Abstract
Cooperative interactions by DNA-binding proteins have been implicated in cell-fate decisions in a variety of organisms. To date, however, there are few examples in which the importance of such interactions has been explicitly tested in vivo. Here, we tested the importance of cooperative DNA binding by the Bicoid protein in establishing a pattern along the anterior–posterior axis of the early Drosophila embryo. We found that bicoid mutants specifically defective in cooperative DNA binding fail to direct proper development of the head and thorax, leading to embryonic lethality. The mutants did not faithfully stimulate transcription of downstream target genes such as hunchback (hb), giant, and Krüppel. Quantitative analysis of gene expression in vivo indicated that bcd cooperativity mutants were unable to accurately direct the extent to which hb is expressed along the anterior–posterior axis and displayed a reduced ability to generate sharp on/off transitions for hb gene expression. These failures in precise transcriptional control demonstrate the importance of cooperative DNA binding for embryonic patterning in vivo.
Keywords: cooperativity, pattern formation, morphogen, transcription
Cooperative DNA binding allows gene regulatory proteins to work at low concentrations and, in some cases, to function as genetic switches to control cell-fate decisions (1). The best-studied example is phage lambda cI repressor, for which small changes in its concentration determine the decision between lysis and lysogeny (2). Cooperative DNA binding can occur between monomers or dimers of a given DNA-binding protein, as is typical in prokaryotes (3), or between two or more different proteins, as is commonly observed in eukaryotes (4, 5). Cooperative DNA binding is distinct from transcriptional synergy (6), a mechanism of gene regulation in which DNA-bound proteins interact cooperatively with other components of the transcription machinery to enhance (or repress) transcription.
We have been studying cooperative interactions by the Bicoid protein, which is a morphogen that controls anterior–posterior (A-P) cell fates during the early development of the Drosophila embryo. Bicoid is present in a concentration gradient and stimulates transcription of individual genes at different positions along the A-P axis (7, 8). For example, in the anterior of the embryo, where its concentration is high, Bicoid activates the gap gene, hunchback (hb), whereas in the posterior, where its concentration is low, Bicoid activates knirps (9, 10). It was originally proposed that the affinity of the binding sites within Bicoid target genes dictates how far along the A-P axis they are activated (11). Although this model is simple and elegant, it does not appear to be sufficient to explain Bicoid function. Additional mechanisms have been identified that affect A-P expression of Bicoid-dependent genes (12–15). It is also not understood how Bicoid activates target genes so precisely within discrete domains having sharply defined posterior boundaries.
Some of these mysteries might be explained on the basis of Bicoid's interactions with other proteins (16–20). For example, Bicoid interacts with the Sap18-Rpd3 histone deacetylase complex, which likely converts Bicoid from an activator to a repressor in the anterior tip of the embryo (14, 18, 21). In addition, Bicoid monomers interact with one another to exhibit cooperative DNA binding (22, 23). Specifically, a Bicoid monomer bound to a strong (high-affinity) binding site lowers the free energy of binding of a second monomer to an adjacent weak (low-affinity) site (23). This result contrasts with other mechanisms of cooperative gene activation that do not rely on direct protein–protein interactions (24). Bicoid cooperative coupling has important implications for Bicoid target gene regulation in the embryo. For example, cooperative DNA binding should affect the extent of target gene expression along the A-P axis and produce a threshold-effect for DNA binding such that sharp on/off borders of target gene expression can be generated.
To test these hypotheses explicitly, we isolated mutations in the bicoid gene (bcd) that encode proteins specifically defective in cooperative DNA binding (25). The mutations were isolated by using a genetic screen in yeast that detected strong-site/weak-site coupling. Some of the mutations map to the DNA-binding domain (homeodomain) of Bicoid but are not in positions known to contact base pairs in the major groove of DNA (26–28). The mutant Bicoid proteins are stable in vivo (in yeast cells) and are not affected for DNA recognition or nuclear entry (25) nor are they defective in transcription activation per se; the mutants activated a reporter gene containing a single binding site to the same level as did wild-type Bicoid.
In this study, Bicoid cooperative DNA binding was further examined in vitro, and cooperativity mutants were introduced into Drosophila, where their ability to direct A-P patterning was tested. We found that Bicoid mutants were defective in anterior patterning, which resulted in a high proportion of embryonic lethality. In mutant embryos, Bicoid-dependent target genes such as hb, giant (gt), and Krüppel (Kr) were not expressed to their proper levels, and/or their normally precise spatial domains of expression along the A-P axis were not correctly established. These results are a direct demonstration of the importance of cooperative DNA binding for pattern formation during embryonic development.
Materials and Methods
Expression of Full-Length Bicoid in Yeast. HA-tagged wild-type and K57R Bicoid proteins were expressed in Saccharomyces cerevisiae strain EGY48 (29) from the GAL1 promoter and induced by using an estradiol hormone system as described in refs. 23 and 30. Yeast extracts containing Bicoid were prepared as described in ref. 31. Concentrations of HA-tagged Bicoid, estimated by Western analysis, ranged from 10 to 30 μM.
Gel-Shift Analysis. Gel-shift titration experiments were performed as described in ref. 23 by using oligonucleotides containing strong Bicoid binding sites (S) of the general sequence 5′-TCGAC{(TCTAATCCC)TA}n-3′ (n = 2 or 3). The sites are spaced 11 bp center-to-center and oriented head-to-tail.
