineering with nucleic acid materials for biosensing applications. Adv Func Mater. 2022;32:2201069. [Google Scholar]
- 7.Mendes BB, Conniot J, Avital A, Yao D, Jiang X, Zhou X, Sharf-Pauker N, Xiao Y, Adir O, Liang H, et al. Nanodelivery of nucleic acids. Nature Rev Methods Primers. 2022;2:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu F, Qing T, Qing Z. DNA-coded metal nano-fluorophores: Preparation, properties and applications in biosensing and bioimaging. Nano Today. 2021;36: 101021. [Google Scholar]
- 9.Chen C, Yang Z, Tang X. Chemical modifications of nucleic acid drugs and their delivery systems for gene-based therapy. Med Res Rev. 2018;38:829–69. [DOI] [PubMed] [Google Scholar]
- 10.Xu W, He W, Du Z, Zhu L, Huang K, Lu Y, Luo Y. Functional nucleic acid nanomaterials: development, properties, and applications. Angew Chem Int Ed Engl. 2021;60:6890–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao X, Zhu L, He W, Luo Y, Xu W. Functional nucleic acids tailoring and its application. TrAC, Trends Anal Chem. 2019;118:138–57. [Google Scholar]
- 12.Shin J, Kang N, Kim B, Hong H, Yu L, Kim J, Kang H, Kim JS. One-dimensional nanomaterials for cancer therapy and diagnosis. Chem Soc Rev. 2023;52:4488–514. [DOI] [PubMed] [Google Scholar]
- 13.Lin M, Gao Y, Hornicek F, Xu F, Lu TJ, Amiji M, Duan Z. Near-infrared light activated delivery platform for cancer therapy. Adv Coll Interface Sci. 2015;226:123–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y, Zuo X, Li Q, Chen F, Chen YR, Deng J, Han D, Hao C, Huang F, Huang Y, et al. Nucleic acids analysis. Sci China Chem. 2021;64:171–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lv Z, Huang M, Li P, Xu M, Yao C, Yang D. Hybridization chain reaction-based DNA nanomaterials for biosensing, bioimaging and therapeutics. Chin Chem Lett. 2024;35: 108601. [Google Scholar]
- 16.Biju V. Chemical modifications and bioconjugate reactions of nanomaterials for sensing, imaging, drug delivery and therapy. Chem Soc Rev. 2014;43:744–64. [DOI] [PubMed] [Google Scholar]
- 17.Kaymaz SV, Nobar HM, Sarıgül H, Soylukan C, Akyüz L, Yüce M. Nanomaterial surface modification toolkit: principles, components, recipes, and applications. Adv Coll Interface Sci. 2023;322: 103035. [DOI] [PubMed] [Google Scholar]
- 18.Do HD, Couillaud BM, Doan B-T, Corvis Y, Mignet N. Advances on non-invasive physically triggered nucleic acid delivery from nanocarriers. Adv Drug Deliv Rev. 2019;138:3–17. [DOI] [PubMed] [Google Scholar]
- 19.Cullis PR, Felgner PL. The 60-year evolution of lipid nanoparticles for nucleic acid delivery. Nat Rev Drug Discovery. 2024;23:709–22. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen QH, Kim MI. Nanomaterial-mediated paper-based biosensors for colorimetric pathogen detection. TrAC, Trends Anal Chem. 2020;132: 116038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang C, Belwal T, Luo Z, Su B, Lin X. Application of nanomaterials in isothermal nucleic acid amplification. Small. 2022;18: e2102711. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y, Li ZH, Zeng X, Zhang XZ. Bacteria-based bioactive materials for cancer imaging and therapy. Adv Drug Deliv Rev. 2023;193: 114696. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Lu L, Song ZL, Song W, Fu Z, Chao Q, Fan GC, Chen Z, Luo X. Covalent amide-bonded nanoflares for high-fidelity intracellular sensing and targeted therapy: a superstable nanosystem free of nonspecific interferences. Anal Chem. 2021;93:7879–88. [DOI] [PubMed] [Google Scholar]
- 24.Li S, Tian J, Zhu L, Huang K, Chu H, Xu W. Functional nucleic acid lateral flow magnetic biosensor based on blocking the super PCR and magnetic test strip for rapid detection of genetically modified maize MON810(†). Anal Chim Acta. 2022;1202: 339660. [DOI] [PubMed] [Google Scholar]
- 25.Samanta A, Deng Z, Liu Y, Yan H. A perspective on functionalizing colloidal quantum dots with DNA. Nano Res. 2013;6:853–70. [Google Scholar]
- 26.Francés-Soriano L, Estebanez N, Pérez-Prieto J, Hildebrandt N. DNA-coated upconversion nanoparticles for sensitive nucleic acid FRET biosensing. Adv Func Mater. 2022;32:2201541. [Google Scholar]
- 27.Zhang D, Peng R, Liu W, Donovan MJ, Wang L, Ismail I, Li J, Li J, Qu F, Tan W. Engineering DNA on the surface of upconversion nanoparticles for bioanalysis and therapeutics. ACS Nano. 2021;15:17257–74. [DOI] [PubMed] [Google Scholar]
- 28.Nair DP, Podgórski M, Chatani S, Gong T, Xi W, Fenoli CR, Bowman CN. The thiol-michael addition click reaction: a powerful and widely used tool in materials chemistry. Chem Mater. 2014;26:724–44. [Google Scholar]
- 29.Di Z, Liu B, Zhao J, Gu Z, Zhao Y, Li L. An orthogonally regulatable DNA nanodevice for spatiotemporally controlled biorecognition and tumor treatment. Sci Adv. 2020;6:eaba9381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leidner A, Weigel S, Bauer J, Reiber J, Angelin A, Grösche M, Scharnweber T, Niemeyer CM. Biopebbles: DNA-functionalized core–shell silica nanospheres for cellular uptake and cell guidance studies. Adv Func Mater. 2018;28:1707572. [Google Scholar]
- 31.Sui N, Wang L, Yan T, Liu F, Sui J, Jiang Y, Wan J, Liu M, Yu WW. Selective and sensitive biosensors based on metal-enhanced fluorescence. Sens Actuators, B Chem. 2014;202:1148–53. [Google Scholar]
- 32.Lee JS, Lytton-Jean AK, Hurst SJ, Mirkin CA. Silver nanoparticle-oligonucleotide conjugates based on DNA with triple cyclic disulfide moieties. Nano Lett. 2007;7:2112–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wen Y, McLaughlin CK, Lo PK, Yang H, Sleiman HF. Stable gold nanoparticle conjugation to internal DNA positions: facile generation of discrete gold nanoparticle-DNA assemblies. Bioconjug Chem. 2010;21:1413–6. [DOI] [PubMed] [Google Scholar]
- 34.Karakoti AS, Shukla R, Shanker R, Singh S. Surface functionalization of quantum dots for biological applications. Adv Colloid Interface Sci. 2015;215:28–45. [DOI] [PubMed] [Google Scholar]
- 35.Liu B, Liu J. Interface-driven hybrid materials based on DNA-functionalized gold nanoparticles. Matter. 2019;1:825–47. [Google Scholar]
- 36.Zhu D, Chao J, Pei H, Zuo X, Huang Q, Wang L, Huang W, Fan C. Coordination-mediated programmable assembly of unmodified oligonucleotides on plasmonic silver nanoparticles. ACS Appl Mater Interfaces. 2015;7:11047–52. [DOI] [PubMed] [Google Scholar]
- 37.Qing Z, Luo G, Xing S, Zou Z, Lei Y, Liu J, Yang R. Pt-S bond-mediated nanoflares for high-fidelity intracellular applications by avoiding thiol cleavage. Angew Chem Int Ed Engl. 2020;59:14044–8. [DOI] [PubMed] [Google Scholar]
- 38.Yang W, Feng L. Mg(2+)-promoted high-efficiency DNA conjugation on polydopamine surfaces for aptamer-based ochratoxin A detection. Anal Chim Acta. 2024;1298: 342382. [DOI] [PubMed] [Google Scholar]
