Abstract
Breast cancer remains one of the most common cancers in women worldwide. Early detection is critical for improving patient outcomes, yet current screening methods have limitations. Therefore, there is a pressing need for more sensitive and specific approaches to detect breast cancer in its earliest stages. Liquid biopsy has emerged as a promising non-invasive method for early cancer detection and management. DNA methylation, an epigenetic alteration that often precedes genetic changes, has been observed in precancerous or early cancer stages, making it a valuable biomarker. This review explores the role of DNA methylation in breast cancer and its potential for developing blood-based tests. We discuss advancements in DNA methylation detection methods, recent discoveries of potential DNA methylation biomarkers from both single-omics and multi-omics integration studies, and the role of machine learning in enhancing diagnostic accuracy. Challenges and future directions are also addressed. Although challenges remain, advances in multi-omics integration and machine learning continue to enhance the clinical potential of methylation-based biomarkers. Ongoing research is crucial to further refine these approaches and improve early detection and patient outcomes.
Keywords: Breast cancer, DNA methylation, Liquid biopsy, Early detection, Biomarker, Multi-omics, Machine learning
Background
Breast cancer is the most commonly diagnosed malignancy among women and remains a leading cause of cancer-related mortality. Early detection is crucial for improving treatment outcomes and survival rates. While mammography remains the gold standard for breast cancer screening, its sensitivity is reduced in women with dense breast tissue, increasing the risk of missed diagnoses [1]. Additionally, mammography can lead to false positives and overdiagnosis, resulting in unnecessary biopsies and overtreatment [2]. These limitations underscore the need for highly sensitive and specific biomarkers to improve early detection and risk assessment.
DNA methylation, a key epigenetic modification, regulates gene expression without altering the DNA sequence and is essential for normal cellular function. However, aberrant DNA methylation plays a pivotal role in cancer development. In breast cancer, hypermethylation of tumor suppressor genes (e.g. BRCA1, ITHI5, RASSF1 A) and global hypomethylation contribute to disease development and progression [3–5]. Methylation changes often arise early in tumorigenesis, preceding genetic mutations, making them an attractive biomarker for early detection [6, 7]. Unlike the heterogeneity of gene mutations, DNA methylation patterns are more consistent within tumor cells of the same type and across larger genomic regions, enabling the use of multiple CpGs for detection [8]. Additionally, distinct methylation signatures can differentiate breast cancer subtypes, such as triple-negative breast cancer (TNBC), aiding in patient stratification and personalized treatment approach [9].
Liquid biopsy has emerged as a promising, minimally invasive alternative to traditional tissue biopsies for detecting tumor-derived biomarkers in bodily fluids such as blood and plasma. Unlike tissue biopsies, which are invasive, difficult to repeat, and subject to sampling bias, liquid biopsy offers a less invasive and easily repeatable testing option [10]. Additionally, it can detect minimal amounts of tumor DNA, which may be missed by traditional imaging techniques. Among the tumor-derived components detectable in liquid biopsy, circulating tumor DNA (ctDNA) has demonstrated strong potential for cancer detection and monitoring [11, 12]. Liquid biopsy-based approaches have further expanded the clinical application of DNA methylation biomarkers, enabling the non-invasive detection of ctDNA methylation, which provides valuable insights into tumor biology without requiring tissue samples. ctDNA, a fraction of cell-free DNA (cfDNA), originates from apoptotic or necrotic tumor cells and carries genetic and epigenetic alterations that mirror the tumor tissue. This makes ctDNA methylation analysis a powerful strategy for early breast cancer detection. However, detecting tumor-specific ctDNA remains challenging, particularly in early-stage cancers due to their low abundance. This necessitates the need for highly sensitive detection technologies capable of distinguishing tumor-derived methylation patterns from background cfDNA.
Advancements in next-generation sequencing (NGS) and PCR-based techniques have significantly improved the detection of ctDNA methylation. PCR-based methods, such as droplet-digital PCR (ddPCR), offer high sensitivity for detecting CpG sites, making them valuable for targeted biomarker validation. In contrast, NGS-based approaches, including whole-genome bisulfite sequencing (WGBS) and reduced representation bisulfite sequencing (RRBS), provide single-base resolution and high-throughput methylation profiling, enabling more comprehensive analysis. To enhance ctDNA methylation detection, various studies have optimized these methods to address challenges such as low ctDNA abundance and high fragmentation [13–18]. Recent advances in bisulfite-free sequencing, such as enzymatic methylation sequencing (EM-seq) and Tet-assisted pyridine borane sequencing (TAPS), have improved DNA integrity preservation and sequencing efficiency, minimizing errors associated with bisulfite conversion [19–22]. Additionally, direct methylation detection technologies such as Single-Molecule Real-Time Sequencing (SMRT-seq) and Oxford Nanopore sequencing eliminate the need for bisulfite treatment, allowing for base modification detection in native DNA molecules [23, 24]. These approaches provide long-read sequencing capabilities, which enhance the resolution of methylation patterns in fragmented ctDNA, improving sensitivity for ctDNA detection. Machine learning algorithms are also increasingly applied to identify cancer-specific methylation patterns and integrate multi-omics data, enhancing biomarker discovery and data interpretation accuracy [25, 26].
In this review, we focus on DNA methylation-based biomarkers for breast cancer, particularly those with early detection or diagnostic potential identified in studies published between 2019 and January 2025. We also explore how multi-omics or integrative analysis approaches have significantly enhanced biomarker discovery. Our comprehensive literature search, conducted on PubMed and Google Scholar, included terms such as “breast cancer,” “DNA methylation,” “liquid biopsy,” “ctDNA,” “multi-omics,” “integrative analysis,” “diagnosis,” “early detection,” and “biomarker.” The final reference list was curated based on originality and relevance to the scope of this review. To provide a structured overview, Table 1 compares various DNA methylation detection methods, outlining their advantages, limitations, and suitability for low-input samples. Table 2 presents DNA methylation biomarkers for breast cancer, including those in clinical trials and ongoing research, while Table 3 summarizes studies that have integrated multi-omics approaches with DNA methylation analysis. Figures 1 and 2 provide visual summaries of key methodologies, with Fig. 1 illustrating various DNA methylation detection methods and Fig. 2 depicting the general workflow for single-omics and multi-omics analyses. This review highlights recent advancements in DNA methylation biomarker research and its integration with multi-omics strategies, emphasizing their potential for early breast cancer detection and diagnosis.
Table 1.
Comparison of DNA methylation detection methods
| Method | Technology | Coverage type | DNA input requirementa | Detection sensitivityb | Throughput | Workflow complexity |
Cost estimate ($–$$$$) |
Optimized for low-input samples | Best for | References |
|---|---|---|---|---|---|---|---|---|---|---|
| Whole-Genome Bisulfite Sequencing (WGBS) | Short-read NGS (Illumina) | Whole-genome (single-base resolution) | ≥ 100 ng |
High (~ 99% sensitivity at ≥ 30x) |
High | High (bisulfite conversion, library preparation, deep sequencing, bioinformatics) | $$$$ | Yes | Comprehensive methylation profiling | [13, 71, 119–121] |
| Reduced Representation Bisulfite Sequencing (RRBS) | Short-read NGS (Illumina) | Epigenome-wide (CpG-rich regions, e.g. promoter, CpG islands, etc.) | ≥ 30 ng |
Moderate (covers ~ 10% of CpGs) |
Moderate | High (enzyme digestion, library preparation, size selection, bisulfite conversion, PCR, sequencing, bioinformatics) | $$$ | Yes | Large-scale, cost-effective methylation analysis | [14, 38, 121–124] |
| Targeted Methylation Sequencing | Short-read NGS (Illumina) (Hybrid capture/Amplicon-based) | Targeted CpG sites (custom panels) | ≥ 100 ng | Moderate (selected cancer-specific regions) | Moderate | High (bisulfite conversion, library preparation, target enrichment (hybrid capture/amplicon-based), sequencing, bioinformatics) | $$–$$$ (varies by panel size) | Yes | Liquid biopsy, cancer biomarker panels | [42, 43, 125] |
|
Enzymatic Methylation Sequencing (EM-Seq) |
Short-read NGS (Illumina) | Whole genome (can be adapted for target enrichment workflow) | ≥ 10 ng | High (~ 99% sensitivity at ≥ 30x) | High (like WGBS as both require deep sequencing for whole methylome coverage) | High (enzymatic conversion, library preparation, sequencing, bioinformatics) | $$$$ | Yes | Bisulfite-free methylation analysis (preserves DNA integrity, suitable for low-input samples) | [19, 20, 44] |
| Tet-Assisted Pyridine Borane Sequencing (TAPS) | Short-read NGS (Illumina) (can be adapted for long-read sequencing) | Epigenome-wide | ≥ 100 ng | High (at ≥ 30x, accurate detection of both 5 mC and 5 hmC) | High | Moderate (enzymatic conversion, library preparation, sequencing, bioinformatics) | $$$ | Yes | Bisulfite-free methylation analysis (less DNA degradation, suitable for low-input samples) | [21, 22, 45, 126, 127] |
|
Methylated DNA Immunoprecipitation Sequencing (MeDIP-Seq) |
Affinity-based NGS (uses 5 mC antibodies to capture methylated DNA) | Epigenome-wide (targets low CpG density regions, covering > 95% of the genome) | ≥ 50 ng | Moderate (bias towards regions with lower CpG density) | Moderate | Moderate (antibody-based enrichment, library preparation, sequencing, bioinformatics) | $$$ | Yes | Suitable for analyzing methylation patterns across large genomic regions | [15, 121, 128, 129] |
| Methylated DNA Capture Sequencing (MethylCap-Seq) | Affinity-based NGS (uses MBD domain to capture methylated DNA) | Epigenome-wide (regions with high CpG density, ~ 50–80% of CpG islands across the genome) | ≥ 1 µg | Moderate | Moderate | Moderate (MBD-based capture, library preparation, sequencing, bioinformatics) | $$$ | Yes | Cost-effective way to analyze methylation patterns in CpG-rich areas | [130–132] |
| Illumina Infinium MethylationEPIC v2.0 | Microarray | Targeted (predefined 930,000 CpG sites) | ≥ 250 ng | Moderate (for targeted CpGs, lower sensitivity for rare methylation events) | Moderate | Low (array hybridization, scanning) | $$ | No | Large scale, epigenome-wide association studies | [34, 36] |
| Pyrosequencing | Sequencing-by-synthesis | Targeted CpG regions | ≥ 20 ng | Low (detects ≥ 5% methylation) | Low (several CpG sites within an amplicon) | Low (direct sequencing, PCR validation) | $ | Yes | Clinical assays, biomarker validation | [133–136] |
| Quantitative Methylation-specific PCR (qMSP) | PCR-based detection | Targeted CpG sites | ≥ 20 ng | High (LOD of 0.1%) | Low (one to a few CpGs) | Low (primer design, bisulfite conversion, qPCR-based method) | $ | Yes | Locus-specific methylation detection, clinical diagnostics, biomarker validation | [27, 137–139] |
| Droplet Digital PCR (ddPCR) | Digital PCR | Targeted CpG sites | ≥ 1 ng | Very high (LOD of 0.001%) | Low (one to a few CpGs) | Low (primer design, bisulfite conversion, digital quantification) | $ | Yes | Ultra-sensitive detection for low-abundance methylation signals, biomarker validation | [13, 140–142] |
|
PacBio Single Molecule Real-time Sequencing (SMRT-Seq) |
Long-read NGS (PacBio) | Epigenome-wide or targeted | ≥ 100 ng | Variable (long-range detection) | Moderate | Moderate (long-read library preparation, sequencing, base calling, bioinformatic) | $$$$ | Yes | Long-range phased methylation analysis | [23, 46, 127, 143, 144] |
| Oxford Nanopore Sequencing | Long-read NGS (Nanopore) | Epigenome-wide or targeted | ≥ 50 ng | Variable (depending on depth) | Scalable | Moderate (library prep, sequencing, base calling, bioinformatics) | $$$$ | Yes | Long-read direct methylation detection | [24, 47, 144, 145] |
LOD limit of detection, MBD methyl-CpG-binding domain, NGS next-generation sequencing
aDNA input requirement listed are estimates for genomic DNA (gDNA). Actual amounts may vary depending on the specific kit and protocol used. Optimized methods for cfDNA/ctDNA or low-input samples require less DNA
bLOD will depend on the specific assay design, target gene, and instrument used
Table 2.