Fly Stocks. Transgenic Drosophila melanogaster lines were generated by using w1118. The bcd null stock (bcdE1) was th1 st1 kniri-1 bcd6 rnroe-1 pp/TM3 Sb1 and was obtained from Bloomington Stock Center (Indiana University). Balancer lines were CyO/nub b Sco lt stw3 and TM3 Sb1/H2.
P-Elements and Generation of Transgenic Flies. Two point mutations, K57R and S35T (homeodomain numbering; ref. 32), were individually engineered into pP{CaSpeR-4} containing a full-length (8.7 kb) genomic bcd rescue fragment (33). Transgenic flies were generated as described in ref. 34. For both bcd K57R and bcd S35T,two independent lines were obtained for each on the second and third chromosomes, respectively. For bcdWT, one insertion on the X chromosome was obtained. The presence of K57R or S35T alleles was confirmed by PCR and DNA sequencing.
Cuticle Preparations. Embryos were aged for 36 h, dechorionated in 50% bleach, devitellinized in methanol, and cleared in Hoyer's mountant/lactic acid (1:1) at 70°C for 3 h (35). First-instar larvae were hand-picked and cleared in Hoyer's.
Whole-Mount in Situ Hybridization. One- to 4-hour-old embryos were collected on apple juice-agar plates, dechorionated, fixed, and devitellinized (36). RNA in situ hybridization (37) was performed by using digoxigenin-labeled antisense RNA probes. Embryos were dehydrated and mounted in JB-4 (Polysciences).
Quantifying Gene Expression. Fluorescent staining for Bcd, Hb, and even-skipped protein (Eve) was performed by using rat anti-Bcd (1:1,000), rabbit anti-Eve (1:2,000), and guinea pig anti-Hb (1:2,000) (38). Secondary antibodies were conjugated with Alexa Fluor 488, 555, and 647 and used at a 1:2,000 dilution (Molecular Probes). Nuclear labeling was obtained by using a monoclonal anti-histone antibody (1:10,000) (Chemicon) and rat biotin-SP-conjugated anti-mouse Ab (1:1,000) (Jackson Laboratories) and Streptavidin Alexa Fluor 700 (1:1,000) (Molecular Probes). Embryos were mounted, scanned by confocal microscopy, and the images were processed and quantified by using the visiquest program (Accusoft, Northborough, MA) according to the methods of Janssens et al. (ref. 39 and Supporting Materials and Methods, which is published as supporting information on the PNAS web site).
Results
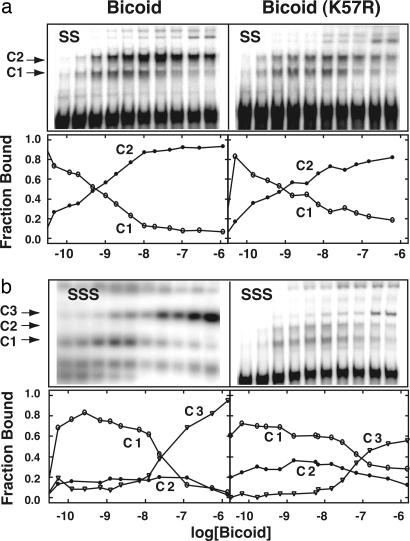
Bicoid Mutants Defective in Cooperative DNA Binding. Previous in vitro analysis of bcd cooperativity mutants used isolated homeodomains (60 aa) (25). Here, we examined cooperative DNA-binding in vitro by using full-length Bicoid from yeast cell extracts. As expected, gel-shift analysis of wild-type Bicoid showed cooperative coupling between monomers bound to adjacent sites (Fig. 1a Left). Binding of a single monomer (C1 complex) is transient, and a doubly bound complex (C2) forms readily on a two-site oligonucleotide template (SS) as the Bicoid concentration is increased. In contrast, the Bicoid(K57R) mutant forms the C2 complex less readily with a substantial proportion remaining in the C1 complex, even at higher protein concentrations (Fig. 1a Right). The averages of several experiments were used to plot the percent abundance of the major bound species as a function of protein concentration (Fig. 1, lower graphs). A decrease in the accumulation of intermediates along a reaction pathway is a hallmark of cooperative systems. The persistence of a substantial population of the C1 complex in the case of Bicoid(K57R) indicates a reduction in cooperative interactions.
Fig. 1.
Cooperative DNA-binding defect in the full-length Bicoid(K57R) mutant. Upper panels show representative gel shifts obtained by using cell extracts containing wild-type Bicoid or Bicoid(K57R). Labeled oligonucleotides carrying either two (SS) or three (SSS) strong (high-affinity) Bicoid binding sites (TCTAATCCC) were used. The single-bound (C1), double-bound (C2), and triple-bound (C3) complexes are indicated. Bicoid concentrations span 5 orders of magnitude. Lower graphs show the fraction of each major DNA-bound complex as a function of total Bicoid concentration. The curves are averages from two or more experiments. Higher-order species, which occur at high Bicoid concentrations (e.g., C4, C5, and C6) were not considered in this analysis.
Similar results were obtained by using a three strong-site oligonucleotide template (SSS) (Fig. 1b). Wild-type readily forms the triply bound complex (C3) compared with the Bicoid(K57R) mutant, which shows more C1 and C2 intermediate complexes. At the highest concentrations of extract, the mutant eventually formed a significant amount of triply bound complex (C3), as expected, because DNA recognition per se is not defective. These data reveal a reduction in cooperative DNA binding by full-length Bicoid(K57R) compared with the wild-type protein, confirming previous results with transcription assays in yeast (25).