- 39.Hu Y, Grösche M, Sheshachala S, Oelschlaeger C, Willenbacher N, Rabe KS, Niemeyer CM. Bottom-up assembly of DNA-silica nanocomposites into micrometer-sized hollow spheres. Angew Chem Int Ed Engl. 2019;58:17269–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Fazio AF, Misatziou D, Baker YR, Muskens OL, Brown T, Kanaras AG. Chemically modified nucleic acids and DNA intercalators as tools for nanoparticle assembly. Chem Soc Rev. 2021;50:13410–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rubner MM, Achatz DE, Mader HS, Stolwijk JA, Wegener J, Harms GS, Wolfbeis OS, Wagenknecht HA. DNA “Nanolamps”: “Clicked” DNA conjugates with photon upconverting nanoparticles as highly emissive biomaterial. ChemPlusChem. 2012;77:129–34. [Google Scholar]
- 42.Siegel N, Hasebe H, Chiarelli G, Garoli D, Sugimoto H, Fujii M, Acuna GP, Kołątaj K. Universal click-chemistry approach for the DNA functionalization of nanoparticles. J Am Chem Soc. 2024;146:17250–60. [DOI] [PubMed] [Google Scholar]
- 43.Luo Y-R. Comprehensive handbook of chemical bond energies. CRC Press; 2007. [Google Scholar]
- 44.Zhang X, Servos MR, Liu J. Surface science of DNA adsorption onto citrate-capped gold nanoparticles. Langmuir. 2012;28:3896–902. [DOI] [PubMed] [Google Scholar]
- 45.Liu C, Zhang P, Zhai X, Tian F, Li W, Yang J, Liu Y, Wang H, Wang W, Liu W. Nano-carrier for gene delivery and bioimaging based on carbon dots with PEI-passivation enhanced fluorescence. Biomaterials. 2012;33:3604–13. [DOI] [PubMed] [Google Scholar]
- 46.Wu Z, Liu GQ, Yang XL, Jiang JH. Electrostatic nucleic acid nanoassembly enables hybridization chain reaction in living cells for ultrasensitive mRNA imaging. J Am Chem Soc. 2015;137:6829–36. [DOI] [PubMed] [Google Scholar]
- 47.Zhuang W-R, Wang Y, Cui P-F, Xing L, Lee J, Kim D, Jiang H-L, Oh Y-K. Applications of π-π stacking interactions in the design of drug-delivery systems. J Control Release. 2019;294:311–26. [DOI] [PubMed] [Google Scholar]
- 48.Peng Y, Huang Y, Zhu Y, Chen B, Wang L, Lai Z, Zhang Z, Zhao M, Tan C, Yang N, et al. Ultrathin two-dimensional covalent organic framework nanosheets: preparation and application in highly sensitive and selective DNA detection. J Am Chem Soc. 2017;139:8698–704. [DOI] [PubMed] [Google Scholar]
- 49.Yerneni SS, Lathwal S, Shrestha P, Shirwan H, Matyjaszewski K, Weiss L, Yolcu ES, Campbell PG, Das SR. Rapid on-demand extracellular vesicle augmentation with versatile oligonucleotide tethers. ACS Nano. 2019;13:10555–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Banga RJ, Chernyak N, Narayan SP, Nguyen ST, Mirkin CA. Liposomal spherical nucleic acids. J Am Chem Soc. 2014;136:9866–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gu P, Li Y, Li L, Deng S, Zhu X, Song Y, Song E, Tan W. Azo reductase activated magnetic resonance tuning probe with “Switch-On” property for specific and sensitive tumor imaging in vivo. ACS Nano. 2023;17:24384–94. [DOI] [PubMed] [Google Scholar]
- 52.Khramtsov P, Kropaneva M, Kalashnikova T, Bochkova M, Timganova V, Zamorina S, Rayev M. Highly stable conjugates of carbon nanoparticles with DNA aptamers. Langmuir. 2018;34:10321–32. [DOI] [PubMed] [Google Scholar]
- 53.Xue Q, Jiang D, Wang L, Jiang W. Quantitative detection of single molecules using enhancement of Dye/DNA conjugate-labeled nanoparticles. Bioconjug Chem. 2010;21:1987–93. [DOI] [PubMed] [Google Scholar]
- 54.D’Agata R, Palladino P, Spoto G. Streptavidin-coated gold nanoparticles: critical role of oligonucleotides on stability and fractal aggregation. Beilstein J Nanotechnol. 2017;8:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vizzini P, Manzano M, Farre C, Meylheuc T, Chaix C, Ramarao N, Vidic J. Highly sensitive detection of Campylobacter spp. In chicken meat using a silica nanoparticle enhanced dot blot DNA biosensor. Biosens Bioelectron. 2021;171:112689. [DOI] [PubMed] [Google Scholar]
- 56.Yao G, Li J, Li Q, Chen X, Liu X, Wang F, Qu Z, Ge Z, Narayanan RP, Williams D, et al. Programming nanoparticle valence bonds with single-stranded DNA encoders. Nat Mater. 2020;19:781–8. [DOI] [PubMed] [Google Scholar]
- 57.Bawazer LA, Newman AM, Gu Q, Ibish A, Arcila M, Cooper JB, Meldrum FC, Morse DE. Efficient selection of biomineralizing DNA aptamers using deep sequencing and population clustering. ACS Nano. 2014;8:387–95. [DOI] [PubMed] [Google Scholar]
- 58.Lu C, Huang Z, Liu B, Liu Y, Ying Y, Liu J. Poly-cytosine DNA as a high-affinity ligand for inorganic nanomaterials. Angew Chem Int Ed Engl. 2017;56:6208–12. [DOI] [PubMed] [Google Scholar]
- 59.Demirbas A, Karaytuğ T, Arabaci N, Yilmaz ES, Ocsoy I, Synthesis of metallic and metal oxide nanomaterials. Green Synthesis of Nanomaterials for Bioenergy Applications 2020:99–123.
- 60.Lundberg H, Tinnis F, Selander N, Adolfsson H. Catalytic amide formation from non-activated carboxylic acids and amines. Chem Soc Rev. 2014;43:2714–42. [DOI] [PubMed] [Google Scholar]
- 61.Sun H, Schanze KS. Functionalization of water-soluble conjugated polymers for bioapplications. ACS Appl Mater Interfaces. 2022;14:20506–19. [DOI] [PubMed] [Google Scholar]
- 62.Allen CL, Williams JM. Metal-catalysed approaches to amide bond formation. Chem Soc Rev. 2011;40:3405–15. [DOI] [PubMed] [Google Scholar]
- 63.Gong H, Zi Y, Kan G, Li L, Shi C, Wang X, Zhong J. Preparation of food-grade EDC/NHS-crosslinked gelatin nanoparticles and their application for pickering emulsion stabilization. Food Chem. 2024;436: 137700. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Y-T, Dong M, Xu P-P, Cai J-H, Liu S-H, Gao Y-B, Wang L-B, Li J, Jiang H, Wang J-D, Wang D-S. Aptamer-mediated DNA concatemer functionalized magnetic nanoparticles for reversible capture and release of circulating tumor cells. Colloids Surf, B. 2022;218: 112733. [DOI] [PubMed] [Google Scholar]
- 65.Qi L, Xiao Y, Fu X, Yang H, Fang L, Xu R, Ping J, Han D, Jiang Y, Fang X. Monodispersed and monofunctionalized DNA-caged Au nano-clusters with enhanced optical properties for STED imaging. Small. 2024;20: e2400238. [DOI] [PubMed] [Google Scholar]
- 66.Koshizuka M, Takahashi N, Shimada N. Organoboron catalysis for direct amide/peptide bond formation. Chem Commun (Camb). 2024;60:11202–22. [DOI] [PubMed] [Google Scholar]
- 67.D’Amaral MC, Jamkhou N, Adler MJ. Efficient and accessible silane-mediated direct amide coupling of carboxylic acids and amines. Green Chem. 2021;23:288–95. [Google Scholar]
- 68.Chatterjee M, Chanda N. Formulation of PLGA nano-carriers: specialized modification for cancer therapeutic applications. Mater Adv. 2022;3:837–58. [Google Scholar]
- 69.Lee JC, Donahue ND, Mao AS, Karim A, Komarneni M, Thomas EE, Francek ER, Yang W, Wilhelm S. Exploring maleimide-based nanoparticle surface engineering to control cellular interactions. ACS Appl Nano Mater. 2020;3:2421–9. [Google Scholar]
- 70.Stasińska AR, Putaj P, Chmielewski MK. Disulfide bridge as a linker in nucleic acids’ bioconjugation. Part I: an overview of synthetic strategies. Bioorg Chem. 2019;92:103223. [DOI] [PubMed] [Google Scholar]
- 71.Hnasko RM. Bioconjugation of antibodies to horseradish peroxidase (HRP). Methods Mol Biol. 2015;1318:43–50. [DOI] [PubMed] [Google Scholar]