Summary of recent liquid biopsy-based DNA methylation studies in breast cancer
| Study (authors) | Cancer types | Markers | Sample | Detection method (s) | No. of cases | No. of controls | AUC | Sens (%) | Stage I/II Sens (%) | Spec (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| For detection | ||||||||||
| Mijnes et al. [31] | Breast | SPAG6, NKX2-6, ITIH5, PER1 | Plasma | 450 K array, pyrosequencing | 111 | 14 | 0.729 | 63 | – | 80 |
| Moss et al. [72] | Breast | KRT19, LMX1B, ZNF296 | Plasma | TBS | 30 | 64 | 0.913 | 80 | – | 97 |
| Liu et al. [64] | Breast |
10 cfDNA markers (RYR2, RYR3, GABRB3, DCDC2 C, AC096570.1, LINC00923) |
Plasma | WGBS | 101 | 102 | 0.81 | – | – | – |
| Wang et al. [28] | Breast | CCDC181, GCM2, ITPRIPL1 | Plasma | EPIC (850 K) array, qMSP | 199 | 247 | 0.961 | 92.9 | – | 87.5 |
| Zhang et al. [43] | Breast | 26 cfDNA markers | Plasma | TBS | 204 | 129 | 0.945 | 87.76 | 87.5 | 92.73 |
| Gao et al. [13] | Breast | 15 ctDNA DMRs | Plasma | WGBS, ddPCR | 123 | 40 | 0.967 | 87 | – | 100 |
| Kresovich et al. [35] | Breast | 19 CpGs & 5 DNA methylation estimators | Blood | 450 K array | 1728 | 1375 | 0.69 | – | – | – |
| Chung et al. [122] | Breast | 49 genomic regions | Buffy coat | RRBS | 340 | 340 | 0.86 | – | – | – |
| Wang et al. [32] | Breast |
4 CpG markers (cg16652347, cg13828440, cg11754974 & cg18637238) |
PBMC | 850 K (EPIC) array, pyrosequencing, TBS | 491 | 290 | 0.918 | 80.3 | 85.7 | 89.1 |
| Lee et al. [34] | Breast | 51 CpG sites | Blood | EPIC array | 256 | 268 | 0.827 | 75 | – | 78 |
| Liu et al. [71] | Multiple (> 50 cancer types, including BC) | A classifier developed based on targeted methylation panel of > 100,000 methylation regions (refer to reference [71]) | Plasma | TBS | 2482 | 4207 | – | 54.9 | 18 (for all cancer types); < 5 (for BC) | 99.3 |
| Klein et al. [40] | Multiple (> 50 cancer types, including BC) | A refined assay and classifiers based on Liu et al. (refer to reference [40]) | Plasma | TBS | 2823 | 1254 | – | 51.5 | 16.8 (for all cancer types); 2.6 (for BC) | 99.5 |
| Jamshidi et al. [82] | Multiple (> 20 cancer types, including BC) | Whole-genome methylation classifier (refer to reference [82]) | Plasma | WGBS | 1628 | 1172 | 0.63 | 34 | – | 98 |
| Gao et al. [13] | Breast | 12 ctDNA DMRs | Plasma | WGBS | 78 (ER +) | 43 (ER−) | 0.780 | 73 | – | 87 |
| Manoochehri et al. [30] | Breast | 3 DMRs overlapping with SPAG6, LINC10606, TBCD/ZNF750 | Plasma | 450 K & EPIC arrays, ddPCR | 262 (TNBC) | 84 | 0.74 | – | – | – |
AUC area under the curve, BC breast cancer, ddPCR droplet digital PCR, DMRs differentially methylated regions, ER estrogen receptor, qMSP quantitative methylation-specific PCR, RRBS reduced representation bisulfite sequencing, Sens sensitivity, Spec specificity, TBS targeted bisulfite sequencing, TNBC triple-negative breast cancer, WGBS whole-genome bisulfite sequencing
Table 3.
Integrative DNA methylation studies for breast cancer detection
| Cancer types | Features/signatures | No. of subjects | Sample types | Detection method (s) | Main findings | AUC | Sens (%) | Stage I/II Sens (%) | BC Sens (%) | Spec (%) | TOO (%) | Ref. | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Breast | GW genotype, DNA methylation, gene expression | 122,977 BC, 105,974 controls | WBC | SNP array, Methylation 450 K array, gene expression profiling | The integrative analyses of genetic, DNA methylation, and gene expression identified 38 CpGs that may affect BC risk through regulating expression of 21 genes | – | – | – | – | – | – | [146] | |
| Breast | Gene expression, DNA methylation, DNA copy number | 40 DCIS, 259 IBC | Tissue | Gene expression profiling, Methylation 450 K array, SNP array | Gene expression-based model features achieved the highest classification accuracy, but incorporating methylation data enhanced the sensitivity in classifying DCIS cases | 0.995 | 86.3 | – | – | 99.6 | – | [78] | |
| Breast | GW methylation, CNA, end motif, fragment length | 239 BC, 278 controls | Plasma | Targeted and shallow WGBS | Integrating cfDNA methylation, CNA, and end motif features could enhance the accuracy of early-stage BC detection | 0.91 | 65 | 84 | – | 96 | – | [70] | |
| Multiple (Breast, CRC, gastric, lung, and liver) | cfDNA methylation, fragmentomics, DNA copy number, end motifs |
223 BC, 159 CRC, 98 gastric, 136 lung, 122 liver, and 1550 controls |
Plasma | Targeted and shallow WGBS | The combination of multimodal analysis of cfDNA signatures and machine learning algorithms accurately detected and localized five types of cancer using low-pass sequencing | 0.95 | 72.4 | 73.9 | 49.3 | 97 | 70 | [83] | |
| Multiple (Breast, CRC, esophagus, liver, lung, pancreas, and gastric) | GW cfDNA methylation, fragmentation, CNA | 66 BC, 150 CRC, 61 esophageal, 113 liver, 157 lung, 119 pancreas, 114 gastric, and 497 controls | Plasma | Shallow WMS | Integrating cfDNA methylation and fragmentation signatures at cancer-specific accessible chromatin regions accurately distinguished cancer patients from controls and identified the TOO | 0.966 | 83 | 73 | – | 99 | 65 | [84] | |
AUC area under the curve, BC breast cancer, CNA copy number alteration, CRC colorectal, DCIS ductal carcinoma in situ, GW genome-wide, IBC invasive breast cancer, Sens sensitivity, SNP single nucleotide polymorphism, Spec specificity, TOO tissue of origin, WBC white blood cells, WGBS whole-genome bisulfite sequencing, WMS whole methylome sequencing
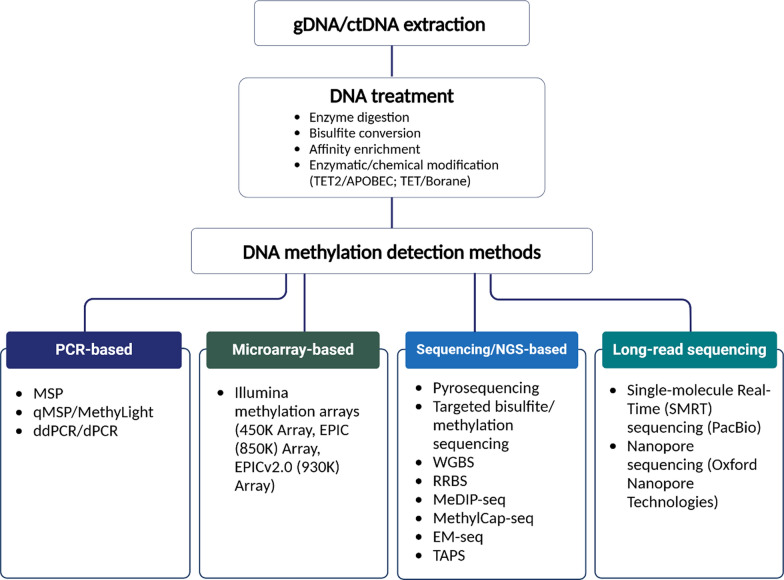
Fig. 1.
Overview of different methods for DNA methylation detection and analysis. This figure presents PCR-based techniques for targeted analysis of specific CpG sites, as well as microarray, next-generation sequencing (NGS), and third-generation sequencing methods that provide a comprehensive view of the methylome, enabling large-scale DNA methylation profiling across the genome. Figure was created with BioRender.com. ctDNA circulating-tumor DNA, ddPCR droplet digital PCR, dPCR digital PCR, EM-seq enzymatic methyl-seq, gDNA genomic DNA, MeDIP-seq methylated DNA immunoprecipitation sequencing, MethylCap-seq methylated DNA capture sequencing, MSP methylation-specific PCR, NGS next generation sequencing, PCR polymerase chain reaction, qMSP quantitative methylation-specific PCR, RRBS reduced representation bisulfite sequencing, TAPS TET-assisted pyridine borane sequencing, WGBS whole-genome bisulfite sequencing
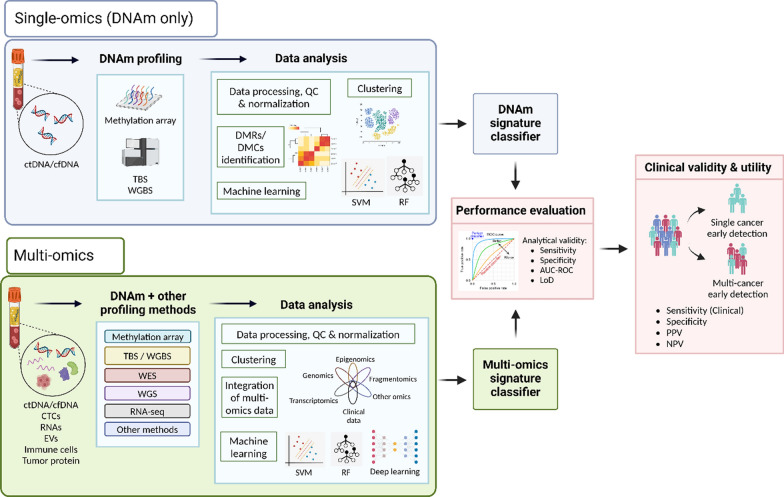
Fig. 2.