Because the binding data were generated by using cell extracts rather than highly purified proteins (purified Bicoid is insoluble), the concentration of active Bicoid is not known. Therefore, it is not possible to directly compare the extent of binding of wild-type Bicoid with that of the K57R mutant nor can the free energies of binding be accurately determined. Instead, the relevant comparison is the proportional amount of each species formed (e.g., C1, C2, or C3) as a function of concentration (lower graphs in Fig. 1). And, although the reduction in free energy of binding of the K57R mutant is likely to be small (D.S.B., unpublished data), small changes in binding affinities often translate into large changes in biological response (1).
Bicoid Cooperativity Mutants in Drosophila. We generated transgenic lines carrying the bcdK57R mutation, another slightly less severe mutation, bcdS35T (25), and a wild-type control (bcdWT). The transgenes were expressed by using a genomic fragment that completely rescues the bcd null phenotype (33). These mutations and the wild-type control were crossed or recombined into a bcd null background (bcdE1/bcdE1). Several independent lines were analyzed. Because bcd is a maternal-effect gene, it was possible to generate bcdE1/bcdE1 flies that carried either one or two copies of each of the three transgenes (bcdK57R, bcdS35T, or bcdWT). The transgenes were expressed to normal levels, and the mRNAs localized correctly in blastoderm-staged embryos (Fig. 6, which is published as supporting information on the PNAS web site, and data not shown). The phenotypes of embryos derived from these transgenic mothers (hereafter referred to as bcd mutant embryos) were examined.
Bicoid Cooperativity Mutants Show Embryonic Lethality. We tested whether bcd cooperativity mutations rescued the embryonic lethality of bcd mutant embryos (Table 1). One copy of the control bcdWT transgene was sufficient to rescue the bcdE1/bcdE1 lethality to levels similar to that of bcdE1/+ embryos (8% vs. 4% unhatched). In contrast, one copy of either the bcdK57R or bcdS35T transgene did not fully rescue, with 69% and 37% of embryos, respectively, showing an unhatched phenotype. Of the hatched embryos, a large percentage showed larval head defects, especially among the bcdK57R embryos (Table 1).
Table 1. Viability of bcd cooperativity mutant embryos.
| Relevant maternal genotype* | % unhatched (n)† | % larval head defects‡ (n) |
|---|---|---|
| w1118 | 2–3 (703) | none |
| bcdE1/+ | 4 (593) | none |
| bcdE1/bcdE1 | 100 (1,238) | n/a |
| bcdWT (1×) | 8 (635) | 3 (162) |
| bcdK57R (1×) | 69 (744) | 50 (252) |
| bcdS35T (1×) | 37 (535) | 11 (121) |
| bcdK57R (2×) | 25 (661) | 36 (409) |
| bcdS35T (2×) | 3 (317) | none |
| bcdK57R bcdS35T | 31 (410) | 26 (538) |
1× and 2× are the number of copies of the transgene. All are in bcdE1/bcdE1 backgrounds
n, number of embryos scored
Scored only in hatched embryos. n/a is not applicable, because no larvae hatched
The more severe lethality associated with the bcdK57R allele relative to the bcdS35T allele parallels the severity of their cooperativity defects as measured by using transcription assays in yeast (25). The embryonic lethality was also dose-dependent; increasing the dosage of the bcdK57R or bcdS35T transgenes from one to two copies reduced the percentage of unhatched embryos to 25% and 3%, respectively. This result is consistent with the fact that cooperative interactions are less critical at higher protein concentrations, and that the defect in Bicoid cooperativity mutants is highly concentration-dependent (25).
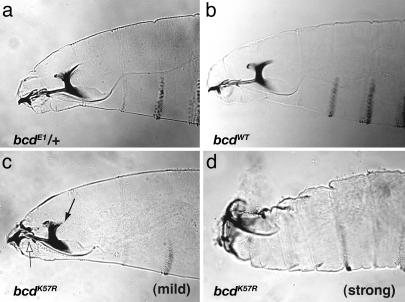
Cooperativity Mutants Show a Range of Head Defects. To determine the cause(s) of the lethality observed in the bcd cooperativity mutants, we examined embryos for bcd-related phenotypes (40). A range of head defects was observed (Fig. 2). For example, bcdK57R mutant embryos (bcdK57R; bcdE1/bcdE1) had mild defects such as reductions in the dorsal arm, dorsal bridge, and labrum (Fig. 2c), or moderate to strong defects such as the absence of discernable head skeletal structures and general disorganization of the head (Fig. 2d). Approximately 10% of the embryos lacked head and thoracic structures entirely and contained a posterior duplication, as in bcd null embryos (data not shown). These embryos lacked detectable Bicoid protein, as also observed for the bcdWT lines, where there was an 8% lethality. The other genotypes, including those of bcdS35T (Table 1), showed less severe phenotypes.
Fig. 2.
bcdK57R mutant embryos show head defects. Cuticle preparations of first-instar larvae are shown. Controls, bcdE1/+ (a), and bcdWT (b) show normal head development. A range of head defects occurs in bcdK57R mutants (one copy of transgene). Two phenotypes are shown, mild head defects (c), which occur in approximately one-half of the progeny, and strong defects (d), which occur in approximately one-third of the progeny. In c, the solid arrow indicates reduced size of the dorsal arm, and the open arrow indicates disorganization of the labrum and epistomal sclerite.