- 72.Derfus AM, Chen AA, Min DH, Ruoslahti E, Bhatia SN. Targeted quantum dot conjugates for siRNA delivery. Bioconjug Chem. 2007;18:1391–6. [DOI] [PubMed] [Google Scholar]
- 73.Zheng Y, Thai T, Reineck P, Qiu L, Guo Y, Bach U. DNA-directed self-assembly of core-satellite plasmonic nanostructures: a highly sensitive and reproducible near-IR SERS sensor. Adv Func Mater. 2013;23:1519–26. [Google Scholar]
- 74.Liu X, Liu M, Chen J, Li Z, Yuan Q. Rational design and biomedical applications of DNA-functionalized upconversion nanoparticles. Chin Chem Lett. 2018;29:1321–32. [Google Scholar]
- 75.Zhang D, Huarng MC, Alocilja EC. A multiplex nanoparticle-based bio-barcoded DNA sensor for the simultaneous detection of multiple pathogens. Biosens Bioelectron. 2010;26:1736–42. [DOI] [PubMed] [Google Scholar]
- 76.Manjappa AS, Chaudhari KR, Venkataraju MP, Dantuluri P, Nanda B, Sidda C, Sawant KK, Ramachandra Murthy RS. Antibody derivatization and conjugation strategies: application in preparation of stealth immunoliposome to target chemotherapeutics to tumor. J Control Release. 2011;150:2–22. [DOI] [PubMed] [Google Scholar]
- 77.Kong L, Li H, Zhang X, Zhuo Y, Chai Y, Yuan R. A novel ratiometric electrochemical biosensor using only one signal tag for highly reliable and ultrasensitive detection of miRNA-21. Anal Chem. 2022;94:5167–72. [DOI] [PubMed] [Google Scholar]
- 78.Srinivasan C, Lee J, Papadimitrakopoulos F, Silbart LK, Zhao M, Burgess DJ. Labeling and intracellular tracking of functionally active plasmid DNA with semiconductor quantum dots. Mol Ther. 2006;14:192–201. [DOI] [PubMed] [Google Scholar]
- 79.Banerjee A, Pons T, Lequeux N, Dubertret B. Quantum dots-DNA bioconjugates: synthesis to applications. Interface Focus. 2016;6:20160064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cal PM, Bernardes GJ, Gois PM. Cysteine-selective reactions for antibody conjugation. Angew Chem Int Ed Engl. 2014;53:10585–7. [DOI] [PubMed] [Google Scholar]
- 81.Szijj PA, Bahou C, Chudasama V. Minireview: addressing the retro-Michael instability of maleimide bioconjugates. Drug Discov Today Technol. 2018;30:27–34. [DOI] [PubMed] [Google Scholar]
- 82.Huang W, Wu X, Gao X, Yu Y, Lei H, Zhu Z, Shi Y, Chen Y, Qin M, Wang W, Cao Y. Maleimide-thiol adducts stabilized through stretching. Nat Chem. 2019;11:310–9. [DOI] [PubMed] [Google Scholar]
- 83.Ahangarpour M, Kavianinia I, Hume PA, Harris PWR, Brimble MA. N-vinyl acrylamides: versatile heterobifunctional electrophiles for thiol-thiol bioconjugations. J Am Chem Soc. 2022;144:13652–62. [DOI] [PubMed] [Google Scholar]
- 84.Vasco AV, Taylor RJ, Méndez Y, Bernardes GJL. On-demand thio-succinimide hydrolysis for the assembly of stable protein-protein conjugates. J Am Chem Soc. 2024;146:20709–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhong X, Li Z, Shi R, Yan L, Zhu Y, Li H. Schiff base-modified nanomaterials for ion detection: a review. ACS Appl Nano Mater. 2022;5:13998–4020. [Google Scholar]
- 86.Lynge ME, van der Westen R, Postma A, Städler B. Polydopamine—a nature-inspired polymer coating for biomedical science. Nanoscale. 2011;3:4916–28. [DOI] [PubMed] [Google Scholar]
- 87.Zandieh M, Liu J. Transition metal-mediated DNA adsorption on polydopamine nanoparticles. Langmuir. 2020;36:3260–7. [DOI] [PubMed] [Google Scholar]
- 88.Yang Y, Wang S, Zhou Z, Zhang R, Shen H, Song J, Su P, Yang Y. Enhanced reusability and activity: DNA directed immobilization of enzyme on polydopamine modified magnetic nanoparticles. Biochem Eng J. 2018;137:108–15. [Google Scholar]
- 89.Liu M, Peng Y, Nie Y, Liu P, Hu S, Ding J, Zhou W. Co-delivery of doxorubicin and DNAzyme using ZnO@polydopamine core-shell nanocomposites for chemo/gene/photothermal therapy. Acta Biomater. 2020;110:242–53. [DOI] [PubMed] [Google Scholar]
- 90.Azadbakht A, Beirnvand S. Voltammetric aptamer-based switch probes for sensing diclofenac using a glassy carbon electrode modified with a composite prepared from gold nanoparticles, carbon nanotubes and amino-functionalized Fe3O4 nanoparticles. Microchim Acta. 2017;184:2825–35. [Google Scholar]
- 91.Moses JE, Moorhouse AD. The growing applications of click chemistry. Chem Soc Rev. 2007;36:1249–62. [DOI] [PubMed] [Google Scholar]
- 92.Chen Z, Sun M, Luo F, Xu K, Lin Z, Zhang L. Stimulus-response click chemistry based aptamer-functionalized mesoporous silica nanoparticles for fluorescence detection of thrombin. Talanta. 2018;178:563–8. [DOI] [PubMed] [Google Scholar]
- 93.Fantoni NZ, El-Sagheer AH, Brown T. A Hitchhiker’s guide to click-chemistry with nucleic acids. Chem Rev. 2021;121:7122–54. [DOI] [PubMed] [Google Scholar]
- 94.Shi W, Tang F, Ao J, Yu Q, Liu J, Tang Y, Jiang B, Ren X, Huang H, Yang W, Huang W. Manipulating the click reactivity of dibenzoazacyclooctynes: from azide click component to caged acylation reagent by silver catalysis. Angew Chem Int Ed Engl. 2020;59:19940–4. [DOI] [PubMed] [Google Scholar]
- 95.Morris W, Briley WE, Auyeung E, Cabezas MD, Mirkin CA. Nucleic acid-metal organic framework (MOF) nanoparticle conjugates. J Am Chem Soc. 2014;136:7261–4. [DOI] [PubMed] [Google Scholar]
- 96.Lee J, Soares G, Doty C, Park J, Hovey J, Schrader A, Han HS. Versatile prepolymer platform for controlled tailoring of quantum dot surface properties. ACS Appl Mater Interfaces. 2024;16:15202–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang Y, Wang Y, Zheng X, Ducrot É, Lee MG, Yi GR, Weck M, Pine DJ. Synthetic strategies toward DNA-coated colloids that crystallize. J Am Chem Soc. 2015;137:10760–6. [DOI] [PubMed] [Google Scholar]
- 98.Serrano CM, Freeman R, Godbe J, Lewis JA, Stupp SI. DNA-peptide amphiphile nanofibers enhance aptamer function. ACS Appl Bio Mater. 2019;2:2955–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Oh JS, Wang Y, Pine DJ, Yi G-R. High-density PEO-b-DNA brushes on polymer particles for colloidal superstructures. Chem Mater. 2015;27:8337–44. [Google Scholar]
- 100.Cutler JI, Auyeung E, Mirkin CA. Spherical nucleic acids. J Am Chem Soc. 2012;134:1376–91. [DOI] [PubMed] [Google Scholar]
- 101.Liu J, Lu Y. Preparation of aptamer-linked gold nanoparticle purple aggregates for colorimetric sensing of analytes. Nat Protoc. 2006;1:246–52. [DOI] [PubMed] [Google Scholar]
- 102.Ding Q, Qiu W, Sun C, Ren H, Liu G: Comparison of DNA-gold nanoparticle conjugation methods: application in lateral flow nucleic acid biosensors. Molecules 2023, 28. [DOI] [PMC free article] [PubMed]
- 103.Zhang X, Servos MR, Liu J. Instantaneous and quantitative functionalization of gold nanoparticles with thiolated DNA using a pH-assisted and surfactant-free route. J Am Chem Soc. 2012;134:7266–9. [DOI] [PubMed] [Google Scholar]
- 104.Liu B, Liu J. Freezing directed construction of bio/nano interfaces: reagentless conjugation, denser spherical nucleic acids, and better nanoflares. J Am Chem Soc. 2017;139:9471–4. [DOI] [PubMed] [Google Scholar]
- 105.Hao Y, Li Y, Song L, Deng Z. Flash synthesis of spherical nucleic acids with record DNA density. J Am Chem Soc. 2021;143:3065–9. [DOI] [PubMed] [Google Scholar]