General workflow of single-omics and multi-omics approaches in genome-wide DNA methylation analysis. The single-omics approach (in blue box) focuses solely on DNA methylation data, while the multi-omics approach (in green box) integrates DNA methylation with other omics data. Figure was created with BioRender.com. AUC-ROC area under the receiver operating characteristic curve, cfDNA cell-free DNA, cfRNA cell-free RNA, CTCs circulating tumor cells, ctDNA circulating-tumor DNA, DMCs differentially methylated CpG sites, DMRs differentially methylated regions, DNAm DNA methylation, EVs extracellular vesicles, LoD limit of detection, NPV negative predictive value, PPV positive predictive value, RF random forest, RNA-seq RNA sequencing, SVM support vector machine, TBS targeted bisulfite sequencing, WES whole-exome sequencing, WGBS whole-genome bisulfite sequencing, WGS whole-genome sequencing
Advances in DNA methylation detection methods
DNA methylation analysis has emerged as a powerful strategy for early breast cancer detection and biomarker discovery. Advances in detection methods over the past decade have improved assay sensitivity, specificity, and clinical feasibility. Approaches range from locus-specific assays targeting predefined CpG sites to genome-wide profiling for comprehensive methylation assessment (Fig. 1). In this section, we categorize these methods into: (i) established methods, covering commonly used non-NGS-based and NGS-based techniques, and (ii) emerging technologies, highlighting novel methods that are shaping the future of methylation-based biomarker discovery and early detection. A comparative table (Table 1) summarizing key aspects of DNA methylation detection methods has been added to facilitate method selection based on coverage, sensitivity, input requirements, throughput, and clinical applicability.
Established DNA methylation detection methods
Locus-specific methylation detection methods (non-NGS-based)
Locus-specific methylation detection methods remain widely used for targeted analysis and biomarker validation. PCR-based approaches, including methylation-specific PCR (MSP), quantitative MSP (qMSP), and MethyLight, enable cost-effective, sensitive detection of hypermethylated CpG sites in genes such as BRCA1, RASSF1 A, and APC, which are frequently altered in breast cancer [27–29]. These methods allow for the rapid assessment of known epigenetic markers but are inherently limited to predefined CpG sites, restricting their use for de novo biomarker discovery. Advanced techniques like droplet digital PCR (ddPCR) and digital PCR (dPCR) further enhance quantification accuracy, particularly for low-abundance methylation signals in liquid biopsies [13, 30]. Pyrosequencing, a sequencing-by-synthesis method, provides real-time, quantitative methylation analysis across multiple CpG sites, making it valuable for biomarker validation [31, 32]. Meanwhile, methylation arrays, such as Illumina Infinium Methylation BeadChips (450 K, EPIC, and EPIC v2.0), enable high-throughput epigenetic profiling of up to 930,000 CpG sites, facilitating large-scale studies and risk prediction models [32–36]. Despite their efficiency, these non-NGS-based focus on predetermined CpG regions, limiting their ability to capture novel or rare methylation changes in breast cancer development and progression.
Epigenome-wide and targeted methylation sequencing methods (NGS-based)
NGS-based methylation sequencing has transformed methylation profiling by providing single-base resolution across the genome. WGBS remains the gold standard for DNA methylation studies, providing comprehensive and unbiased methylome-wide coverage, however, its high cost and computational complexity limits its routine use [37]. RRBS lowers cost by selectively targeting CpG-rich regions, particularly promoters and enhancers, but its dependence on restriction enzyme digestion results in incomplete genome coverage [38]. Targeted methylation sequencing, using hybrid capture or amplicon-based enrichment, focuses on clinically relevant CpG sites, making it a practical and scalable approach for liquid biopsy applications [39]. These NGS-based methods are gaining traction in multi-cancer early detection (MCED) assays, such as Galleri (GRAIL). However, sensitivity for early-stage breast cancer detection remains a challenge, with detection rates as low as 16.8% for stage I cancers [40]. Additionally, cost, scalability, and ctDNA enrichment continue to be major limitations.
Emerging technologies for liquid biopsy-based breast cancer detection
Low-input and low-pass WGBS
Traditional WGBS requires substantial DNA input, posing challenges for ctDNA analysis in early breast cancer. To overcome this limitation, Gao and colleagues developed an improved ctDNA-WGBS method that generates high-quality methylation profiles using as little as 1 ng of ctDNA, significantly enhancing its feasibility for liquid biopsies [13]. Similarly, low-pass WGBS, which sequences cfDNA at lower depths, has been shown to identify epigenome-wide fragmentation patterns, improving cancer detection sensitivity [41]. cfDNA-adapted methods such as cf-RRBS, cfMeDIP-seq, and cfMethyl-Seq further optimize methylation profiling for fragmented ctDNA, improving sensitivity and specificity in breast cancer detection [14–16].
Targeted methylation sequencing
Targeted methylation sequencing enhances early breast cancer detection by focusing on tumor-specific methylation patterns. By enriching breast cancer-specific epigenetic alterations, this approach improves the detection of ctDNA in liquid biopsy samples, thereby enhancing the sensitivity and specificity of screening assays [42]. Zhang and colleagues developed the AnchorIRIS™ assay, which profiles tumor-derived methylation signatures from low-input cfDNA, achieving 89.37% sensitivity and 100% specificity [43]. More recently, Enhanced Linear-Splinter Amplification Sequencing (ELSA-seq) has improved early cancer detection by increasing methylation signal recovery, achieving 52–81% sensitivity and 96% specificity [17, 18]. By integrating machine learning, ELSA-seq enhances ctDNA signal differentiation, improving early detection accuracy. These findings highlight the growing potential of targeted methylation sequencing in biomarker-driven early detection strategies for breast cancer. However, low ctDNA levels in early-stage cancers reduce detection reliability, highlighting the need for optimized methylation panels and ultra-sensitive detection technologies to improve early breast cancer screening and risk assessment [41].
Bisulfite-free methylation detection
Bisulfite-free methylation detection methods address the limitations of bisulfite conversion, which can degrade DNA and introduce sequencing bias. Enzymatic Methylation Sequencing (EM-Seq) and TET-assisted Pyridine Borane Sequencing (TAPS) preserve DNA integrity and sequencing accuracy by eliminating the need for harsh bisulfite conversion. EM-Seq utilizes enzyme-based modifications to selectively protect methylated cytosines while deaminating unmethylated cytosines [19, 20]. An advanced high-multiplex methylation detection technique, IMPRESS, integrates EM-Seq with single-molecule Molecular Inversion Probes (smMIP) to further enhance methylation detection from low-input cfDNA (5 ng), achieving 92% sensitivity and 98% specificity for breast cancer-specific markers [44]. TAPS, on the other hand, utilizes oxidation–reduction chemistry to convert 5-methylcytosine (5 mC) into thymine without DNA fragmentation, offering improved methylation resolution for cfDNA analysis [21, 22]. A recent advancement in this method enabled deep sequencing (80x) of cfDNA, supporting multi-modal analysis of copy number aberrations, somatic mutations, and DNA methylation, achieving an AUC of 86% at ctDNA fractions as low as 0.7% [45]. Additionally, bisulfite-free methods have demonstrated enhanced compatibility with long-read sequencing platforms such as Oxford Nanopore and PacBio SMRT sequencing (see section on ‘Direct Methylation Detection via Long-Read Sequencing’), allowing for simultaneous methylation and mutation analysis in breast cancer liquid biopsy samples. By overcoming the limitations of bisulfite conversion, these emerging techniques offer a more reliable and scalable approach for detecting methylation-based biomarkers, paving the way for improved breast cancer screening and personalized treatment strategies.
Direct methylation detection via long-read sequencing
Long-read sequencing technologies, such as PacBio SMRT sequencing and Oxford Nanopore, have advanced direct methylation detection by enabling real-time identification of 5 mC and 5 hmC modifications without requiring bisulfite conversion. SMRT sequencing detects polymerase kinetics to identify methylation status, allowing simultaneous methylation and mutation analysis [23]. Circular consensus sequencing (CCS) in PacBio SMRT, enhances read accuracy beyond 99.9%, improving methylation profiling from low input cfDNA [46]. Oxford Nanopore sequencing directly measures 5 mC and 5 hmC in real time by detecting ionic current changes, making it a promising tool for liquid biopsy-based methylation analysis [24]. A recent advancement by Lau et al. introduced a nanopore-based single-molecule sequencing approach that preserves native cfDNA integrity while using machine learning to enhance direct methylation profiling [47]. This method improves sequencing throughput, generating up to 200 million reads per cfDNA sample and distinguishing tumor-derived cfDNA methylation patterns from immune cell-derived cfDNA. While these technologies offer unparalleled advantages in long-range methylation analysis and structural variant detection, challenges such as sequencing error rates and computational demands still limit their widespread clinical adoption [48]. Continued optimization in accuracy, cost reduction, and bioinformatics pipelines will be essential for their full integration into breast cancer early detection and biomarker discovery.
Computational strategies and machine learning
Low-pass WGBS reduces sequencing costs but compromises single-base resolution, necessitating computational approaches to reconstruct missing methylation data. Advanced bioinformatics tools such as METHimpute and Methylation Rate Modeling (MRM) mitigate this limitation by inferring methylation status in low-coverage regions using Hidden Markov Models and smoothed methylation rates, respectively, thereby enhancing tumor-specific methylation detection in cfDNA [49, 50]. Beyond imputation, computational methods play a critical role in differential methylation analysis and integrating results with clinical data to better understand epigenetic alterations in breast cancer [51]. As methylation datasets grow, robust bioinformatics pipelines and statistical frameworks are increasingly essential for accurate interpretation [52–54].
Machine learning (ML) has become a powerful tool for cancer detection, facilitating biomarker discovery and uncovering links between methylation patterns and disease phenotypes [25]. Feature selection techniques have been employed to identify informative CpG sites, reducing dimensionality while preserving classification accuracy [55]. Recent ML-driven approaches have further refined DNA methylation-based classifiers, with deep-learning models such as attention-based neural networks enhancing predictive power in breast cancer detection [56]. Additionally, deep embedded clustering techniques have demonstrated high precision in distinguishing breast cancer subtypes based on DNA methylation signatures, supporting their utility in refining diagnostic models [57]. Furthermore, integrative multi-omics analyses, which combine methylation data with transcriptomics, proteomics, and other molecular profiles, leverage ML to improve predictive accuracy in cancer detection [26]. Ensemble learning techniques, which integrate multiple classification models, enhance model performance and robustness, refining classification models for early breast cancer detection and risk assessment [51]. These computational advancements enhance the feasibility and accuracy of emerging DNA methylation analysis methods, advancing non-invasive breast cancer screening and the evolution of methylation-based biomarker discovery.
NGS-based methylation sequencing in breast cancer
NGS has revolutionized DNA methylation analysis, enabling high-throughput epigenetic profiling for breast cancer detection, monitoring, and treatment response. Methylation sequencing is categorized into tissue-based and liquid biopsy-based approaches. While tissue-based sequencing provides high-resolution tumor methylation profiles, it is invasive and unsuitable for repeated monitoring. In contrast, liquid biopsy-based sequencing allows non-invasive ctDNA detection, making it valuable for early detection and longitudinal disease surveillance.
Liquid biopsy approaches are further divided into tumor-informed and tumor-naïve strategies. Tumor-informed methods leverage known tumor methylation signatures to improve specificity but are limited when prior tumor data is unavailable. Tumor-naïve methods identify cancer-associated methylation markers de novo using machine learning, offering broader applicability but require large training datasets for accuracy.
Tissue-based methylation sequencing
Tissue-based NGS remains the gold standard for high-resolution methylation profiling, directly analyzing tumor DNA from biopsies or resections with high sensitivity and specificity [58]. This method has been instrumental in identifying hypermethylated tumor suppressor genes and hypomethylated oncogenes that serve as early diagnostic markers, contributing to the development of methylation-based screening assays [59].
