biochemistry. For the article “Assembly of the bacteriophage T4 primosome: Single-molecule and ensemble studies,” by Zhiquan Zhang, Michelle M. Spiering, Michael A. Trakselis, Faoud T. Ishmael, Jun Xi, Stephen J. Benkovic, and Gordon G. Hammes, which appeared in issue 9, March 1, 2005, of Proc. Natl. Acad. Sci. USA (102, 3254–3259; first published February 22, 2005; 10.1073/pnas.0500327102), the authors note that Figs. 1 A, 2, and 3 were prepared incorrectly and are not accurate representations of the data. The corrected figures and their legends appear below. These errors do not alter the conclusions of the article.
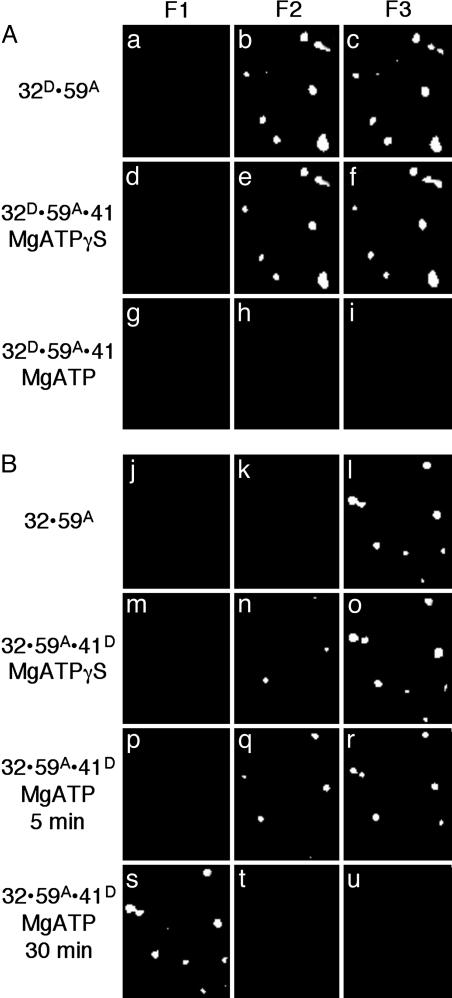
Fig. 1.
Protein–protein interactions between gp32 and gp59 on fDNA. (A) The fluorescence from individual molecules of fDNA with the proteins bound in the order as indicated at the side of each row. The gp32 protein is labeled with A488 (gp32D) and the gp59 protein is labeled with A555 (gp59A). The filter sets are described in Experimental Methods: F1 is for A488 emission, F2 for FRET between A488 and A555, and F3 for A555 emission. (B) Ensemble FRET studies of Oregon-green-488-maleimide-labeled gp59 titrated into a solution of 400 nM CPM-labeled gp32 and 100 nM fDNA. The fluorescence spectra of 400 nM CPM-gp32 alone (black line), the endpoint of the titration at 1 μM Oregon-green-488-maleimide-gp59 (dark gray line), and several intermediate spectra (light gray lines) are shown. (C) Analysis of the donor quenching and acceptor sensitization plotted against the gp59 concentration determines the stoichiometry among gp32, gp59, and fDNA to be 1:1:1 with a calculated binding constant of ≈40 nM.
Fig. 2.
The ATP-dependent loading of gp41 by gp59 onto fDNA coated with gp32. The fluorescence from individual molecules of fDNA with the proteins bound in the order as indicated at the side of each row. (A) The gp32 protein is labeled with A488 (gp32D), the gp59 protein is labeled with A555 (gp59A), and the gp41 protein is unlabeled. MgATPγS (500 μM) is present for the sample in row 2, and 500 μM MgATP is present for the sample in row 3. (B) The gp32 protein is unlabeled, the gp59 protein is labeled with A555 (gp59A), and the gp41 is labeled with A488 (gp41D). MgATPγS (500 μM) is present for the sample in row 2, and 500 μM MgATP is present for the sample in row 3 (5 min after addition of gp41 and nucleotide) and in row 4 (30 min after addition of gp41 and nucleotide). The filter sets are described in the legend to Fig. 1.
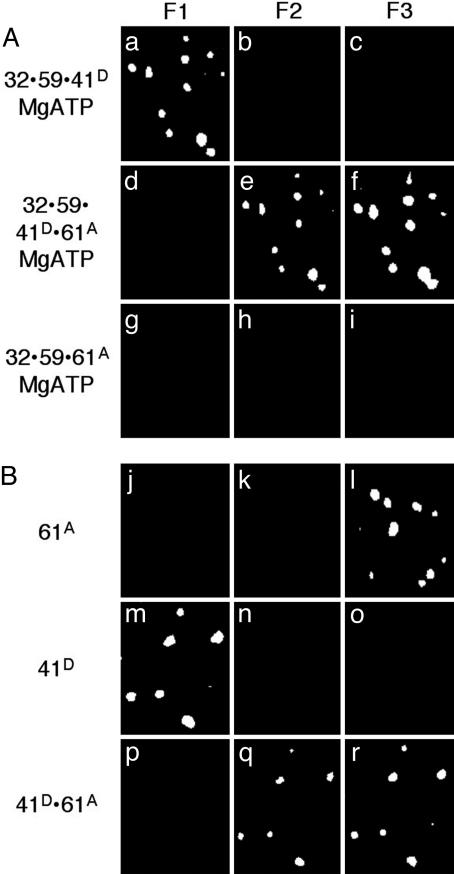
Fig. 3.
Completed primosome formation by the addition of gp61 or an alternative, direct formation of the gp41/gp61 primosome. The fluorescence from individual molecules of fDNA with the proteins bound in the order as indicated at the side of each row. (A) The gp32 and gp59 proteins are unlabeled, the gp41 protein is labeled with A488 (gp41D), and the gp61 protein is labeled with A555 (gp61A) in the presence of 500 μM MgATP. (B) The gp41 protein is labeled with A488 (gp41D), and the gp61 protein is labeled with A555 (gp61A) in the absence of MgATP. The filter sets are described in the legend to Fig. 1.