- 106.Huang M, Xiong E, Wang Y, Hu M, Yue H, Tian T, Zhu D, Liu H, Zhou X. Fast microwave heating-based one-step synthesis of DNA and RNA modified gold nanoparticles. Nat Commun. 2022;13:968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jian H, Xu G, Yi Y, Hao Y, Wang Y, Xiong L, Wang S, Liu S, Meng C, Wang J, et al. The origin and impeded dissemination of the DNA phosphorothioation system in prokaryotes. Nat Commun. 2021;12:6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhou W, Wang F, Ding J, Liu J. Tandem phosphorothioate modifications for DNA adsorption strength and polarity control on gold nanoparticles. ACS Appl Mater Interfaces. 2014;6:14795–800. [DOI] [PubMed] [Google Scholar]
- 109.Saran R, Huang Z, Liu J. Phosphorothioate nucleic acids for probing metal binding, biosensing and nanotechnology. Coord Chem Rev. 2021;428: 213624. [Google Scholar]
- 110.Xiao F, Chen Z, Wei Z, Tian L. Hydrophobic Interaction: A Promising Driving Force for the Biomedical Applications of Nucleic Acids. Adv Sci (Weinh). 2020;7:2001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li D, Song S, Fan C. Target-responsive structural switching for nucleic acid-based sensors. Acc Chem Res. 2010;43:631–41. [DOI] [PubMed] [Google Scholar]
- 112.Li F, Pei H, Wang L, Lu J, Gao J, Jiang B, Zhao X, Fan C. Nanomaterial-based fluorescent DNA analysis: a comparative study of the quenching effects of graphene oxide, carbon nanotubes, and gold nanoparticles. Adv Func Mater. 2013;23:4140–8. [Google Scholar]
- 113.Ma X, Kou X, Xu Y, Yang D, Miao P. Colorimetric sensing strategy for heparin assay based on PDDA-induced aggregation of gold nanoparticles. Nanoscale Adv. 2019;1:486–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Goyal G, Ammanath G, Palaniappan A, Liedberg B. Stoichiometric tuning of PNA probes to Au(0.8)Ag(0.2) alloy nanoparticles for visual detection of nucleic acids in plasma. ACS Sens. 2020;5:2476–85. [DOI] [PubMed] [Google Scholar]
- 115.Yan N, Wang X, Lin L, Song T, Sun P, Tian H, Liang H, Chen X. Gold nanorods electrostatically binding nucleic acid probe for in vivo microRNA amplified detection and photoacoustic imaging-guided photothermal therapy. Adv Func Mater. 2018;28:1800490. [Google Scholar]
- 116.Mignon P, Loverix S, Steyaert J, Geerlings P. Influence of the pi-pi interaction on the hydrogen bonding capacity of stacked DNA/RNA bases. Nucleic Acids Res. 2005;33:1779–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhao M, Wang Y, Ma Q, Huang Y, Zhang X, Ping J, Zhang Z, Lu Q, Yu Y, Xu H, et al. Ultrathin 2D metal-organic framework nanosheets. Adv Mater. 2015;27:7372–8. [DOI] [PubMed] [Google Scholar]
- 118.Lu C, Liu Y, Ying Y, Liu J. Comparison of MoS(2), WS(2), and graphene oxide for DNA adsorption and sensing. Langmuir. 2017;33:630–7. [DOI] [PubMed] [Google Scholar]
- 119.Zhao Z, Fan H, Zhou G, Bai H, Liang H, Wang R, Zhang X, Tan W. Activatable fluorescence/MRI bimodal platform for tumor cell imaging via MnO2 nanosheet-aptamer nanoprobe. J Am Chem Soc. 2014;136:11220–3. [DOI] [PubMed] [Google Scholar]
- 120.Meyer EE, Rosenberg KJ, Israelachvili J. Recent progress in understanding hydrophobic interactions. Proc Natl Acad Sci U S A. 2006;103:15739–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Alshaer W, Hillaireau H, Vergnaud J, Mura S, Deloménie C, Sauvage F, Ismail S, Fattal E. Aptamer-guided siRNA-loaded nanomedicines for systemic gene silencing in CD-44 expressing murine triple-negative breast cancer model. J Control Release. 2018;271:98–106. [DOI] [PubMed] [Google Scholar]
- 122.Zou J, Shi M, Liu X, Jin C, Xing X, Qiu L, Tan W. Aptamer-functionalized exosomes: elucidating the cellular uptake mechanism and the potential for cancer-targeted chemotherapy. Anal Chem. 2019;91:2425–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Banga RJ, Meckes B, Narayan SP, Sprangers AJ, Nguyen ST, Mirkin CA. Cross-linked micellar spherical nucleic acids from thermoresponsive templates. J Am Chem Soc. 2017;139:4278–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tang J, Huang C, Shu J, Zheng J, Ma D, Li J, Yang R. Azoreductase and target simultaneously activated fluorescent monitoring for cytochrome c release under hypoxia. Anal Chem. 2018;90:5865–72. [DOI] [PubMed] [Google Scholar]
- 125.Weber PC, Ohlendorf DH, Wendoloski JJ, Salemme FR. Structural origins of high-affinity biotin binding to streptavidin. Science. 1989;243:85–8. [DOI] [PubMed] [Google Scholar]
- 126.Zhang Y, Lu F, Yager KG, van der Lelie D, Gang O. A general strategy for the DNA-mediated self-assembly of functional nanoparticles into heterogeneous systems. Nat Nanotechnol. 2013;8:865–72. [DOI] [PubMed] [Google Scholar]
- 127.He Q, Wu Q, Feng X, Liao Z, Peng W, Liu Y, Peng D, Liu Z, Mo M. Interfacing DNA with nanoparticles: surface science and its applications in biosensing. Int J Biol Macromol. 2020;151:757–80. [DOI] [PubMed] [Google Scholar]
- 128.Ye Y, Hou S, Wu X, Cheng X, He S. Freeze-driven adsorption of poly-A DNA on gold nanoparticles: from a stable biointerface to plasmonic dimers. Langmuir. 2022;38:4625–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhu D, Li J, Wang L, Li Q, Wang L, Song B, Zhou R, Fan C. Hydrophobic collapse-driven nanoparticle coating with poly-adenine adhesives. Chem Commun (Camb). 2021;57:3801–4. [DOI] [PubMed] [Google Scholar]
- 130.Wang X, Yang Z, Li Z, Huang K, Cheng N, Liu J. Rapid thermal drying synthesis of nonthiolated spherical nucleic acids with stability rivaling thiolated DNA. Angew Chem Int Ed Engl. 2024;63: e202410353. [DOI] [PubMed] [Google Scholar]
- 131.Yang J, Pong BK, Lee JY, Too HP. Dissociation of double-stranded DNA by small metal nanoparticles. J Inorg Biochem. 2007;101:824–30. [DOI] [PubMed] [Google Scholar]
- 132.Ranganathan SV, Halvorsen K, Myers CA, Robertson NM, Yigit MV, Chen AA. Complex thermodynamic behavior of single-stranded nucleic acid adsorption to graphene surfaces. Langmuir. 2016;32:6028–34. [DOI] [PubMed] [Google Scholar]
- 133.Iliafar S, Mittal J, Vezenov D, Jagota A. Interaction of single-stranded DNA with curved carbon nanotube is much stronger than with flat graphite. J Am Chem Soc. 2014;136:12947–57. [DOI] [PubMed] [Google Scholar]
- 134.Campuzano S, Yáñez-Sedeño P, Pingarrón JM. Nanoparticles for nucleic-acid-based biosensing: opportunities, challenges, and prospects. Anal Bioanal Chem. 2019;411:1791–806. [DOI] [PubMed] [Google Scholar]
- 135.Park JS, Na HK, Min DH, Kim DE. Desorption of single-stranded nucleic acids from graphene oxide by disruption of hydrogen bonding. Analyst. 2013;138:1745–9. [DOI] [PubMed] [Google Scholar]
- 136.Li Y, Zhang Z, Liu B, Liu J. Adsorption of DNA oligonucleotides by boronic acid-functionalized hydrogel nanoparticles. Langmuir. 2019;35:13727–34. [DOI] [PubMed] [Google Scholar]
- 137.Gao X, Yao X, Zhong Z, Jia L. Rapid and sensitive detection of Staphylococcus aureus assisted by polydopamine modified magnetic nanoparticles. Talanta. 2018;186:147–53. [DOI] [PubMed] [Google Scholar]