However, the invasive nature of tissue biopsies makes them unsuitable for routine screening and repeated sampling, particularly for early-stage cancers where non-invasive methods are preferable. Tumor heterogeneity also results in methylation variability across different tumor regions, meaning a single biopsy may not capture the full epigenetic landscape [10]. These challenges highlight the need for liquid biopsy-based methylation sequencing for real-time, non-invasive tumor monitoring.
Liquid biopsy-based methylation sequencing
Liquid biopsy-based NGS provides a non-invasive alternative by analyzing ctDNA methylation from blood or other bodily fluids. Methylation changes often arise early in tumorigenesis, sometimes preceding genetic mutations, making ctDNA a promising biomarker for early-stage detection [7]. Unlike tissue-based sequencing, which provides a static snapshot of tumor methylation, liquid biopsy enables real-time tracking of tumor-derived epigenetic changes, improving risk assessment and early intervention.
However, early-stage breast cancer detection using ctDNA remains challenging due to the low ctDNA abundance, especially when tumors are small and confined to the breast [60]. Additionally, ctDNA is highly fragmented, typically ranging between 50–200 base pairs, necessitating ultra-sensitive sequencing technologies to accurately detect and quantify tumor-specific methylation changes [61]. Unlike tissue-based NGS, which directly analyses tumor DNA, liquid biopsy must contend with background noise from non-tumor-derived cfDNA, complicating tumor signal extraction and data interpretation [62, 63].
Tumor-informed liquid biopsy methylation sequencing
Tumor-informed methods integrate known tumor methylation profiles (e.g. TCGA or matched tumor biopsies) to improve ctDNA detection [64]. By leveraging differentially methylated regions (DMRs) associated with breast cancer, these methods enhance specificity and reduce false positives. Despite these advantages, tumor-informed methods face challenges in early-stage breast cancer, where low tumor-derived ctDNA levels and high background cfDNA reduce detection sensitivity [65]. Furthermore, because all tissues release cfDNA, effective tumor signal detection requires broader deconvolution strategies rather than relying solely on predefined tumor methylation patterns [66, 67]. Methylation heterogeneity within tumors can also result in inconsistent cfDNA signatures, limiting model performance [64]. These challenges, along with the impracticality of obtaining tumor biopsy data in screening settings, reduce the applicability of tumor-informed approaches for population-wide screening [62].
Tumor-naïve liquid biopsy methylation sequencing
Tumor-naïve method enables the de novo identification of breast cancer-specific methylation signatures without relying on predefined tumor profiles from matched tumor tissues. WGBS and cfMeDIP-seq have been instrumental in unbiasedly profiling breast cancer-associated DNA methylation alterations. Studies demonstrate that WGBS on cfDNA can identify novel hypermethylated regions in metastatic breast cancer, distinguishing tumor-derived signals from normal cfDNA [68]. Similarly, cfMeDIP-seq enables ctDNA quantification using methylation signatures independent of tumor tissue. This approach also identifies cancer-specific methylation signatures and fragment-length score from cfDNA, which have been shown to predict treatment outcomes in pembrolizumab-treated cancer patients [69].
Tumor-naïve approaches have shown strong performance in early detection. Pham et al. developed a tumor-naïve cfDNA methylation test that identifies breast cancer-specific methylation markers directly from plasma, without relying on predefined tumor profiles. By analyzing de novo cfDNA methylation features, the test achieved an AUC of 0.91 for early-stage breast cancer detection, demonstrating the feasibility of unbiased methylation biomarker discovery [70]. Further supporting this approach, the Circulating Cell-free Genome Atlas (CCGA) study demonstrated that cfDNA methylation sequencing combined with machine learning could detect multiple cancer types, including breast cancer, with high specificity (~ 99%) [71]. Unlike tumor-informed methods, tumor-naïve strategies detect cancer in individuals without prior tumor data, making them highly suitable for early screening and diagnosis.
DNA methylation biomarkers for early breast cancer diagnosis
Potential biomarkers for distinguishing early breast cancer
In recent years, DNA methylation markers have garnered significant interest as potential biomarkers in liquid biopsies for various cancers. However, despite extensive research, very few blood-based DNA methylation markers have been clinically approved for the early detection and diagnosis of breast cancer, primarily due to technical limitations, high validation costs, and lack of standardization, which leave many promising markers still in the research phase. A notable exception is the Galleri test by GRAIL, a MCED test that analyses ctDNA methylation patterns in blood and can detect more than 50 types of cancer, including breast cancer [40, 71]. More details about this test will be discussed in a later section on the “Performance of MCED tests in breast cancer” and is summarized in Table 2.
In response to the need for more effective non-invasive diagnostic markers, researchers have expanded their focus from single-gene targets to panels of multiple methylation markers. This shift, driven by the potential to enhance diagnostic accuracy beyond what single-gene assays achieve, aims to leverage the synergistic effects of multiple biomarkers. This approach greatly improves both the sensitivity and specificity of the assays, as demonstrated in various studies (Table 2). For example, Moss et al. showed that a panel consisting of three breast cancer DNA markers (KRT19, LMX1B, and ZNF296) could effectively identify breast cancer patients [72]. The combination of all three markers, with an 80% sensitivity, significantly outperformed the sensitivity of any single marker alone, which had individual sensitivities of 60% for KRT19, 40% for LMX1B, and 63% for ZNF296. Further advancing this field, Liu et al. developed a predictive model using ten cfDNA methylation markers from four genes (RYR2, RYR3, GABRB3, and DCDC2 C) and two long non-coding RNAs (AC096570.1 and LINC00923) [64]. This model differentiates between malignant and benign breast cancer samples with an area under the curve (AUC) of 0.81, outperforming the AUCs of traditional diagnostic methods such as mammography, ultrasound, and tumor markers CA153 and CEA. When combined with mammography and ultrasound, the model significantly enhances diagnostic performance, achieving high detection rates of over 93.3% for early-stage breast cancer.
Additionally, a recent study by Wang et al. developed a multiplex methylation-specific quantitative PCR assay utilizing four breast cancer-specific CpG methylation markers [32]. This assay was able to distinguish early-stage breast cancer patients from normal controls with high overall detection rate of 82%. This performance is significantly higher than that of traditional tumor markers CA153 (5.3%), CEA (6.0%), and CA125 (3.5%). In another study, Zhang and colleagues evaluated a panel of 26 methylation markers that showed higher diagnostic power compared to mammography alone [43]. When combined with mammography, this panel achieved exceptional diagnostic outcomes, with an AUC of 0.995. Notably, the combined model showed significant improvements, with sensitivity increasing by 6.25% and specificity by 3.89% over the methylation model alone. These findings highlight the potential of DNA methylation markers to enhance early breast cancer detection, with their combination with established diagnostics offering improved sensitivity and accuracy.
Clinical stage is a crucial factor in cancer detection. For a diagnostic or screening biomarker to be clinically applicable, it must reliably identify tumors at early stages (Stage 0-II), where early intervention can significantly improve patient outcomes. However, many methylation markers for cancer detection lack performance evaluations for early-stage cancers, highlighting a significant gap. Among the studies reviewed in Table 2, only four assessed their assays’ performance in early-stage breast cancer [32, 40, 43, 71].
Potential biomarkers for early-stage stratification of breast cancer subtypes
Studies have revealed that different breast cancer subtypes exhibit distinct methylation patterns [73, 74]. A diagnostic biomarker capable of distinguishing between normal and cancer patients, as well as stratifying breast cancer into specific subtypes, can significantly enhance early-stage disease management. Such stratification would allow for personalized treatment plans tailored to the molecular characteristics of each subtype. A recent study using whole-genome methylation profiling of ctDNA identified a diagnostic signature comprising 15 ctDNA markers that effectively classified healthy individuals and early-stage breast cancer patients, achieving an AUC of 0.967 [13]. Additionally, a 12-marker ctDNA methylation signature was identified for differentiating estrogen receptor (ER) status, with an AUC of 0.984 in the training cohort (n = 60) and 0.780 in the test cohort (n = 61).
However, despite the growing interest in DNA methylation in breast cancer, no studies to date have identified specific methylation biomarkers for early-stage stratification of other breast cancer subtypes, except for TNBC. Most existing studies, particularly those on HER2-positive breast cancer, have primarily focused on its prognostic value or predictive role in treatment response, rather than its potential for early detection or subtype stratification. Meanwhile, TNBC has gained significant research attention, with several studies exploring methylation-based stratification of TNBC subtypes, highlighting its potential for more precise classification and risk assessment. Notably, epigenetic alterations in TNBC occur more frequently than in other breast cancer subtypes, further driving research interest in TNBC-specific methylation signatures [75].
TNBC remains one of the most challenging breast cancer subtypes to manage due to its aggressive phenotype and limited treatment options. Several research groups have characterized TNBC-specific methylation signatures to support early detection and provide more efficient treatment options. Manoochehri and colleagues evaluated genome-wide DNA methylation from breast tissues, peripheral blood, and plasma samples from TBNC patients and healthy controls, selecting the top six DMRs for clinical validation in cfDNA samples using droplet digital PCR (ddPCR) [30]. They found that the methylation score, calculated based on the top three significant DMRs overlapping with SPAG6, LINC10606, and TBCD/ZNF750 genes, was significantly higher in TNBC patients than in healthy individuals, with an AUC of 0.74, indicating its potential to discriminate TNBC cases from healthy controls. However, as this study was retrospective, future prospective studies are needed to confirm the assay’s applicability for early detection of TNBC. Another research group developed a multiplexed-targeted NGS approach known as mDETECT for detecting and monitoring metastatic TNBC [76]. This assay uses 54 PCR probes from 47 regions, including a BRCA1 methylation probe due to its frequent occurrence and predictive value in TNBC. The mDETECT assay demonstrated high efficacy in detecting tumors in serum samples from metastatic TNBC patients, achieving an AUC of 0.97, with a sensitivity of 93% and a specificity of 100%. Given that BRCA1 carriers are at high risk of developing TNBC, the mDETECT assay may also play a role in the early detection of this population. However, despite the inclusion of the BRCA1 probe, the study found its clinical significance unclear due to its association with abnormal mammograms and previous breast biopsies in the control group.
Integrative multi-omics approaches for early breast cancer detection
While single-omics data provides valuable insights, they often fail to capture the full complexities of cancer. Multi-omics approaches, integrating data from genomics, epigenomics, fragmentomics, transcriptomics, proteomics, and metabolomics, offer a more comprehensive understanding of the interactions between genotype, phenotype, and environment (Fig. 2). By uncovering the intricate molecular mechanisms driving cancer, multi-omics can enhance cancer detection sensitivity (Table 3). This review focuses on studies that integrate DNA methylation data with other omics layers to improve diagnostic accuracy.
Computational and machine learning approaches in multi-omics
Integrating multi-omics data is challenging due to biological variability, computational complexity, data noise, and the limitations of traditional analytical methods in handling high-dimensional datasets [77]. To address these issues, advanced computational techniques, particularly ML and artificial intelligence (AI), are increasingly employed to extract meaningful insights and improve biomarker discovery (Fig. 2). These approaches can efficiently analyze large, complex datasets, identifying patterns and associations that may be overlooked by conventional methods.