Cooperativity Mutants Display Aberrant hb mRNA Expression. Cooperative gene activation is thought to play an important role in the ability of Bicoid to activate genes along the A-P axis of the embryo (8, 9) Cooperative DNA binding, in particular, was predicted to be important for helping to define the precise spatial domain of expression of the zygotic gap gene, hb, which contains multiple Bicoid binding sites of varying affinities (9, 11, 22, 23). Based on these predictions, we compared the pattern of hb expression in wild-type and in bcd cooperativity mutant embryos.
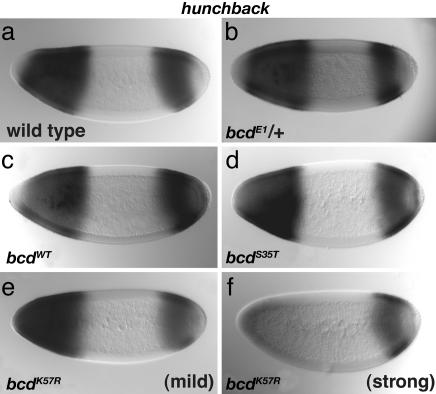
Cooperativity mutant embryos showed changes in levels of hb expression (Fig. 3). The most pronounced effects were found in the single-copy bcdK57R mutant background, where zygotic hb expression in the anterior was reduced and variable. In approximately half of the embryos, Bicoid-dependent hb expression appeared relatively normal (Fig. 3e) or only mildly reduced, whereas in the remaining embryos, anterior hb expression was significantly reduced (Fig. 3f). The variability of hb expression mirrors the variability of mutant phenotypes observed for bcdK57R embryos (Table 1). Expression of hb was mostly normal in bcdS35T (Fig. 3d) and two-copy bcdK57R embryos (data not shown), as expected given their milder mutant phenotypes.
Fig. 3.
Anterior hb expression is reduced in bcdK57R mutants. In situ hybridization reveals hb mRNA expression patterns in blastoderm-staged embryos. Control wild-type (a), bcdE1 heterozygous mutant (b), and bcdWT (c) embryos all show the expected patterns of hb expression. Normal to mild phenotypes were observed in bcdS35T mutants (d). Mild to strong defects in hb expression were observed for bcdK57R mutants (e and f, respectively).
Quantitation Reveals Spatial-Control Defects in hb Expression. To better understand the consequences of bcd cooperativity mutations, we quantitated their effects on hb expression. We used an enhanced fluorescence imaging technique (41, 42), in which signals from antibody staining are digitized and their intensities are normalized by using histone nuclear staining, to produce an accurate, quantitative measure of gene expression (in individual nuclei) within individual embryos.
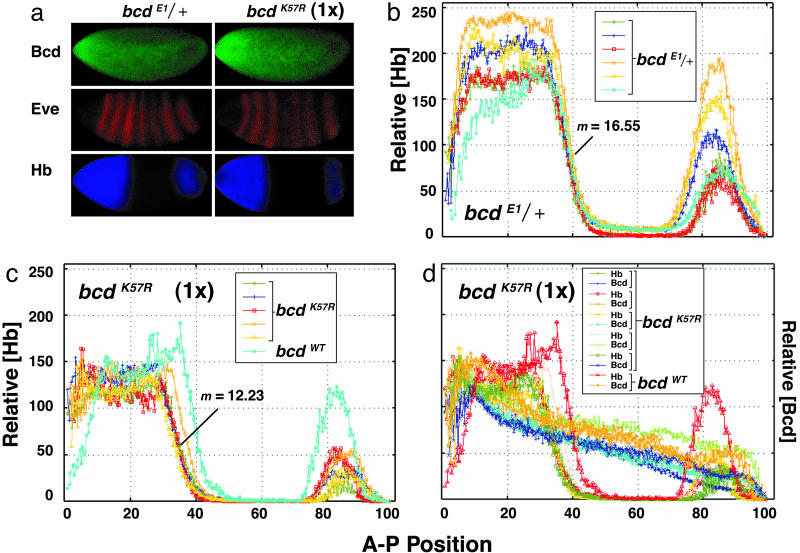
Examples of staining of wild-type and mutant embryos are shown (Fig. 4a). Staining for Eve helped identify embryos of the correct developmental stage (mid-stage 14), and served as an internal control for efficient staining. The intensity of the hb protein signal (Hb) was plotted as a function of distance along the A-P axis (Fig. 4 b–d). The overall magnitude of staining (signal height) varied with experiment and is not considered significant for this analysis. Using this approach, we monitored two key spatial parameters of hb expression in wild-type and in mutant embryos: (i) the extent along the A-P axis to which hb is expressed, and (ii) the sharpness of the posterior border of hb expression.
Fig. 4.
Quantitative analysis of hb expression in bcd cooperativity mutants. (a) Fluorescence immunostaining for Bcd, Eve, and Hb in control (bcdE1/+)or bcd cooperativity mutant (bcdK57R) embryos. In b–d, the relative Hb signal is plotted as a function of distance along the A-P axis, with each curve (color coded) representing data from an individual embryo staged ≈30 min after the onset of stage 14. In d, the data are from individual embryos double-stained for Bcd and Hb. In c and d, data for control embryos are also included. m is the average slope of the intensity of Hb staining for four to five embryos.