- 138.Zhan L, Peng L, Huang C-Z. Stable silver nanoparticles–aptamer bioconjugates for cellular prion protein imaging. Chin Sci Bull. 2014;59:964–70. [Google Scholar]
- 139.Zhong D, Yang K, Wang Y, Yang X. Dual-channel sensing strategy based on gold nanoparticles cooperating with carbon dots and hairpin structure for assaying RNA and DNA. Talanta. 2017;175:217–23. [DOI] [PubMed] [Google Scholar]
- 140.You M, Jia P, He X, Wang Z, Feng S, Ren Y, Li Z, Cao L, Gao B, Yao C, et al. Quantifying and adjusting plasmon-driven nano-localized temperature field around gold nanorods for nucleic acids amplification. Small Methods. 2021;5: e2001254. [DOI] [PubMed] [Google Scholar]
- 141.Zhang F, Wang S, Liu J. Gold nanoparticles adsorb DNA and aptamer probes too strongly and a comparison with graphene oxide for biosensing. Anal Chem. 2019;91:14743–50. [DOI] [PubMed] [Google Scholar]
- 142.Xu L, Zhang P, Liu Y, Fang X, Zhang Z, Liu Y, Peng L, Liu J. Continuously tunable nucleotide/lanthanide coordination nanoparticles for DNA adsorption and sensing. ACS Omega. 2018;3:9043–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ma H, Li Z, Xue N, Cheng Z, Miao X. A gold nanoparticle based fluorescent probe for simultaneous recognition of single-stranded DNA and double-stranded DNA. Mikrochim Acta. 2018;185:93. [DOI] [PubMed] [Google Scholar]
- 144.Liu B, Salgado S, Maheshwari V, Liu J. DNA adsorbed on graphene and graphene oxide: fundamental interactions, desorption and applications. Curr Opin Colloid Interface Sci. 2016;26:41–9. [Google Scholar]
- 145.Chen X, Chen T, Ren L, Chen G, Gao X, Li G, Zhu X. Triplex DNA nanoswitch for pH-sensitive release of multiple cancer drugs. ACS Nano. 2019;13:7333–44. [DOI] [PubMed] [Google Scholar]
- 146.Wang L, Yan Y. A review of pH-responsive organic-inorganic hybrid nanoparticles for RNAi-based therapeutics. Macromol Biosci. 2021;21:2100183. [DOI] [PubMed] [Google Scholar]
- 147.Han G, You CC, Kim BJ, Turingan RS, Forbes NS, Martin CT, Rotello VM. Light-regulated release of DNA and its delivery to nuclei by means of photolabile gold nanoparticles. Angew Chem. 2006;118:3237–41. [DOI] [PubMed] [Google Scholar]
- 148.O’Hagan MP, Duan Z, Huang F, Laps S, Dong J, Xia F, Willner I. Photocleavable ortho-nitrobenzyl-protected DNA architectures and their applications. Chem Rev. 2023;123:6839–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wu M, Kempaiah R, Huang PJ, Maheshwari V, Liu J. Adsorption and desorption of DNA on graphene oxide studied by fluorescently labeled oligonucleotides. Langmuir. 2011;27:2731–8. [DOI] [PubMed] [Google Scholar]
- 150.Li F, Zhang H, Dever B, Li XF, Le XC. Thermal stability of DNA functionalized gold nanoparticles. Bioconjug Chem. 2013;24:1790–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Goodman AM, Hogan NJ, Gottheim S, Li C, Clare SE, Halas NJ. Understanding resonant light-triggered DNA release from plasmonic nanoparticles. ACS Nano. 2017;11:171–9. [DOI] [PubMed] [Google Scholar]
- 152.Morgan E, Wupperfeld D, Morales D, Reich N. Shape matters: gold nanoparticle shape impacts the biological activity of siRNA delivery. Bioconjug Chem. 2019;30:853–60. [DOI] [PubMed] [Google Scholar]
- 153.Yang J, Su Q, Song C, Luo H, Jiang H, Ni M, Meng F. A comprehensive adsorption and desorption study on the interaction of DNA oligonucleotides with TiO(2) nanolayers. Phys Chem Chem Phys. 2024;26:22681–95. [DOI] [PubMed] [Google Scholar]
- 154.Li P, Li M, Zhang F, Wu M, Jiang X, Ye B, Zhao Z, Yue D, Fan Q, Chen H. High-efficient nucleic acid separation from animal tissue samples via surface modified magnetic nanoparticles. Sep Purif Technol. 2021;262: 118348. [Google Scholar]
- 155.Bai Y, Roncancio D, Suo Y, Shao Y, Zhang D, Zhou C. A method based on amino-modified magnetic nanoparticles to extract DNA for PCR-based analysis. Colloids Surf B Biointerfaces. 2019;179:87–93. [DOI] [PubMed] [Google Scholar]
- 156.Li H, Zhang Y, Wang L, Tian J, Sun X. Nucleic acid detection using carbon nanoparticles as a fluorescent sensing platform. Chem Commun (Camb). 2011;47:961–3. [DOI] [PubMed] [Google Scholar]
- 157.Liu B, Sun Z, Zhang X, Liu J. Mechanisms of DNA sensing on graphene oxide. Anal Chem. 2013;85:7987–93. [DOI] [PubMed] [Google Scholar]
- 158.Wen L, Wang M. Functionalities of pH-responsive DNA nanostructures in tumor-targeted strategies. J Mater Chem B. 2024;12:12174–90. [DOI] [PubMed] [Google Scholar]
- 159.Lei Y, Li C, Ji X, Sun H, Liu X, Mao Z, Chen W, Qing Z, Liu J. Lowering entropic barriers in triplex DNA switches facilitating biomedical applications at physiological pH. Angew Chem Int Ed. 2024;63: e202402123. [DOI] [PubMed] [Google Scholar]
- 160.Liu J, Huang Y, Kumar A, Tan A, Jin S, Mozhi A, Liang X-J. pH-Sensitive nano-systems for drug delivery in cancer therapy. Biotechnol Adv. 2014;32:693–710. [DOI] [PubMed] [Google Scholar]
- 161.Li L, Xing H, Zhang J, Lu Y. Functional DNA molecules enable selective and stimuli-responsive nanoparticles for biomedical applications. Acc Chem Res. 2019;52:2415–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Li J, Wang X, Liang X. Modification of nucleic acids by azobenzene derivatives and their applications in biotechnology and nanotechnology. Chem Asian J. 2014;9:3344–58. [DOI] [PubMed] [Google Scholar]
- 163.Wang C, O’Hagan MP, Li Z, Zhang J, Ma X, Tian H, Willner I. Photoresponsive DNA materials and their applications. Chem Soc Rev. 2022;51:720–60. [DOI] [PubMed] [Google Scholar]
- 164.Herdt AR, Drawz SM, Kang Y, Taton TA. DNA dissociation and degradation at gold nanoparticle surfaces. Colloids Surf B Biointerfaces. 2006;51:130–9. [DOI] [PubMed] [Google Scholar]
- 165.Hastman DA, Oh E, Melinger JS, Green CM, Thielemann AJP, Medintz IL, Díaz SA. Smaller gold nanoparticles release DNA more efficiently during fs laser pulsed optical heating. Small. 2024;20: e2303136. [DOI] [PubMed] [Google Scholar]
- 166.Chen CC, Lin YP, Wang CW, Tzeng HC, Wu CH, Chen YC, Chen CP, Chen LC, Wu YC. DNA-gold nanorod conjugates for remote control of localized gene expression by near infrared irradiation. J Am Chem Soc. 2006;128:3709–15. [DOI] [PubMed] [Google Scholar]
- 167.Barhoumi A, Huschka R, Bardhan R, Knight MW, Halas NJ. Light-induced release of DNA from plasmon-resonant nanoparticles: Towards light-controlled gene therapy. Chem Phys Lett. 2009;482:171–9. [Google Scholar]
- 168.Huschka R, Zuloaga J, Knight MW, Brown LV, Nordlander P, Halas NJ. Light-induced release of DNA from gold nanoparticles: nanoshells and nanorods. J Am Chem Soc. 2011;133:12247–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Riley RS, Dang MN, Billingsley MM, Abraham B, Gundlach L, Day ES. Evaluating the mechanisms of light-triggered siRNA release from nanoshells for temporal control over gene regulation. Nano Lett. 2018;18:3565–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sanchis A, Salvador JP, Marco MP. Light-induced mechanisms for nanocarrier’s cargo release. Colloids Surf B Biointerfaces. 2019;173:825–32. [DOI] [PubMed] [Google Scholar]