Recent studies have demonstrated the effectiveness of ML-driven approaches in integrating DNA methylation with additional omics layers, significantly enhancing cancer classification models. For example, Xu et al. demonstrated that integrating genome-wide methylation profiles with transcriptomic and metabolic data using supervised ML and feature selection techniques significantly improved the sensitivity of distinguishing ductal carcinoma in situ (DCIS) from invasive breast cancer (IBC), achieving an AUROC of 0.95 [78]. Similarly, deep learning has also proven effective in integrating multi-omics data to enhance breast cancer subtype classification and improve prognosis prediction. A deep learning model trained on multi-omics datasets, including mRNA, miRNA, gene mutations, DNA methylation, and magnetic resonance imaging (MRI) images, achieved an impressive accuracy of up to 98.0% in TNBC subtype classification, surpassing traditional single-omics approaches [79].
The role of fragmentomics in multi-omics
An emerging omic approach gaining traction is fragmentomics, which analyzes the fragmentation patterns and size profiles of cfDNA/ctDNA, providing an additional layer of information for early cancer detection [80]. Tumor-derived ctDNA fragments exhibit unique fragmentation characteristics, including shorter fragment sizes, altered nucleosome positioning, and distinct fragment end motifs, which can serve as additional biomarkers for cancer detection [41, 61].
Integrating cfDNA methylation with fragmentomic features, researchers have significantly enhanced detection accuracy, with ML-driven models outperforming single-feature approaches [81]. A recent study by Pham et al. demonstrated that the integration of ctDNA methylation and fragmentomic features (fragment length and end motif) using ML algorithms significantly improved breast cancer detection accuracy, achieving an AUC of 0.91 compared to single-feature models [70]. These findings highlight the importance of leveraging multiple tumor-associated signals, such as epigenetic modifications and structural DNA features, to enhance sensitivity and specificity in liquid biopsy-based early detection.
The role of DNA methylation in MCED and tumor origin identification
DNA methylation is a key biomarker in MCED assays, offering higher predictive performance compared to other genomic and epigenomic features. The CCGA study systematically evaluated various cfDNA characteristics, including whole-genome methylation, single nucleotide variants, somatic copy number alterations, fragment endpoints, fragment lengths and allelic imbalance, to assess their utility in MCED [82]. Their findings confirmed that whole-genome methylation patterns achieved the highest predictive accuracy among all tested modalities. Notably, while integrating fragmentomics and genomic variants led to minor improvements, it did not significantly enhance sensitivity, underscoring DNA methylation as a key biomarker for early cancer detection.
Beyond detection, DNA methylation plays a crucial role in identifying the tissue or cancer cell origin (TOO/CSO), which is particularly important for MCED tests. Since cfDNA is derived from multiple tissues, determining the primary tumor site is essential for guiding downstream diagnostic workups and clinical intervention. Jamshidi et al. found that whole-genome methylation feature was the most effective in predicting TOO, with a methylation-based classifier accurately predicting TOO in 75% of samples [82]. Independent validation studies have reported accuracy rates as high as 93% for methylation-based TOO/CSO classifier [40, 71]. Additionally, TOO can be further refined by incorporating cfDNA fragmentation profiles, end motifs, and copy number alterations (Table 3) [83, 84]. Collectively, these findings underscore the value of integrating DNA methylation with other cfDNA-derived features to enhance both the detection and localization of early-stage malignancies.
Multi-omics applications in MCED and cancer detection
Advanced multi-omics and computational methods have also been applied to improve the accuracy and sensitivity of MCED tests, including those designed for breast cancer detection. For instance, Bie et al. employed low-pass whole-methylome sequencing on plasma cfDNA samples from 497 healthy controls and 780 patients across seven cancer types (breast, colorectal, esophageal, liver, lung, pancreatic and gastric cancer) [84]. Their computational analysis examined cancer-associated features such as DNA methylation, fragment size, fragment end motifs, and copy number alterations. Among these features, the methylated fragment ratio performed the best (AUC = 0.947), followed by fragment end motif (AUC = 0.935), fragment size index (AUC = 0.906), with copy number alterations showing the lowest AUC of 0.861. To improve detection performance, they developed THEMIS, an ensemble ML classifier that integrates all four modalities to distinguish cancer patients from healthy controls. THEMIS outperformed individual modalities with an overall AUC of 0.966. THEMIS also demonstrated high AUCs in detecting all seven cancer types, ranging from 0.938 for breast and liver cancer to 0.990 for colorectal cancer.
Similarly, Nguyen et al. demonstrated that integrating methylation, fragmentomics, DNA copy number alterations, and fragment end motifs from shallow genome-wide sequencing of ctDNA improved MCED performance across breast, colorectal, gastric, lung, and liver cancer [83]. Their stacking ensemble ML model classified cancer status with an AUC of 0.95, outperforming single-feature models (AUC = 0.81–0.92). The model achieved a sensitivity of 72.4% at 97% specificity in the validation cohort, with the highest detection rate observed for liver cancer (91.1%), while breast cancer had the lowest detection rate (49.3%). Despite this limitation, both studies underscored the potential of integrating DNA methylation and fragmentation profiles from cfDNA sequencing to improve MCED assays.
Performance of MCED tests in breast cancer
Currently, most MCED tests in development detect mutations and fragmentation patterns in cfDNA, but only Galleri (GRAIL) and PanSEER (Singlera Genomics) focus specifically on detecting cancer-specific DNA methylation patterns in blood [40, 71, 85]. The Galleri test screens over 50 cancer types, including breast cancer, whereas PanSEER focuses on colorectal, esophageal, liver, lung and stomach cancers.
The CCGA study (NCT02889978) is a landmark prospective study that laid the groundwork for the development of the MCED Galleri test. This multicentre study, involving over 15,000 participants, was divided into three pre-specified sub-studies: (1) evaluation of cfDNA features using prototype assays and machine-learning classifiers to identify the most promising approach for developing an MCED test, (2) prospective training and validation of the chosen methylation-based assay, and (3) clinical validation [40, 71, 82]. In the clinical validation phase, the test demonstrated an overall sensitivity of 51.5% and a specificity of 99.5%. However, its sensitivity for breast cancer was notably low, with a detection rate of only 30.5%. Sensitivity varied significantly by stage—2.6% for Stage I, 47.5% for Stage II, 85.5% for Stage III, and 90.9% for Stage IV, highlighting the significant challenges in early-stage breast cancer detection.
The PATHFINDER study (NCT04241796) further assessed the Galleri test in a real-world setting by screening 6662 cancer-free individuals aged 50 and older [86]. Of the 92 positive cancer signals detected, 35 were true positives, while 57 were false positives. Among the true positives, 48% (14/29) of non-recurrent cancers were detected at early stages (Stage I–II), though none of these were early-stage breast cancer. Additionally, the test missed 17 cases of breast cancer, including 13 in early stages, which were instead detected through standard screening methods. Despite this, the test successfully identified five cases of metastatic breast cancer relapse, suggesting its potential for monitoring cancer recurrence.
Single cancer screening tests often have high false positive rates, resulting in low positive predictive values (PPVs) of approximately 5–10%. In contrast, MCED tests, which screen for multiple cancers from a single blood draw, can achieve higher PPVs of up to 40–50% [87]. The CCGA study, based on SEER (Surveillance, Epidemiology, and End Results Program) cancer incidence and stage distribution in individuals aged 50–79 years, reported a PPV of 44.4% [40]. However, the subsequent PATHFINDER study showed a lower overall PPV of 38%, indicating that Galleri test sensitivity may be lower in real-world screening populations than in controlled trial settings [86].
Despite these limitations, GRAIL continues the STRIVE study (NCT03085888), a large-scale prospective cohort trial involving approximately 100,000 women undergoing mammography screenings, aimed at assessing the test’s performance and clinical utility [71]. However, recent scrutiny has raised concerns about Galleri’s effectiveness and the ethical implications. A BMJ investigation revealed that Galleri’s sensitivity for stage I cancers is only 16.8%, questioning its suitability for large-scale deployment [88]. Additionally, concerns about regulatory transparency, the NHS’s financial commitment, and potential conflicts of interest have fueled debate over whether Galleri should be used as a screening tool for early cancer. These findings underscore the need for rigorous, independent validation of MCED tests before widespread adoption.
Clinical validation and utility of methylation-based liquid biopsy assays
The clinical validation of methylation-based liquid biopsy assays is essential to determine their real-world effectiveness in early breast cancer detection. Despite significant research efforts, no plasma- or blood-based DNA methylation markers have been approved for breast cancer screening or diagnosis. Over 40 studies have evaluated more than 400 DNA methylation-based biomarkers for early detection, yet most of these markers have shown limited diagnostic performance, with little overlap between reported CpG sites, making standardization and validation challenging [89]. The only clinical trial that specifically assessed ctDNA methylation for early breast cancer detection (NCT03480659) was prematurely terminated due to technical issues with sample collection, highlighting the logistical difficulties associated with ensuring reliable sample integrity in real-world settings [90].
Beyond individual trial limitations, methylation-based MCED tests, such as Galleri (GRAIL), have demonstrated suboptimal performance for early-stage breast cancer. The PATHFINDER study, a prospective trial evaluating Galleri’s clinical performance, reported an overall sensitivity of only 29% for detecting known cancers, indicating that the sensitivity for early-stage breast cancer may be significantly lower [86]. The sensitivity for stage I breast cancer detection has been reported as low as 2.6%, raising concerns about the feasibility of using these assays as standalone screening tools [40, 88]. The limited detection rates emphasize the need for strategies to enhance assay sensitivity, such as optimizing ctDNA enrichment, refining bioinformatics pipelines, and integrating additional molecular features to improve early-stage detection.
To improve clinical applicability, multi-omics approaches integrating methylation data with fragmentomics, copy number alterations, or proteomics have been proposed (see section on ‘Integrative Multi-omics Approaches for Early Breast Cancer Detection’). Integrating these complementary molecular signals has the potential to increase the accuracy of liquid biopsy-based tests by reducing false positives and enhancing early-stage cancer detection sensitivity. However, these approaches remain in the experimental phase, requiring further validation through large-scale prospective clinical trials to assess their added value over existing screening methods.
Challenges and limitations
Methylation-based liquid biopsy holds significant promise for early breast cancer detection, but several challenges must be addressed before these assays can be fully integrated into clinical practice. These challenges span technical limitations, biological variability, and regulatory hurdles, all of which influence the feasibility and real-world applicability of methylation-based diagnostic assays.
Sensitivity, specificity, and overdiagnosis risks
Achieving high sensitivity and specificity is a major challenge in methylation-based assays, particularly in early-stage breast cancer, where ctDNA levels are extremely low due to limited tumor shedding and the short half-life of ctDNA [82, 91]. This makes capturing tumor-derived methylation signals difficult, impacting detection accuracy. Additionally, ctDNA shedding varies by breast cancer subtype, with TNBC shedding more ctDNA than hormone receptor-positive subtypes, further complicating the development of a universal detection approach [92]. While highly sensitive assays are necessary for detecting low ctDNA levels, excessive sensitivity raises concerns about overdiagnosis. Detecting methylated ctDNA from indolent tumors could lead to unnecessary interventions and overtreatment, increasing healthcare costs without substantial clinical benefits [93].
Reproducibility and standardization issues
Assay reproducibility remains a key challenge due to pre-analytical and methodological variabilities, affecting sensitivity, specificity, and cross-study comparability. Pre-analytical factors, such as sample collection, processing, and storage, can impact ctDNA integrity. Plasma collected in EDTA tubes must be processed within four hours to prevent genomic DNA contamination, posing logistical challenges for real-world implementation [94]. While ultra-low temperature storage generally preserves global methylation profiles, methylation levels at individual CpG sites may still be affected over time [95–97]. Specialized tubes, like Streck’s cfDNA BCT help stabilize cfDNA, but variability in sample handling and processing across laboratories remains a concern [98].