For control embryos, bcdE1/+ (Fig. 4b) and bcdWT (Fig. 7, which is published as supporting information on the PNAS web site) the Hb expression in individual embryos is similar, with the position at which half-maximal signal is measured at 39–40% egg length (measured from the anterior). In contrast, Hb expression in individual bcdK57R mutant embryos is shifted toward the anterior to 34–36% egg-length (Fig. 4c). The posterior border of Hb expression also shifts toward the anterior in bcdS35T embryos (Fig. 7), although as expected, this shift is smaller (to 36–38% egg length).
The second parameter, sharpness of the Hb posterior border, had been difficult to assess by using traditional in situ hybridization. However, using the enhanced imaging technique, the sharpness of this border can be quantified by the slope (m)ofthe fluorescence intensity. The sharpness is reduced in bcd cooperativity mutants. The slope of the posterior Hb signal was 12.23 ± 1.35 for the bcdK57R embryos (Fig. 4c) compared to 16.55 ± 1.69 and 16.71 ± 1.89 for the bcdE1/+ and bcdWT control embryos, respectively (Figs. 4b and 7). This difference is most easily seen in Fig. 4c, where data for the bcdWT control are plotted alongside that of the bcdK57R mutant embryos. As expected, the difference in slope was allele-specific, with bcdK57R embryos having a stronger defect than bcdS35T embryos (m = 15.63 ± 1.75; Fig. 7).
We also examined Bicoid protein expression in bcd mutant embryos to confirm that the gradient was correctly established. The levels of the mutant Bicoid protein in bcdK57R embryos were essentially normal compared with those of control embryos (Fig. 4 a and d). However, as reported in ref. 13, Bicoid levels (both mutant and wild-type) were more variable than Hb levels.
Altered Expression of gt and Kr. The expression of other Bicoid target-genes was also examined in bcd cooperativity mutants. No changes were detected for the head-specific genes, empty-spiracles (ems), buttonhead (btd), orthodenticle (otd) and cap n' collar (cnc), whose expression domains are limited to anterior regions with the highest concentrations of Bicoid protein (data not shown). However, changes were observed for the expression of gap genes, gt and Kr, which are expressed further down the Bicoid gradient (Fig. 5).
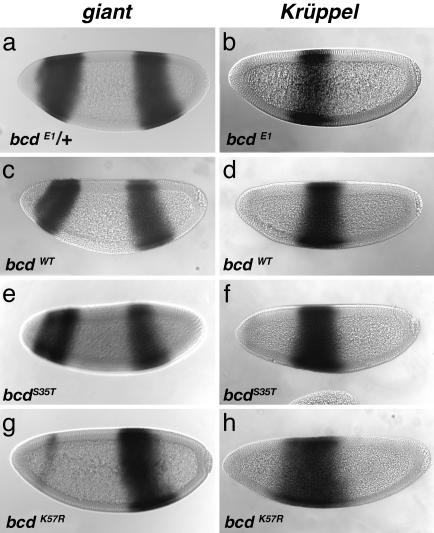
Fig. 5.
Expression of giant and kr are defective in bcdK57R mutants. In situ hybridization reveals gt (a, c, e, and g) and Kr (b, d, f, and h) mRNA expression. Control embryos bcdE1/+ (a) and bcdWT (c) as well as bcdS35T (e) show the expected patterns of gt expression. In contrast, more than half of the bcdK57R (one copy of transgene) embryos show a severe reduction in the anterior (bcd-dependent) expression of gt (g). Kr expression is as expected in control embryos, bcdE1/+ (b) and bcdWT (d). In contrast, the Kr domain is expanded anteriorly in bcdS35T and bcdK57R embryos (one copy of each transgene) (f and h).
For gt, which is directly activated by Bicoid (43), bcdK57R embryos showed a dramatic reduction in expression. In some bcdK57R embryos, anterior gt staining was nearly absent (Fig. 5g), whereas in others, it was only moderately reduced (data not shown). The results for gt are similar to those described above for hb, i.e., that the effects of bcd cooperativity mutations were variable, allele-specific (bcdK57R stronger effect than bcdS35T), and dose-dependent. As expected, there was no effect on the posterior gt stripe, because its expression is not Bicoid-dependent (43).
The bcd cooperativity mutants also showed a derepression of Kr, such that Kr expression expanded anteriorly (Fig. 5 f and h and Table 2). The position of the anterior border of the Kr domain was different between the bcdK57R and bcdS35T mutants and control embryos (bcdWT and bcdE1/+) (P < 0.001 for each). The expansion was greatest in the bcdK57R embryos. The anterior expansion of Kr appears to be at odds with the previous findings that showed Bicoid activates Kr expression (44). However, bcd also indirectly regulates Kr through repression by hb and gt (43). Thus, reduction of hb and gt in the bcd cooperativity mutants would likely explain the expansion of the Kr expression domain.