- 171.Lei J, Ju H. Signal amplification using functional nanomaterials for biosensing. Chem Soc Rev. 2012;41:2122–34. [DOI] [PubMed] [Google Scholar]
- 172.Su Y, Chu H, Tian J, Du Z, Xu W. Insight into the nanomaterials enhancement mechanism of nucleic acid amplification reactions. TrAC, Trends Anal Chem. 2021;137: 116221. [Google Scholar]
- 173.Li M, Lin YC, Wu CC, Liu HS. Enhancing the efficiency of a PCR using gold nanoparticles. Nucl Acids Res. 2005;33: e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Li H, Huang J, Lv J, An H, Zhang X, Zhang Z, Fan C, Hu J. Nanoparticle PCR: nanogold-assisted PCR with enhanced specificity. Angew Chem Int Ed Engl. 2005;44:5100–3. [DOI] [PubMed] [Google Scholar]
- 175.Tian L, Cronin TM, Weizmann Y. Enhancing-effect of gold nanoparticles on DNA strand displacement amplifications and their application to an isothermal telomerase assay. Chem Sci. 2014;5:4153–62. [Google Scholar]
- 176.Lou X, Zhang Y. Mechanism studies on NanoPCR and applications of gold nanoparticles in genetic analysis. ACS Appl Mater Interfaces. 2013;5:6276–84. [DOI] [PubMed] [Google Scholar]
- 177.Sedighi A, Oberc C, Whitehall V, Li PC. NanoHDA: a nanoparticle-assisted isothermal amplification technique for genotyping assays. Nano Res. 2017;10:12–21. [Google Scholar]
- 178.Xue Z, You M, Peng P, Tong H, He W, Li A, Mao P, Xu T, Xu F, Yao C. Taqman-MGB nanoPCR for highly specific detection of single-base mutations. Int J Nanomedicine. 2021. 10.2147/IJN.S310254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ye X, Fang X, Li X, Kong J. Gold nanoparticle-mediated nucleic acid isothermal amplification with enhanced specificity. Anal Chim Acta. 2018;1043:150–7. [DOI] [PubMed] [Google Scholar]
- 180.Upadhyay A, Yang H, Zaman B, Zhang L, Wu Y, Wang J, Zhao J, Liao C, Han Q, ZnO Nanolower-based NanoPCR as an efficient diagnostic tool for quick diagnosis of canine vector-borne pathogens. Pathogens 2020, 9. [DOI] [PMC free article] [PubMed]
- 181.Sun C, Cheng Y, Pan Y, Yang J, Wang X, Xia F. Efficient polymerase chain reaction assisted by metal–organic frameworks. Chem Sci. 2020;11:797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhu HH, Li Y, Wu LX, Wang KS, Zhang Y, Fan QY, Ming ZZ, Chen WQ, Liu WW. Internal heating method of loop-mediated isothermal amplification for detection of HPV-6 DNA. Mikrochim Acta. 2022;189:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yüce M, Uysal E, Budak H. Amplification yield enhancement of short DNA templates using bulk and surface-attached amine-functionalized single-wall carbon nanotubes. Appl Surf Sci. 2015;349:147–55. [Google Scholar]
- 184.Hwang SH, Im SG, Hah SS, Cong VT, Lee EJ, Lee YS, Lee GK, Lee DH, Son SJ. Effects of upconversion nanoparticles on polymerase chain reaction. PLoS ONE. 2013;8: e73408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Xue Z, You M, Peng P, Tong H, He W, Li A, Mao P, Xu T, Xu F, Yao C. Taqman-MGB nanoPCR for highly specific detection of single-base mutations. Int J Nanomedicine. 2021;16:3695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sang F, Yang Y, Yuan L, Ren J, Zhang Z. Development of a high-throughput real time PCR based on a hot-start alternative for Pfu mediated by quantum dots. Nanoscale. 2015;7:15852–62. [DOI] [PubMed] [Google Scholar]
- 187.Cao X, Chen J, Wen S, Peng C, Shen M, Shi X. Effect of surface charge of polyethyleneimine-modified multiwalled carbon nanotubes on the improvement of polymerase chain reaction. Nanoscale. 2011;3:1741–7. [DOI] [PubMed] [Google Scholar]
- 188.Liang G, Ma C, Zhu Y, Li S, Shao Y, Wang Y, Xiao Z. Enhanced specificity of multiplex polymerase chain reaction via CdTe quantum dots. Nanoscale Res Lett. 2011;6:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Rehman A, Sarwar Y, Raza ZA, Hussain SZ, Mustafa T, Khan WS, Ghauri MA, Haque A, Hussain I. Metal nanoparticle assisted polymerase chain reaction for strain typing of Salmonella Typhi. Analyst. 2015;140:7366–72. [DOI] [PubMed] [Google Scholar]
- 190.Zhang Z, Shen C, Wang M, Han H, Cao X. Aqueous suspension of carbon nanotubes enhances the specificity of long PCR. Biotechniques. 2008;44:537–8. [DOI] [PubMed] [Google Scholar]
- 191.Chen P, Pan D, Fan C, Chen J, Huang K, Wang D, Zhang H, Li Y, Feng G, Liang P, et al. Gold nanoparticles for high-throughput genotyping of long-range haplotypes. Nat Nanotechnol. 2011;6:639–44. [DOI] [PubMed] [Google Scholar]
- 192.Tian J, Chu H, Zhang Y, Li K, Tian H, Zhang X, Xu W. TiO(2) nanoparticle-enhanced linker recombinant strand displacement amplification (LRSDA) for universal label-free visual bioassays. ACS Appl Mater Interfaces. 2019;11:46504–14. [DOI] [PubMed] [Google Scholar]
- 193.Abdul Khaliq R, Kafafy R, Salleh HM, Faris WF. Enhancing the efficiency of polymerase chain reaction using graphene nanoflakes. Nanotechnology. 2012;23: 455106. [DOI] [PubMed] [Google Scholar]
- 194.Wan W, Yeow JT. The effects of gold nanoparticles with different sizes on polymerase chain reactionefficiency. Nanotechnology. 2009;20: 325702. [DOI] [PubMed] [Google Scholar]
- 195.Zeng R, Du Z, Ma H, Meng X, Li E, Li J. The 60 nm gold nanoparticles improve qPCR amplification efficiency through specific palindromic sequences (GGATCC or ACCGGT) in primers. Biochimica et Biophysica Acta General Sub. 2024;1868:130560. [DOI] [PubMed] [Google Scholar]
- 196.Vanzha E, Pylaev T, Khanadeev V, Konnova S, Fedorova V, Khlebtsov N. Gold nanoparticle-assisted polymerase chain reaction: effects of surface ligands, nanoparticle shape and material. RSC Adv. 2016;6:110146–54. [Google Scholar]
- 197.Evanko D. Hybridization chain reaction. Nat Methods. 2004;1:186–186. [Google Scholar]
- 198.Li H, Wang X, Wei S, Zhao C, Song X, Xu K, Li J, Pang B, Wang J. Applications of hybridization chain reaction optical detection incorporating nanomaterials: a review. Anal Chim Acta. 2022;1190: 338930. [DOI] [PubMed] [Google Scholar]
- 199.Li T, Zou L, Zhang J, Li G, Ling L. Non-invasive diagnosis of bladder cancer by detecting telomerase activity in human urine using hybridization chain reaction and dynamic light scattering. Anal Chim Acta. 2019;1065:90–7. [DOI] [PubMed] [Google Scholar]
- 200.Park C, Song Y, Jang K, Choi C-H, Na S. Target switching catalytic hairpin assembly and gold nanoparticle colorimetric for EGFR mutant detection. Sens Actuators, B Chem. 2018;261:497–504. [Google Scholar]
- 201.Quan K, Huang J, Yang X, Yang Y, Ying L, Wang H, Wang K. An enzyme-free and amplified colorimetric detection strategy: assembly of gold nanoparticles through target-catalytic circuits. Analyst. 2015;140:1004–7. [DOI] [PubMed] [Google Scholar]
- 202.Wang G, Li J, He Y, Liu J, Yu M, Wang G. Establishment of a universal and sensitive plasmonic biosensor platform based on the hybridization chain reaction (HCR) amplification induced by a triple-helix molecular switch. Analyst. 2020;145:3864–70. [DOI] [PubMed] [Google Scholar]