Methodological inconsistencies further contribute to variability in methylation analysis. Differences in DNA extraction methods, bisulfite conversion protocols, sequencing depth, and bioinformatics pipelines can introduce discrepancies in detection sensitivity and limit reproducibility across platforms [99, 100]. To address these challenges, the American Society of Clinical Oncology (ASCO) and the College of American Pathologists (CAP) emphasize the importance of standardized protocols and proficiency testing to ensure the analytical validity of ctDNA assays [101]. Key performance metrics such as limit of detection (LoD), clinical sensitivity and specificity, and assay robustness must be rigorously evaluated before widespread clinical adoption [101, 102]. However, many early breast cancer ctDNA studies have inadequately reported analytical validity, often prioritizing sensitivity to detect the low levels of ctDNA in early-stage breast cancer. Without robust standardization, the reproducibility of methylation-based liquid biopsy assays remains a significant barrier to their clinical scalability and integration into routine practice.
Biological variability and confounding factors
Beyond technical challenges, biological variability introduces additional complexity in methylation analysis. DNA methylation patterns are influenced by age, comorbidities (e.g., hypertension, cardiovascular disease, diabetes, obesity), as well as environmental exposures and lifestyle factors, which can lead to false positives or misclassification [103–107]. Without well-matched controls, these confounding variables can reduce specificity and limit assay accuracy, making robust validation studies essential prior to clinical implementation.
Regulatory challenges and barriers to clinical implementation
The regulatory landscape presents additional barriers to clinical adoption of methylation-based liquid biopsy assays. Agencies such as the U.S. Food and Drug Administration (FDA) and the European Medicines Agency (EMA) require rigorous validation before these assays can be integrated into routine clinical use. Approval depends on demonstrating analytical validity (precision, reproducibility, and sensitivity), clinical validity (the ability to distinguish cancer from non-cancer samples), and clinical utility (demonstrated improvement in patient outcomes) [101, 108]. However, no methylation-based liquid biopsy test has yet received FDA approval for early breast cancer detection, reflecting the ongoing challenges in meeting these regulatory benchmarks.
A major regulatory hurdle is the lack of standardized performance criteria for these assays. Unlike colorectal cancer screening tools, breast cancer-specific methylation tests remain largely investigational, requiring further validation before regulatory approval [109]. Variability in methylation detection methodologies and lack of consensus on performance benchmarks make cross-study comparisons difficult. Industry-wide guidelines and proficiency testing programs, as recommended by CAP and ASCO, are essential to improve assay comparability and facilitate regulatory approval.
Reimbursement policies remain a major obstacle to the adoption of methylation-based liquid biopsy assays, as many insurers classify them as experimental, limiting patient access and clinical integration [110]. Unlike mammography, which has clear reimbursement frameworks, these assays lack standardized billing codes, complicating coverage. Companion diagnostics for treatment selection are more likely to be reimbursed than early detection tests [111]. Additionally, publicly funded systems require cost-effectiveness analyses, often delaying adoption despite clinical promise. To address this, some tests are launched as laboratory-developed tests (LDTs) while gathering real-world data to support future reimbursement claims. This adaptive approach allows companies to demonstrate clinical utility through post-market studies while working toward broader payer acceptance. However, securing reimbursement requires proving not only clinical accuracy but also long-term economic value from early detection [111].
Further barriers to adoption include high costs, specialized laboratory infrastructure, and bioinformatics expertise, which many clinical centers lack [112]. Advanced sequencing-based assays have long turnaround times and significant cost burdens, posing economic challenges for both healthcare systems and patients [113, 114]. Moreover, limited awareness among clinicians and patients regarding the utility of methylation-based tests may slow their integration into standard screening programs. Addressing these issues will require clear performance benchmarks, standardized analytical workflows, and expanded reimbursement policies to facilitate the clinical translation of methylation-based liquid biopsy tests for early breast cancer detection.
Conclusions and future perspectives
Methylation-based biomarkers show significant potential for early cancer detection, offering a non-invasive alternative or complement to traditional screening methods. Advances in liquid biopsy technologies, multi-omics integration and ML-driven analysis have improved assay performance and led to the discovery of numerous potential DNA methylation biomarkers. However, key challenges remain, including assay validation, low ctDNA abundance, and variability in clinical performance.
One major limitation of liquid biopsy technologies is the low levels of ctDNA, particularly in early-stage cancers, which reduce detection sensitivity. To address this, researchers at the Broad Institute and Massachusetts Institute of Technology (MIT) have developed injectable ‘priming agents’ that temporarily increase cfDNA levels in the blood by slowing its clearance, potentially enhancing the effectiveness of liquid biopsies for cancer diagnostics [115]. Additionally, the detection of ctDNA in breast milk offers a novel approach for early breast cancer diagnosis in the postpartum period, suggesting the potential of exploring alternative body fluids for ctDNA detection [116].
Future advancements in methylation-based liquid biopsy assays will likely focus on multi-omics integration to enhance detection accuracy and clinical utility. Emerging evidence suggests that integrating DNA methylation and ctDNA fragmentation profiles can further improve assay performance [70, 83, 84]. Moreover, combining multiple biomarkers, such as protein-DNA mutations or DNA methylation with gene expression data, has shown potential for enhancing assay sensitivity in early-stage cancers [78, 117]. Incorporating proteomics, metabolomics, exosomes, or imaging data, may further boost diagnostic accuracy and facilitate personalized treatment strategies. However, while multi-omics integration generally improves predictive performance, it also introduces challenges related to data complexity, cost, and computational requirements. Advanced AI and ML techniques are also expected to play a crucial role in analyzing tumor-derived methylation patterns in real time, improving precision and scalability in methylation-based biomarker discovery [118].
Acknowledgements
The figures were created with BioRender.com.
Abbreviations
- 5 hmC
5-Hydroxymethylcytosine
- 5 mC
5-Methylcytosines
- AI
Artificial intelligence
- ASCO
American Society of Clinical Oncology
- AUC
Area under the curve
- AUROC
Area under the receiver operating characteristic curve
- CAP
College of American Pathologists
- CCGA
Circulating Cell-free Genome Atlas
- cfDNA
Cell-free DNA
- cfMeDIP-seq
Cell-free methylated DNA immunoprecipitation sequencing
- cfMethyl-Seq
Cell-free DNA methylation sequencing
- CSO
Cancer cell origin
- CTCs
Circulating tumor cells
- ctDNA
Circulating tumor DNA
- DCIS
Ductal carcinoma in situ
- ddPCR
Droplet digital PCR
- DMRs
Differentially methylated regions
- dPCR
Digital PCR
- EM-seq
Enzymatic methyl-seq
- ER
Estrogen receptor
- FDA
The U.S. Food and Drug Administration
- IBC
Invasive breast cancer
- LoD
Limit of detection
- MCED
Multi-cancer early detection
- ML
Machine learning
- MSP
Methylation-specific PCR
- NGS
Next-generation sequencing
- PPVs
Positive predictive values
- qMSP
Quantitative methylation-specific PCR
- RRBS
Reduced representation bisulfite sequencing
- SMRT-seq
Single-Molecule Real-Time Sequencing
- TAPS
TET-assisted pyridine borane sequencing
- TNBC
Triple negative breast cancer
- TOO
Tissue of origin
- WGBS
Whole-genome bisulfite sequencing
- WGS
Whole-genome sequencing
Author contributions
MH and ASGL conceptualized the review. MH wrote the manuscript and prepared the tables and figures. AGSL obtained the funding. All authors reviewed, revised, and approved the final manuscript.
Funding
This study was funded by the National Medical Research Council of Singapore (MOH-OFIRG19nov-0019).
Availability of data and materials
Not applicable. No datasets were generated or analyzed in this study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mann RM, Athanasiou A, Baltzer PAT, Camps-Herrero J, Clauser P, Fallenberg EM, et al. Breast cancer screening in women with extremely dense breasts recommendations of the European Society of Breast Imaging (EUSOBI). Eur Radiol. 2022;32(6):4036–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pace LE. False-positive results of mammography screening in the era of digital breast tomosynthesis. JAMA Netw open. 2022;5(3): e222445. [DOI] [PubMed] [Google Scholar]
- 3.Glodzik D, Bosch A, Hartman J, Aine M, Vallon-Christersson J, Reuterswärd C, et al. Comprehensive molecular comparison of BRCA1 hypermethylated and BRCA1 mutated triple negative breast cancers. Nat Commun. 2020;11(1):3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kloten V, Becker B, Winner K, Schrauder MG, Fasching PA, Anzeneder T, et al. Promoter hypermethylation of the tumor-suppressor genes ITIH5, DKK3, and RASSF1A as novel biomarkers for blood-based breast cancer screening. Breast Cancer Res. 2013;15(1):R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pappalardo XG, Barra V. Losing DNA methylation at repetitive elements and breaking bad. Epigenet Chromatin. 2021;14(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parashar S, Cheishvili D, Mahmood N, Arakelian A, Tanvir I, Khan HA, et al. DNA methylation signatures of breast cancer in peripheral T-cells. BMC Cancer. 2018;18(1):574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baylin SB, Jones PA. A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer. 2011;11(10):726–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roy D, Tiirikainen M. Diagnostic power of DNA methylation classifiers for early detection of cancer. Trends Cancer. 2020;6(2):78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shoukat I, Mueller CR. Searching for DNA methylation in patients triple-negative breast cancer: a liquid biopsy approach. Expert Rev Mol Diagn. 2023;23(1):41–51. [DOI] [PubMed] [Google Scholar]
- 10.Gilson P, Merlin J-L, Harlé A. Deciphering tumour heterogeneity: from tissue to liquid biopsy. Cancers (Basel). 2022;14(6):1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32(6):579–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo H, Wei W, Ye Z, Zheng J, Xu R-H. Liquid biopsy of methylation biomarkers in cell-free DNA. Trends Mol Med. 2021;27(5):482–500. [DOI] [PubMed] [Google Scholar]
- 13.Gao Y, Zhao H, An K, Liu Z, Hai L, Li R, et al. Whole-genome bisulfite sequencing analysis of circulating tumour DNA for the detection and molecular classification of cancer. Clin Transl Med. 2022;12(8): e1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Wilde J, Van Paemel R, De Koker A, Roelandt S, Van de Velde S, Callewaert N, et al. A fast, affordable, and minimally invasive diagnostic test for cancer of unknown primary using DNA methylation profiling. Lab Invest. 2024;104(8): 102091. [DOI] [PubMed] [Google Scholar]
- 15.Shen SY, Singhania R, Fehringer G, Chakravarthy A, Roehrl MHA, Chadwick D, et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature. 2018;563(7732):579–83. [DOI] [PubMed] [Google Scholar]