Table 2. Kr expansion in bcd cooperativity mutants.
| Maternal genotype* | Anterior border of Kr domain,† % (n)‡ | Posterior border of Kr domain,† % (n)‡ |
|---|---|---|
| W1118 | 57.34 ± 1.56 (11) | 42.29 ± 0.97 (11) |
| bcdE1/+ | 63.5 ± 2.38 (15) | 47.65 ± 2.56 (15) |
| bcdWT (1×) | 63.97 ± 2.79 (44) | 46.04 ± 1.88 (44) |
| bcdK57R (1×) | 70.08 ± 2.97 (15) | 47.34 ± 3.76 (15) |
| bcdK57R (2×) | 62.6 ± 4.66 (23) | 44.54 ± 3.31 (23) |
| bcdS35T (1×) | 66.45 ± 3.02 (44) | 47.0 ± 2.14 (44) |
Results are ± standard deviation from the mean.
Relevant genotype as in Table 1
Percent egg length, measured from the posterior
n, total number of embryos scored
Discussion
To date, there have been very few demonstrations of the importance of cooperative DNA binding in vivo (3, 24, 45, 46) and still fewer in which cooperativity mutations have been tested for the ability to carry out their normal function (47–49). Even in well studied bacteriophage and yeast systems, cooperativity mutations in repressor proteins (cI and α2) have typicially been assayed by using artificial reporter genes, rather than being reincorporated into the genomes and tested directly for their effects on the lysis-lysogeny decision or mating type (50). The present study is distinguished by the fact that Bicoid cooperativity mutants were tested for their ability to carry out their normal in vivo function; in this case, to determine cell-fate decisions by directing pattern formation during development.
Our results show that bcdK57R and bcdS35T mutants defective in cooperative DNA binding show severe defects during embryonic development. The head-specific phenotypes and the alterations in Bicoid target-gene expression indicate that cooperative DNA-binding by Bicoid is critical for proper embryonic patterning. In particular, the bcd mutants failed to establish proper expression domains for the gap genes hb, gt, and Kr. Other bcd target genes, namely, the head-gap genes, whose expression is limited to the anterior tip of the embryo, were not visibly affected.
The phenotypes observed in bcd cooperativity mutants were, unexpectedly, somewhat variable. From a genetic standpoint, these bcd alleles showed both incomplete penetrance and variable expressivity. We do not think that this variability can be attributed to gross differences in the amount of mutant protein in individual embryos. Even in wild-type embryos, there is an inherent variability in the Bicoid protein gradient (refs. 13 and 51 and data not shown). Instead, the varying phenotypes might be the result of stochastic processes. That is, the mutant Bicoid proteins might have activities at or near a critical threshold, such that slight changes in their concentration result in large changes in transcription output. Loss of Bicoid cooperativity might therefore eliminate a “buffering mechanism” so that Hb expression would now more closely parallel Bicoid levels. This conclusion might explain the various head phenotypes observed in the bcd cooperativity mutants. In the most severe embryos, genetic “backup systems” (see below) may not have been as robust.
Our results are generally consistent with the original gradient-affinity model, where it was proposed that the distance along the A-P axis to which different Bicoid-dependent genes are expressed would depend on the affinity of the Bicoid-binding sites in their upstream regulatory regions (11). In the case of Bicoid cooperativity mutants, their overall affinity for multisite DNA target genes is lowered, which likely restricts their ability to activate genes further along the A-P axis, consistent with the original model. However, more is likely to be involved than DNA site occupancy, because the affinity of some Bicoid-site enhancers does not strictly correlate with the A-P expression (23) and because other transcription regulators play a critical role in determining A-P expression (15). Critical differences in the arrangement of Bicoid binding sites within an enhancer also effect synergistic interactions by Bicoid with coactivators, such as CREB-BP, thus altering transcription output (20). Such mechanisms are independent of Bicoid site-occupancy per se.
Expression of genes nearest the anterior, where Bicoid concentrations are the highest, was not affected in our mutants (e.g., ems, btd, otd), whereas, hb, which is normally expressed further down the gradient, was strongly affected. All of these genes carry multiple Bicoid binding sites, but the affinities of their enhancers have not been experimentally compared. Bicoid cooperativity mutants were also unable to generate the correct amount of hb expression within the spatially restricted domain, i.e., the intensity of hb expression was often weaker and more variable. This pattern is consistent with a failure of the mutant proteins to cooperatively load the multiple binding sites typicially found in Bicoid-dependent genes.
Previous work in which hb was misexpressed by using cisacting regulatory elements of bcd indicated that the sharp posterior border of hb expression is necessary for proper segmentation (52). It has been hypothesized that this sharp border is generated from the broad positional information inherent in the Bicoid protein gradient by a mechanism that relies, at least in part, on cooperative DNA binding (22, 23). A simple prediction of our model was that this sharp posterior border would be disrupted in bcd cooperativity mutant embryos, which was indeed what we observed.
The magnitude of this disruption, however, was rather modest, indicating that the Bicoid K57R and S35T mutants may not be completely deficient in cooperative interactions or that additional mechanisms are involved in setting this border. For example, we did not explicity test transcriptional synergy (6, 53), which may be important for Bicoid activity (17, 20, 54). Also, in Drosophila embryos, and perhaps in other organisms, there may be independent mechanisms (e.g., stau-dependent) to ensure that morphogenetic gradients undergo “error correction” to enhance both accuracy and precision in their interpretation (13, 55). Thus, in addition to properties intrinsic to Bicoid, such as DNA binding cooperativity, additional refinement mechanisms may help sharpen the borders of Bicoid-dependent gene expression.