- 203.Chen W, Zhang X, Li J, Chen L, Wang N, Yu S, Li G, Xiong L, Ju H. Colorimetric detection of nucleic acids through triplex-hybridization chain reaction and DNA-controlled growth of platinum nanoparticles on graphene oxide. Anal Chem. 2020;92:2714–21. [DOI] [PubMed] [Google Scholar]
- 204.Tang S, Li Y, Zhu A, Yao Y, Sun J, Zheng F, Lin Z, Shen W. A triple-amplification strategy based on the formation of peroxidase-like two-dimensional DNA/Fe(3)O(4) networks initiated by the hybridization chain reaction for highly sensitive detection of microRNA. Chem Commun (Camb). 2019;55:8386–9. [DOI] [PubMed] [Google Scholar]
- 205.De M, Chou SS, Dravid VP. Graphene oxide as an enzyme inhibitor: modulation of activity of α-chymotrypsin. J Am Chem Soc. 2011;133:17524–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Luo Y, Chen J, Liang J, Liu Y, Liu C, Liu Y, Xu T, Zhang X. Ultrasound-enhanced catalytic hairpin assembly capable of ultrasensitive microRNA biosensing for the early screening of Alzheimer’s disease. Biosens Bioelectron. 2023;242: 115746. [DOI] [PubMed] [Google Scholar]
- 207.Li Z, Xue N, Ma H, Cheng Z, Miao X. An ultrasensitive and switch-on platform for aflatoxin B(1) detection in peanut based on the fluorescence quenching of graphene oxide-gold nanocomposites. Talanta. 2018;181:346–51. [DOI] [PubMed] [Google Scholar]
- 208.Liu Q, Liu M, Jin Y, Li B. Rapid and enzyme-free signal amplification for fluorescent detection of microRNA via localized catalytic hairpin assembly on gold nanoparticles. Talanta. 2022;242: 123142. [DOI] [PubMed] [Google Scholar]
- 209.Yu B, Li F, Zhao T, Li F, Zhou B, Xu H. Hybridization chain reaction-based flow cytometric bead sensor for the detection of emetic Bacillus cereus in milk. Sens Actuators, B Chem. 2018;256:624–31. [Google Scholar]
- 210.Li Z, Mao G, Du M, Tian S, Niu L, Ji X, He Z. A fluorometric turn-on aptasensor for mucin 1 based on signal amplification via a hybridization chain reaction and the interaction between a luminescent ruthenium(II) complex and CdZnTeS quantum dots. Mikrochim Acta. 2019;186:233. [DOI] [PubMed] [Google Scholar]
- 211.Lu Q, Xie L, Yin S, Chen F, Wu C, Liu M, Li H, Zhang Y. Ultrasensitive detection of microRNA-10b through target-triggered catalytic hairpin assembly and upconversion nanoparticles-based luminescence resonance energy transfer. Talanta. 2023;253: 124032. [Google Scholar]
- 212.Wang X, Hou T, Lin H, Lv W, Li H, Li F. In situ template generation of silver nanoparticles as amplification tags for ultrasensitive surface plasmon resonance biosensing of microRNA. Biosens Bioelectron. 2019;137:82–7. [DOI] [PubMed] [Google Scholar]
- 213.Feliu N, Hassan M, Garcia Rico E, Cui D, Parak W, Alvarez-Puebla R. SERS quantification and characterization of proteins and other biomolecules. Langmuir. 2017;33:9711–30. [DOI] [PubMed] [Google Scholar]
- 214.Shao H, Lin H, Guo Z, Lu J, Jia Y, Ye M, Su F, Niu L, Kang W, Wang S, et al. A multiple signal amplification sandwich-type SERS biosensor for femtomolar detection of miRNA. Biosens Bioelectron. 2019;143: 111616. [DOI] [PubMed] [Google Scholar]
- 215.Luo X, Tan R, Xia Z, Yue W, Peng J, Yuan X, Wang M, Li P, He Y. Catalytic hairpin assembly-mediated SERS biosensor for double detection of MiRNAs using gold nanoclusters-doped COF substrate. Sens Actuators, B Chem. 2024;401: 134953. [Google Scholar]
- 216.Xu M, Tang D. Recent advances in DNA walker machines and their applications coupled with signal amplification strategies: a critical review. Anal Chim Acta. 2021;1171: 338523. [DOI] [PubMed] [Google Scholar]
- 217.Yehl K, Mugler A, Vivek S, Liu Y, Zhang Y, Fan M, Weeks ER, Salaita K. High-speed DNA-based rolling motors powered by RNase H. Nat Nanotechnol. 2016;11:184–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Yuan A, Xiao H, Yang F, Hao H, Wang X, Li J, Jin M, Zhao Q, Sha R, Deng Z. DNA walker for signal amplification in living cells. TrAC, Trends Anal Chem. 2023;158: 116870. [Google Scholar]
- 219.Liu C, Wu T, Deng L, Li X, Fu X, Liao S, Ma W, Zou G, Yang H. Programmed DNA walkers for biosensors. Chin Chem Lett. 2024;35: 109307. [Google Scholar]
- 220.Zhang H, Lai M, Zuehlke A, Peng H, Li XF, Le XC. Binding-induced DNA nanomachines triggered by proteins and nucleic acids. Angew Chem Int Ed Engl. 2015;54:14326–30. [DOI] [PubMed] [Google Scholar]
- 221.Zhang X, Wu T, Yang Y, Wen Y, Wang S, Xu L-P. Superwettable electrochemical biosensor based on a dual-DNA walker strategy for sensitive E. coli O157: H7 DNA detection. Sensors Actuators B: Chem. 2020;321:128472. [Google Scholar]
- 222.Walsh CT, Moore BS. Enzymatic cascade reactions in biosynthesis. Angew Chem Int Ed. 2019;58:6846–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Cheng X, Bao Y, Liang S, Li B, Liu Y, Wu H, Ma X, Chu Y, Shao Y, Meng Q. Flap endonuclease 1-assisted DNA walkers for sensitively and specifically sensing ctDNAs. Anal Chem. 2021;93:9593–601. [DOI] [PubMed] [Google Scholar]
- 224.Simmel FC, Yurke B, Singh HR. Principles and applications of nucleic acid strand displacement reactions. Chem Rev. 2019;119:6326–69. [DOI] [PubMed] [Google Scholar]
- 225.Li W, Wang L, Jiang W. A catalytic assembled enzyme-free three-dimensional DNA walker and its sensing application. Chem Commun. 2017;53:5527–30. [DOI] [PubMed] [Google Scholar]
- 226.Song L, Zhuge Y, Zuo X, Li M, Wang F. DNA walkers for biosensing development. Adv Sci. 2022;9:2200327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Ye M, Kong Y, Zhang C, Lv Y, Cheng S, Hou D, Xian Y. Near-infrared light controllable DNA walker driven by endogenous adenosine triphosphate for in situ spatiotemporal imaging of intracellular MicroRNA. ACS Nano. 2021;15:14253–62. [DOI] [PubMed] [Google Scholar]
- 228.Wang Z, Zhang J, Ekman JM, Kenis PJ, Lu Y. DNA-mediated control of metal nanoparticle shape: one-pot synthesis and cellular uptake of highly stable and functional gold nanoflowers. Nano Lett. 2010;10:1886–91. [DOI] [PubMed] [Google Scholar]
- 229.Wu J, Tan LH, Hwang K, Xing H, Wu P, Li W, Lu Y. DNA sequence-dependent morphological evolution of silver nanoparticles and their optical and hybridization properties. J Am Chem Soc. 2014;136:15195–202. [DOI] [PubMed] [Google Scholar]
- 230.Sun W, Shen J, Zhao Z, Arellano N, Rettner C, Tang J, Cao T, Zhou Z, Ta T, Streit JK, et al. Precise pitch-scaling of carbon nanotube arrays within three-dimensional DNA nanotrenches. Science. 2020;368:874–7. [DOI] [PubMed] [Google Scholar]
- 231.Zhou W, Li Y, Je K, Vo T, Lin H, Partridge BE, Huang Z, Glotzer SC, Mirkin CA. Space-tiled colloidal crystals from DNA-forced shape-complementary polyhedra pairing. Science. 2024;383:312–9. [DOI] [PubMed] [Google Scholar]
- 232.Li Z, Lim Y, Tanriover I, Zhou W, Li Y, Zhang Y, Aydin K, Glotzer SC, Mirkin CA. DNA-mediated assembly of Au bipyramids into anisotropic light emitting kagome superlattices. Sci Adv. 2024;10:eadp3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Zhang Y, Xu DD, Tanriover I, Zhou W, Li Y, López-Arteaga R, Aydin K, Mirkin CA, Nonlinear optical colloidal metacrystals. Nature Photon, 2024.