- 16.Stackpole ML, Zeng W, Li S, Liu C-C, Zhou Y, He S, et al. Author correction: cost-effective methylome sequencing of cell-free DNA for accurately detecting and locating cancer. Nat Commun. 2024;15(1):3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang N, Li B, Jia Z, Wang C, Wu P, Zheng T, et al. Ultrasensitive detection of circulating tumour DNA via deep methylation sequencing aided by machine learning. Nat Biomed Eng. 2021;5(6):586–99. [DOI] [PubMed] [Google Scholar]
- 18.Gao Q, Lin YP, Li BS, Wang GQ, Dong LQ, Shen BY, et al. Unintrusive multi-cancer detection by circulating cell-free DNA methylation sequencing (THUNDER): development and independent validation studies. Ann Oncol Off J Eur Soc Med Oncol. 2023;34(5):486–95. [DOI] [PubMed] [Google Scholar]
- 19.Sun Z, Vaisvila R, Hussong L-M, Yan B, Baum C, Saleh L, et al. Nondestructive enzymatic deamination enables single-molecule long-read amplicon sequencing for the determination of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Genome Res. 2021;31(2):291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaisvila R, Ponnaluri VKC, Sun Z, Langhorst BW, Saleh L, Guan S, et al. Enzymatic methyl sequencing detects DNA methylation at single-base resolution from picograms of DNA. Genome Res. 2021;31(7):1280–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Siejka-Zielińska P, Velikova G, Bi Y, Yuan F, Tomkova M, et al. Bisulfite-free direct detection of 5-methylcytosine and 5-hydroxymethylcytosine at base resolution. Nat Biotechnol. 2019;37(4):424–9. [DOI] [PubMed] [Google Scholar]
- 22.Siejka-Zielińska P, Cheng J, Jackson F, Liu Y, Soonawalla Z, Reddy S, et al. Cell-free DNA TAPS provides multimodal information for early cancer detection. Sci Adv. 2021;7(36):eabh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7(6):461–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simpson JT, Workman RE, Zuzarte PC, David M, Dursi LJ, Timp W. Detecting DNA cytosine methylation using nanopore sequencing. Nat Methods. 2017;14(4):407–10. [DOI] [PubMed] [Google Scholar]
- 25.Rauschert S, Raubenheimer K, Melton PE, Huang RC. Machine learning and clinical epigenetics: a review of challenges for diagnosis and classification. Clin Epigenet. 2020;12(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Im YR, Tsui DWY, Diaz LA, Wan JCM. Next-generation liquid biopsies: embracing data science in oncology. Trends Cancer. 2021;7(4):283–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28(8):E32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S-C, Liao L-M, Ansar M, Lin S-Y, Hsu W-W, Su C-M, et al. Automatic detection of the circulating cell-free methylated DNA pattern of GCM2, ITPRIPL1 and CCDC181 for detection of early breast cancer and surgical treatment response. Cancers (Basel). 2021;13(6):1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salta S, P Nunes S, Fontes-Sousa M, Lopes P, Freitas M, Caldas M, et al. A DNA methylation-based test for breast cancer detection in circulating cell-free DNA. J Clin Med. 2018;7(11):420. [DOI] [PMC free article] [PubMed]
- 30.Manoochehri M, Borhani N, Gerhäuser C, Assenov Y, Schönung M, Hielscher T, et al. DNA methylation biomarkers for noninvasive detection of triple-negative breast cancer using liquid biopsy. Int J cancer. 2023;152(5):1025–35. [DOI] [PubMed] [Google Scholar]
- 31.Mijnes J, Tiedemann J, Eschenbruch J, Gasthaus J, Bringezu S, Bauerschlag D, et al. SNiPER: a novel hypermethylation biomarker panel for liquid biopsy based early breast cancer detection. Oncotarget. 2019;10(60):6494–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang T, Li P, Qi Q, Zhang S, Xie Y, Wang J, et al. A multiplex blood-based assay targeting DNA methylation in PBMCs enables early detection of breast cancer. Nat Commun. 2023;14(1):4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pidsley R, Zotenko E, Peters TJ, Lawrence MG, Risbridger GP, Molloy P, et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016;17(1):208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee NY, Hum M, Tan GP, Seah AC, Ong P-Y, Kin PT, et al. Machine learning unveils an immune-related DNA methylation profile in germline DNA from breast cancer patients. Clin Epigenet. 2024;16(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kresovich JK, Xu Z, O’Brien KM, Shi M, Weinberg CR, Sandler DP, et al. Blood DNA methylation profiles improve breast cancer prediction. Mol Oncol. 2022;16(1):42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noguera-Castells A, García-Prieto CA, Álvarez-Errico D, Esteller M. Validation of the new EPIC DNA methylation microarray (900K EPIC v2) for high-throughput profiling of the human DNA methylome. Epigenetics. 2023;18(1):2185742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desmedt C, Voet T, Sotiriou C, Campbell PJ. Next-generation sequencing in breast cancer: first take home messages. Curr Opin Oncol. 2012;24(6):597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu H, Bock C, Mikkelsen TS, Jäger N, Smith ZD, Tomazou E, et al. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat Methods. 2010;7(2):133–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen M, Zhao H. Next-generation sequencing in liquid biopsy: cancer screening and early detection. Hum Genomics. 2019;13(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klein EA, Richards D, Cohn A, Tummala M, Lapham R, Cosgrove D, et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann Oncol. 2021;32(9):1167–77. [DOI] [PubMed] [Google Scholar]
- 41.Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, Bruhm DC, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature. 2019;570(7761):385–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu L, Toung JM, Jassowicz AF, Vijayaraghavan R, Kang H, Zhang R, et al. Targeted methylation sequencing of plasma cell-free DNA for cancer detection and classification. Ann Oncol. 2018;29(6):1445–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, Zhao D, Yin Y, Yang T, You Z, Li D, et al. Circulating cell-free DNA-based methylation patterns for breast cancer diagnosis. NPJ Breast Cancer. 2021;7(1):106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vandenhoeck J, Neefs I, Vanpoucke T, Ibrahim J, Suls A, Peeters D, et al. IMPRESS: improved methylation profiling using restriction enzymes and smMIP sequencing, combined with a new biomarker panel, creating a multi-cancer detection assay. Br J Cancer. 2024;131(7):1224–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vavoulis DV, Cutts A, Thota N, Brown J, Sugar R, Rueda A, et al. Multimodal cell-free DNA whole-genome TAPS is sensitive and reveals specific cancer signals. Nat Commun. 2025;16(1):430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wenger AM, Peluso P, Rowell WJ, Chang P-C, Hall RJ, Concepcion GT, et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat Biotechnol. 2019;37(10):1155–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lau BT, Almeda A, Schauer M, McNamara M, Bai X, Meng Q, et al. Single-molecule methylation profiles of cell-free DNA in cancer with nanopore sequencing. Genome Med. 2023;15(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oehler JB, Wright H, Stark Z, Mallett AJ, Schmitz U. The application of long-read sequencing in clinical settings. Hum Genomics. 2023;17(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taudt A, Roquis D, Vidalis A, Wardenaar R, Johannes F, Colomé-Tatché M. METHimpute: imputation-guided construction of complete methylomes from WGBS data. BMC Genomics. 2018;19(1):444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu F, Xu C, Deng H-W, Shen H. A novel computational strategy for DNA methylation imputation using mixture regression model (MRM). BMC Bioinform. 2020;21(1):552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sahoo K, Sundararajan V. Methods in DNA methylation array dataset analysis: a review. Comput Struct Biotechnol J. 2024;23:2304–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang P, Sinha S, Aldape K, Hannenhalli S, Sahinalp C, Ruppin E. Big data in basic and translational cancer research. Nat Rev Cancer. 2022;22(11):625–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Merkel A, Esteller M. Experimental and bioinformatic approaches to studying DNA methylation in cancer. Cancers (Basel). 2022;14(2):349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu Z, Sandler DP, Taylor JA. Blood DNA methylation and breast cancer: a prospective case-cohort analysis in the sister study. J Natl Cancer Inst. 2020;112(1):87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gomes R, Paul N, He N, Huber AF, Jansen RJ. Application of feature selection and deep learning for cancer prediction using DNA methylation markers. Genes (Basel). 2022;13(9):1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang C, Zhao N, Yuan L, Liu X. Computational detection of breast cancer invasiveness with DNA methylation biomarkers. Cells. 2020;9(2):326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.del Amor R, Colomer A, Monteagudo C, Naranjo V. A deep embedded refined clustering approach for breast cancer distinction based on DNA methylation. Neural Comput Appl. 2022;34(13):10243–55. [Google Scholar]
- 58.Ma L, Guo H, Zhao Y, Liu Z, Wang C, Bu J, et al. Liquid biopsy in cancer current: status, challenges and future prospects. Signal Transduct Target Therapy. 2024;9(1):336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lokk K, Modhukur V, Rajashekar B, Märtens K, Mägi R, Kolde R, et al. DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome Biol. 2014;15(4): r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Panet F, Papakonstantinou A, Borrell M, Vivancos J, Vivancos A, Oliveira M. Use of ctDNA in early breast cancer: analytical validity and clinical potential. NPJ Breast Cancer. 2024;10(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mouliere F, Chandrananda D, Piskorz AM, Moore EK, Morris J, Ahlborn LB, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med. 2018;10(466):eaat4921. [DOI] [PMC free article] [PubMed]
- 62.Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019;20(2):71–88. [DOI] [PubMed] [Google Scholar]
- 63.Xu J, Gao H, Guan X, Meng J, Ding S, Long Q, et al. Circulating tumor DNA: from discovery to clinical application in breast cancer. Front Immunol. 2024;15:1355887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu J, Zhao H, Huang Y, Xu S, Zhou Y, Zhang W, et al. Genome-wide cell-free DNA methylation analyses improve accuracy of non-invasive diagnostic imaging for early-stage breast cancer. Mol Cancer. 2021;20:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mouliere F, Thierry AR. The importance of examining the proportion of circulating DNA originating from tumor, microenvironment and normal cells in colorectal cancer patients. Expert Opin Biol Ther. 2012;12(Suppl 1):S209–15. [DOI] [PubMed] [Google Scholar]
- 66.Barefoot ME, Loyfer N, Kiliti AJ, McDeed AP, Kaplan T, Wellstein A. Detection of cell types contributing to cancer from circulating, cell-free methylated DNA. Front Genet. 2021;12: 671057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li S, Zeng W, Ni X, Liu Q, Li W, Stackpole ML, et al. Comprehensive tissue deconvolution of cell-free DNA by deep learning for disease diagnosis and monitoring. Proc Natl Acad Sci U S A. 2023;120(28): e2305236120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Legendre C, Gooden GC, Johnson K, Martinez RA, Liang WS, Salhia B. Whole-genome bisulfite sequencing of cell-free DNA identifies signature associated with metastatic breast cancer. Clin Epigenet. 2015;7(1):100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stutheit-Zhao EY, Sanz-Garcia E, Liu ZA, Wong D, Marsh K, Abdul Razak AR, et al. Early changes in tumor-naive cell-free methylomes and fragmentomes predict outcomes in pembrolizumab-treated solid tumors. Cancer Discov. 2024;14(6):1048–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pham TMQ, Phan TH, Jasmine TX, Tran TTT, Huynh LAK, Vo TL, et al. Multimodal analysis of genome-wide methylation, copy number aberrations, and end motif signatures enhances detection of early-stage breast cancer. Front Oncol. 2023;13:1127086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu MC, Oxnard GR, Klein EA, Swanton C, Seiden MV, CCGA Consortium. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol. 2020;31(6):745–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moss J, Zick A, Grinshpun A, Carmon E, Maoz M, Ochana BL, et al. Circulating breast-derived DNA allows universal detection and monitoring of localized breast cancer. Ann Oncol. 2020;31(3):395–403. [DOI] [PubMed] [Google Scholar]