Supplementary Material
Acknowledgments
We thank C. Desplan (New York University, New York), P. Gergen (Stony Brook University), M. Levine (University of California, Berkeley), J. Mohler (Barnard College, New York), S. Small (New York University), and E. Wimmer (University of Goettingen, Goettingen, Germany) for valuable reagents and the Wadsworth Center Molecular Genetics Core, Media, and Microscope Facilities, N. Singh, and W. J. Wolfgang for comments on the manuscript. E.S. and D.S.B. were supported by the Albany College of Pharmacy, F.J.P.L. and J.R. by National Institutes of Health Grants RR07801 and TW01147, and C.E.V.-A. by the Brazilian National Council of Technological and Scientific Development. We are especially grateful for support from March of Dimes Grant RG-1-FY01-14 and American Cancer Society Grants RPG-DB128 and RSG-9508506-DDC (to S.D.H.). S.D.H. is an American Cancer Society Research Scholar.
Author contributions: F.J.P.L., C.E.V.-A., D.S.B., and S.D.H. designed research; D.L., M.F., E.S., F.J.P.L., and C.E.V.-A. performed research; D.L., M.F., F.J.P.L., C.E.V.-A., J.R., D.S.B., and S.D.H. analyzed data; and D.L., C.E.V.-A., D.S.B., and S.D.H. wrote the paper.
Abbreviations: A-P, anterior–posterior; bcd, bicoid; hb, hunchback; Kr, Krüppel.
References
- 1.Ptashne, M. (2004) A Genetic Switch: Phage Lambda Revisited (Cold Spring Harbor Lab. Press, Plainview, NY).
- 2.Johnson, A. D., Meyer, B. J. & Ptashne, M. (1979) Proc. Natl. Acad. Sci. USA 76, 5061-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koudelka, G. B. (2000) Curr. Biol. 10, R704-R707. [DOI] [PubMed] [Google Scholar]
- 4.Johnson, A. D. & Herskowitz, I. (1985) Cell 42, 237-247. [DOI] [PubMed] [Google Scholar]
- 5.Courey, A. J. (2001) Curr. Biol. 11, R250-R252. [DOI] [PubMed] [Google Scholar]
- 6.Carey, M., Lin, Y. S., Green, M. & Ptashne, M. (1990) Nature 345, 361-364. [DOI] [PubMed] [Google Scholar]
- 7.Driever, W. & Nusslein-Volhard, C. (1988) Cell 54, 83-93. [DOI] [PubMed] [Google Scholar]
- 8.Driever, W. & Nüsslein-Volhard, C. (1988) Cell 54, 95-104. [DOI] [PubMed] [Google Scholar]
- 9.Driever, W. & Nüsslein-Volhard, C. (1989) Nature 337, 138-143. [DOI] [PubMed] [Google Scholar]
- 10.Rivera-Pomar, R., Lu, X., Perrimon, N., Taubert, H. & Jäckle, H. (1995) Nature 376, 253-256. [DOI] [PubMed] [Google Scholar]
- 11.Driever, W., Thoma, G. & Nüsslein-Volhard, C. (1989) Nature 340, 363-367. [DOI] [PubMed] [Google Scholar]
- 12.Ronchi, E., Treisman, J., Dostatni, N., Struhl, G. & Desplan, C. (1993) Cell 74, 347-355. [DOI] [PubMed] [Google Scholar]
- 13.Houchmandzadeh, B., Wieschaus, E. & Leibler, S. (2002) Nature 415, 798-802. [DOI] [PubMed] [Google Scholar]
- 14.Singh, N., Zhu, W. & Hanes, S. D. (2005) Dev. Biol. 278, 242-254. [DOI] [PubMed] [Google Scholar]
- 15.Ochoa-Espinosa, A., Yucel, G., Kaplan, L., Pare, A., Pura, N., Oberstein, A., Papatsenko, D. & Small, S. (2005) Proc. Natl. Acad. Sci. USA 102, 4960-4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu, W. & Hanes, S. D. (2000) Gene 245, 329-339. [DOI] [PubMed] [Google Scholar]
- 17.Simpson-Brose, M., Treisman, J. & Desplan, C. (1994) Cell 78, 855-865. [DOI] [PubMed] [Google Scholar]
- 18.Zhao, C., Fu, D., Dave, V. & Ma, J. (2003) J. Biol. Chem. 278, 43901-43909. [DOI] [PubMed] [Google Scholar]
- 19.Torigoi, E., Bennani-Baiti, I. M., Rosen, C., Gonzalez, K., Morcillo, P., Ptashne, M. & Dorsett, D. (2000) Proc. Natl. Acad. Sci. USA 97, 2686-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu, D., Wen, Y. & Ma, J. (2004) J. Biol. Chem. 279, 48725-48733. [DOI] [PubMed] [Google Scholar]
- 21.Zhu, W., Foehr, M., Jaynes, J. B. & Hanes, S. D. (2001) Dev. Genes Evol. 211, 109-117. [DOI] [PubMed] [Google Scholar]
- 22.Ma, X., Yuan, D., Diepold, K., Scarborough, T. & Ma, J. (1996) Development (Cambridge, U.K.) 122, 1195-1206. [DOI] [PubMed] [Google Scholar]
- 23.Burz, D. S., Rivera-Pomar, R., Jackle, H. & Hanes, S. D. (1998) EMBO J. 17, 5998-6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vashee, S., Melcher, K., Ding, W.V., Johnston, S. A. & Kodadek, T. (1998) Curr. Biol. 8, 452-458. [DOI] [PubMed] [Google Scholar]
- 25.Burz, D. S. & Hanes, S. D. (2001) J. Mol. Biol. 305, 219-230. [DOI] [PubMed] [Google Scholar]
- 26.Hanes, S. D. & Brent, R. (1989) Cell 57, 1275-1283. [DOI] [PubMed] [Google Scholar]
- 27.Hanes, S. D. & Brent, R. (1991) Science 251, 426-430. [DOI] [PubMed] [Google Scholar]
- 28.Kissinger, C. R., Liu, B., Martin-Blanco, E., Kornberg, T. B. & Pabo, C. O. (1990) Cell 63, 579-590. [DOI] [PubMed] [Google Scholar]
- 29.Gyuris, J., Golemis, E., Chertkov, H. & Brent, R. (1993) Cell 75, 791-803. [DOI] [PubMed] [Google Scholar]
- 30.Louvion, J.-F., Havaux-Copf, B. & Picard, D. (1993) Gene 131, 129-134. [DOI] [PubMed] [Google Scholar]
- 31.Golemis, E. A. & Brent, R. (1992) Mol. Cell. Biol. 12, 3006-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bürglin, T. (1993) in Guidebook to the Homeobox Genes, ed. Duboule, D. (Sambrook and Tooze Scientific in association with Oxford Univ. Press, New York) Vol. 3.