- 234.Zhou W, Lim Y, Lin H, Lee S, Li Y, Huang Z, Du JS, Lee B, Wang S, Sánchez-Iglesias A, et al. Colloidal quasicrystals engineered with DNA. Nat Mater. 2024;23:424–8. [DOI] [PubMed] [Google Scholar]
- 235.Paunovska K, Loughrey D, Dahlman JE. Drug delivery systems for RNA therapeutics. Nat Rev Genet. 2022;23:265–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Shang S, Li X, Wang H, Zhou Y, Pang K, Li P, Liu X, Zhang M, Li W, Li Q, Chen X. Targeted therapy of kidney disease with nanoparticle drug delivery materials. Bioactive Mater. 2024;37:206–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Tao Z, Zhang H, Wu S, Zhang J, Cheng Y, Lei L, Qin Y, Wei H, Yu CY. Spherical nucleic acids: emerging amplifiers for therapeutic nanoplatforms. Nanoscale. 2024;16:4392–406. [DOI] [PubMed] [Google Scholar]
- 238.Valatabar N, Oroojalian F, Kazemzadeh M, Mokhtarzadeh AA, Safaralizadeh R, Sahebkar A. Recent advances in gene delivery nanoplatforms based on spherical nucleic acids. J Nanobiotechnol. 2024;22:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Gilam A, Conde J, Weissglas-Volkov D, Oliva N, Friedman E, Artzi N, Shomron N. Local microRNA delivery targets Palladin and prevents metastatic breast cancer. Nat Commun. 2016;7:12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Valimukhametova AR, Lee BH, Topkiran UC, Gries K, Gonzalez-Rodriguez R, Coffer JL, Akkaraju G, Naumov A. Cancer therapeutic siRNA delivery and imaging by nitrogen- and neodymium-doped graphene quantum dots. ACS Biomater Sci Eng. 2023;9:3425–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Chang Q, Liu C, Xie Z, Shu Q, Xie Y, Su Q, Deng XJC. Polyethylenimine functionalized ultrasmall mesoporous silica nanoparticles for siRNA delivery. ChemNanoMat. 2022;8: e202100453. [Google Scholar]
- 242.Yang S, Chen Y, Gu J, Harris A, Su R-C, Ho EA. pH-sensitive dual-preventive siRNA-based nanomicrobicide reactivates autophagy and inhibits HIV infection in vaginal CD4+ cells. J Contr Release Offi J Contr Release Soc. 2024;366:849–63. [DOI] [PubMed] [Google Scholar]
- 243.Kubczak M, Michlewska S, Karimov M, Ewe A, Noske S, Aigner A, Bryszewska M. Ionov MJIJoP: unmodified and tyrosine-modified polyethylenimines as potential carriers for siRNA: biophysical characterization and toxicity. Int J Pharm. 2022;614: 121468. [DOI] [PubMed] [Google Scholar]
- 244.Shi L, Zhang J, Zhao M, Tang S, Cheng X, Zhang W, Li W, Liu X, Peng H, Wang Q. Effects of polyethylene glycol on the surface of nanoparticles for targeted drug delivery. Nanoscale. 2021;13:10748–64. [DOI] [PubMed] [Google Scholar]
- 245.Ledesma F, Ozcan B, Sun X, Medina SM, Landry MP. Nanomaterial strategies for delivery of therapeutic cargoes. Adv Func Mater. 2022;32:2107174. [Google Scholar]
- 246.Yi Y, Yu M, Feng C, Hao H, Zeng W, Lin C, Chen H, Lv F, Zhu D, Ji XJM. Transforming “cold” tumors into “hot” ones via tumor-microenvironment-responsive siRNA micelleplexes for enhanced immunotherapy. Matter. 2022;5:2285–305. [Google Scholar]
- 247.Gao Y, Jia L, Wang Q, Hu H, Zhao X, Chen D, Qiao M. pH/Redox dual-responsive polyplex with effective endosomal escape for codelivery of siRNA and doxorubicin against drug-resistant cancer cells. ACS Appl Mater Interfaces. 2019;11:16296–310. [DOI] [PubMed] [Google Scholar]
- 248.Nie JJ, Qiao B, Duan S, Xu C, Chen B, Hao W, Yu B, Li Y, Du J, Xu FJ. Unlockable nanocomplexes with self-accelerating nucleic acid release for effective staged gene therapy of cardiovascular diseases. Adv Mater. 2018;30:1801570. [DOI] [PubMed] [Google Scholar]
- 249.Sun Y, Liu L, Zhou L, Yu S, Lan Y, Liang Q, Liu J, Cao A, Liu Y. Tumor microenvironment-triggered charge reversal polymetformin-based nanosystem co-delivered doxorubicin and IL-12 cytokine gene for chemo–gene combination therapy on metastatic breast cancer. ACS Appl Mater Interfaces. 2020;12:45873–90. [DOI] [PubMed] [Google Scholar]
- 250.Lo Y-L, Chang C-H, Wang C-S, Yang M-H, Lin AM-Y, Hong C-J, Tseng W-H. PEG-coated nanoparticles detachable in acidic microenvironments for the tumor-directed delivery of chemo-and gene therapies for head and neck cancer. Theranostics. 2020;10:6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Lin M, Qi X. Advances and challenges of stimuli-responsive nucleic acids delivery system in gene therapy. Pharmaceutics. 2023;15:1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Ren R, Xiong B, Zhu J. surface modification of gold nanorods: multifunctional strategies and application prospects. Chem A Eur J. 2024;30:e202400851. [DOI] [PubMed] [Google Scholar]
- 253.Schmitz FRW, Cesca K, Valério A, de Oliveira D, Hotza D. Colorimetric detection of Pseudomonas aeruginosa by aptamer-functionalized gold nanoparticles. Appl Microbiol Biotechnol. 2023;107:71–80. [DOI] [PubMed] [Google Scholar]
- 254.Liu S, Fang L, Wang Y, Wang L. Universal dynamic DNA assembly-programmed surface hybridization effect for single-step, reusable, and amplified electrochemical nucleic acid biosensing. Anal Chem. 2017;89:3108–15. [DOI] [PubMed] [Google Scholar]
- 255.Wang Z-Y, Li P, Cui L, Qiu J-G, Jiang B. Zhang C-y: Integration of nanomaterials with nucleic acid amplification approaches for biosensing. TrAC Trends Anal Chem. 2020;129:115959. [Google Scholar]
- 256.Koo KM, Carrascosa LG, Shiddiky MJ, Trau M. Poly (A) extensions of miRNAs for amplification-free electrochemical detection on screen-printed gold electrodes. Anal Chem. 2016;88:2000–5. [DOI] [PubMed] [Google Scholar]
- 257.Zhang Z, Wang Y, Zhang N, Zhang S. Self-assembly of nucleic acid molecular aggregates catalyzed by a triple-helix probe for miRNA detection and single cell imaging. Chem Sci. 2016;7:4184–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Su A, Liu Y, Sun W, Liang C, Xu W, Rodger A, Piper J, Wang Y, Xu S. Silver nanoparticles with dual-recognition via CRISPR/dCas9 for SERS identification of two KRAS mutations in nucleic acid targets. ACS Appl Nano Mater. 2024;7:9800–8. [Google Scholar]
- 259.Chen H, Cai S, Luo J, Liu X, Ou L, Zhang Q, Liedberg B, Wang Y. Colorimetric biosensing assays based on gold nanoparticles functionalized/combined with non-antibody recognition elements. TrAC, Trends Anal Chem. 2024;173: 117654. [Google Scholar]
- 260.Jiang Z, Feng B, Xu J, Qing T, Zhang P, Qing Z. Graphene biosensors for bacterial and viral pathogens. Biosens Bioelectron. 2020;166: 112471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.