- 33.Berleth, T., Burri, M., Thoma, G., Bopp, D., Richstein, S., Frigerio, G., Noll, M. & Nusslein-Volhard, C. (1988) EMBO J. 7, 1749-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujioka, M., Jaynes, J. B., Bejsovec, A. M. & Weir, M. (2000) Production of Transgenic Drosophila in Methods in Molecular Biology, Vol. 136, Developmental Biology Protocols, eds. Tuan, R. & Lo, C. (Humana Press, Totowa, NJ), Vol. 2, pp. 353-364. [DOI] [PubMed] [Google Scholar]
- 35.Stern, D. L. & Sucena, E. (2000) in Drosophila Protocols, ed. Hawley, S. R. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 602-614.
- 36.Wieschaus, E. & Nüsslein-Volhard, C. (1986) in Drosophila: A Practical Approach, ed. Roberts, D. B. (IRL, Oxford), pp. 199-227.
- 37.Tautz, D. & Pfeifle, C. (1989) Chromosoma 98, 81-85. [DOI] [PubMed] [Google Scholar]
- 38.Kosman, D., Reinitz, J. & Sharp, D. H. (1998) Pac. Symp. Biocomput., 6-17. [PubMed]
- 39.Janssens, H., Kosman, D., Vanario-Alonso, C. E., Jaeger, J., Samsonova, M. & Reinitz, J. (2005) Dev. Genes Evol. 215, 374-381. [DOI] [PubMed] [Google Scholar]
- 40.Frohnhöfer, H. G. & Nüsslein-Volhard, C. (1986) Nature 324, 120-125. [Google Scholar]
- 41.Poustelnikova, E., Pisarev, A., Blagov, M., Samsonova, M. & Reinitz, J. (2004) Bioinformatics 20, 2212-2221. [DOI] [PubMed] [Google Scholar]
- 42.Reinitz, J., Mjolsness, E. & Sharp, D. H. (1995) J. Exp. Zool. 271, 47-56. [DOI] [PubMed] [Google Scholar]
- 43.Kraut, R. & Levine, M. (1991) Development (Cambridge, U.K.) 111, 601-609. [DOI] [PubMed] [Google Scholar]
- 44.Hoch, M., Seifert, E. & Jackle, H. (1991) EMBO J. 10, 2267-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Astromoff, A. & Ptashne, M. (1995) Proc. Natl. Acad. Sci. USA 92, 8110-8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barbaric, S., Munsterkotter, M., Goding, C. & Horz, W. (1998) Mol. Cell. Biol. 18, 2629-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Filutowicz, M., York, D. & Levchenko, I. (1994) Nucleic Acids Res. 22, 4211-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith, T. L. & Sauer, R. T. (1995) J. Mol. Biol. 249, 729-742. [DOI] [PubMed] [Google Scholar]
- 49.Gottfried, P., Kolot, M., Silberstein, N. & Yagil, E. (2004) FEBS Lett. 577, 17-20. [DOI] [PubMed] [Google Scholar]
- 50.Dodd, I. B., Shearwin, K. E., Perkins, A. J., Burr, T., Hochschild, A. & Egan, J. B. (2004) Genes Dev. 18, 344-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spirov, A. V. & Holloway, D. M. (2003) In Silico Biol. 3, 89-100. [PubMed] [Google Scholar]
- 52.Schulz, C. & Tautz, D. (1994) Development (Cambridge, U.K.) 120, 3043-3049. [DOI] [PubMed] [Google Scholar]
- 53.Chi, T., Lieberman, P., Ellwood, K. & Carey, M. (1995) Nature 377, 254-257. [DOI] [PubMed] [Google Scholar]
- 54.Sauer, F., Hansen, S. K. & Tjian, R. (1995) Science 270, 1783-1788. [DOI] [PubMed] [Google Scholar]
- 55.Aegerter-Wilmsen, T., Aegerter, C. M. & Bisseling, T. (2005) J. Theor. Biol. 234, 13-19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.