- 73.Fleischer T, Tekpli X, Mathelier A, Wang S, Nebdal D, Dhakal HP, et al. DNA methylation at enhancers identifies distinct breast cancer lineages. Nat Commun. 2017;8(1):1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen L, Zeng T, Pan X, Zhang Y-H, Huang T, Cai Y-D. Identifying methylation pattern and genes associated with breast cancer subtypes. Int J Mol Sci. 2019;20(17):4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vietri MT, D’Elia G, Benincasa G, Ferraro G, Caliendo G, Nicoletti GF, et al. DNA methylation and breast cancer: a way forward (review). Int J Oncol. 2021;59(5):98. [DOI] [PubMed] [Google Scholar]
- 76.Cristall K, Bidard F-C, Pierga J-Y, Rauh MJ, Popova T, Sebbag C, et al. A DNA methylation-based liquid biopsy for triple-negative breast cancer. NPJ Precis Oncol. 2021;5(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Picard M, Scott-Boyer M-P, Bodein A, Périn O, Droit A. Integration strategies of multi-omics data for machine learning analysis. Comput Struct Biotechnol J. 2021;19:3735–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu H, Lien T, Bergholtz H, Fleischer T, Djerroudi L, Vincent-Salomon A, et al. Multi-omics marker analysis enables early prediction of breast tumor progression. Front Genet. 2021;12: 670749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang S, Wang Z, Wang C, Li C, Wang B. Comparative evaluation of machine learning models for subtyping triple-negative breast cancer: a deep learning-based multi-omics data integration approach. J Cancer. 2024;15(12):3943–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thierry AR. Circulating DNA fragmentomics and cancer screening. Cell Genomics. 2023;3(1): 100242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lo YMD, Han DSC, Jiang P, Chiu RWK. Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science. 2021;372(6538):eaaw3616. [DOI] [PubMed] [Google Scholar]
- 82.Jamshidi A, Liu MC, Klein EA, Venn O, Hubbell E, Beausang JF, et al. Evaluation of cell-free DNA approaches for multi-cancer early detection. Cancer Cell. 2022;40(12):1537-1549.e12. [DOI] [PubMed] [Google Scholar]
- 83.Nguyen VTC, Nguyen TH, Doan NNT, Pham TMQ, Nguyen GTH, Nguyen TD, et al. Multimodal analysis of methylomics and fragmentomics in plasma cell-free DNA for multi-cancer early detection and localization. Elife. 2023;11:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bie F, Wang Z, Li Y, Guo W, Hong Y, Han T, et al. Multimodal analysis of cell-free DNA whole-methylome sequencing for cancer detection and localization. Nat Commun. 2023;14(1):6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen X, Gole J, Gore A, He Q, Lu M, Min J, et al. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nat Commun. 2020;11(1):3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schrag D, Beer TM, McDonnell CH, Nadauld L, Dilaveri CA, Reid R, et al. Blood-based tests for multicancer early detection (PATHFINDER): a prospective cohort study. Lancet (London, England). 2023;402(10409):1251–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hackshaw A, Clarke CA, Hartman A-R. New genomic technologies for multi-cancer early detection: rethinking the scope of cancer screening. Cancer Cell. 2022;40(2):109–13. [DOI] [PubMed] [Google Scholar]
- 88.McCartney M, Cohen D. Galleri promises to detect multiple cancers-but new evidence casts doubt on this much hyped blood test. BMJ. 2024;7(386): q1706. [DOI] [PubMed] [Google Scholar]
- 89.Guan Z, Yu H, Cuk K, Zhang Y, Brenner H. Whole-blood DNA methylation markers in early detection of breast cancer: a systematic literature review. Cancer Epidemiol Biomark Prev a Publ Am Assoc Cancer Res cosponsored by Am Soc Prev Oncol. 2019;28(3):496–505. [DOI] [PubMed] [Google Scholar]
- 90.Gianni C, Palleschi M, Merloni F, Di Menna G, Sirico M, Sarti S, et al. Cell-free DNA fragmentomics: a promising biomarker for diagnosis, prognosis and prediction of response in breast cancer. Int J Mol Sci. 2022;23(22):14197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):22424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Magbanua MJM, Brown Swigart L, Ahmed Z, Sayaman RW, Renner D, Kalashnikova E, et al. Clinical significance and biology of circulating tumor DNA in high-risk early-stage HER2-negative breast cancer receiving neoadjuvant chemotherapy. Cancer Cell. 2023;41(6):1091-1102.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic—implementation issues and future challenges. Nat Rev Clin Oncol. 2021;18(5):297–312. [DOI] [PubMed] [Google Scholar]
- 94.Meddeb R, Pisareva E, Thierry AR. Guidelines for the preanalytical conditions for analyzing circulating cell-free DNA. Clin Chem. 2019;65(5):623–33. [DOI] [PubMed] [Google Scholar]
- 95.Lee NY, Hum M, Tan GP, Seah AC, Kin PT, Tan NC, et al. Degradation of methylation signals in cryopreserved DNA. Clin Epigenetics. 2023;15(1):147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schröder C, Steimer W. gDNA extraction yield and methylation status of blood samples are affected by long-term storage conditions. PLoS ONE. 2018;13(2): e0192414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bulla A, De Witt B, Ammerlaan W, Betsou F, Lescuyer P. Blood DNA yield but not integrity or methylation is impacted after long-term storage. Biopreserv Biobank. 2016;14(1):29–38. [DOI] [PubMed] [Google Scholar]
- 98.Diaz IM, Nocon A, Held SAE, Kobilay M, Skowasch D, Bronkhorst AJ, et al. Pre-analytical evaluation of streck cell-free DNA blood collection tubes for liquid profiling in oncology. Diagnostics (Basel, Switzerland). 2023;13(7):1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Olova N, Krueger F, Andrews S, Oxley D, Berrens RV, Branco MR, et al. Comparison of whole-genome bisulfite sequencing library preparation strategies identifies sources of biases affecting DNA methylation data. Genome Biol. 2018;19(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sun J, Su M, Ma J, Xu M, Ma C, Li W, et al. Cross-platform comparisons for targeted bisulfite sequencing of MGISEQ-2000 and NovaSeq6000. Clin Epigenet. 2023;15(1):130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Merker JD, Oxnard GR, Compton C, Diehn M, Hurley P, Lazar AJ, et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol. 2018;36(16):1631–41. [DOI] [PubMed] [Google Scholar]
- 102.Deveson IW, Gong B, Lai K, LoCoco JS, Richmond TA, Schageman J, et al. Evaluating the analytical validity of circulating tumor DNA sequencing assays for precision oncology. Nat Biotechnol. 2021;39(9):1115–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Richard MA, Huan T, Ligthart S, Gondalia R, Jhun MA, Brody JA, et al. DNA methylation analysis identifies loci for blood pressure regulation. Am J Hum Genet. 2017;101(6):888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fernández-Sanlés A, Sayols-Baixeras S, Subirana I, Degano IR, Elosua R. Association between DNA methylation and coronary heart disease or other atherosclerotic events: a systematic review. Atherosclerosis. 2017;263:325–33. [DOI] [PubMed] [Google Scholar]
- 105.Chambers JC, Loh M, Lehne B, Drong A, Kriebel J, Motta V, et al. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinol. 2015;3(7):526–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hillary RF, McCartney DL, Smith HM, Bernabeu E, Gadd DA, Chybowska AD, et al. Blood-based epigenome-wide analyses of 19 common disease states: a longitudinal, population-based linked cohort study of 18,413 Scottish individuals. PLoS Med. 2023;20(7): e1004247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wahl S, Drong A, Lehne B, Loh M, Scott WR, Kunze S, et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541(7635):81–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Feilotter H, Bruce C, Diamandis EP, Chatanaka MK, Yousef GM. Guidance for securing approvals for new biomarkers: from discovery to clinical implementation. Crit Rev Clin Lab Sci. 2025;62(1):1–8. [DOI] [PubMed] [Google Scholar]
- 109.Rendek T, Pos O, Duranova T, Saade R, Budis J, Repiska V, et al. Current challenges of methylation-based liquid biopsies in cancer diagnostics. Cancers (Basel). 2024;16(11):2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.ADVI. Payer coverage policies of tumor biomarker and pharmacogenomic testing. American Cancer Society Cancer Action Network. 2023 [cited 2025 Mar 6]. Available from: https://www.fightcancer.org/policy-resources/payer-coverage-policies-tumor-biomarker-and-pharmacogenomic-testing.
- 111.IJzerman MJ, de Boer J, Azad A, Degeling K, Geoghegan J, Hewitt C, et al. Towards routine implementation of liquid biopsies in cancer management: it is always too early, until suddenly it is too late. Diagnostics (Basel, Switzerland). 2021;11 (1):103. [DOI] [PMC free article] [PubMed]
- 112.Sharma M, Verma RK, Kumar S, Kumar V. Computational challenges in detection of cancer using cell-free DNA methylation. Comput Struct Biotechnol J. 2022;20:26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chowdhury B, Cho I-H, Irudayaraj J. Technical advances in global DNA methylation analysis in human cancers. J Biol Eng. 2017;11:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ho H-Y, Chung K-SK, Kan C-M, Wong S-CC. Liquid biopsy in the clinical management of cancers. Int J Mol Sci. 2024;25 (16):8594. [DOI] [PMC free article] [PubMed]
- 115.Martin-Alonso C, Tabrizi S, Xiong K, Blewett T, Sridhar S, Crnjac A, et al. Priming agents transiently reduce the clearance of cell-free DNA to improve liquid biopsies. Science. 2024;383(6680):eadf2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Saura C, Ortiz C, Matito J, Arenas EJ, Suñol A, Martín Á, et al. Early-stage breast cancer detection in breast milk. Cancer Discov. 2023;13(10):2180–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cohen JD, Javed AA, Thoburn C, Wong F, Tie J, Gibbs P, et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc Natl Acad Sci U S A. 2017;114(38):10202–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lipkova J, Chen RJ, Chen B, Lu MY, Barbieri M, Shao D, et al. Artificial intelligence for multimodal data integration in oncology. Cancer Cell. 2022;40(10):1095–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gong T, Borgard H, Zhang Z, Chen S, Gao Z, Deng Y. Analysis and performance assessment of the whole genome bisulfite sequencing data workflow: currently available tools and a practical guide to advance DNA methylation studies. Small Methods. 2022;6(3): e2101251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hai L, Li L, Liu Z, Tong Z, Sun Y. Whole-genome circulating tumor DNA methylation landscape reveals sensitive biomarkers of breast cancer. MedComm. 2022;3(3): e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Beck D, Ben Maamar M, Skinner MK. Genome-wide CpG density and DNA methylation analysis method (MeDIP, RRBS, and WGBS) comparisons. Epigenetics. 2022;17(5):518–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chung FF-L, Maldonado SG, Nemc A, Bouaoun L, Cahais V, Cuenin C, et al. Buffy coat signatures of breast cancer risk in a prospective cohort study. Clin Epigenet. 2023;15(1):102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lak NSM, van Zogchel LMJ, Zappeij-Kannegieter L, Javadi A, van Paemel R, Vandeputte C, et al. Cell-free DNA as a diagnostic and prognostic biomarker in pediatric rhabdomyosarcoma. JCO Precis Oncol. 2023;7: e2200113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Van Paemel R, De Koker A, Vandeputte C, van Zogchel L, Lammens T, Laureys G, et al. Minimally invasive classification of paediatric solid tumours using reduced representation bisulphite sequencing of cell-free DNA: a proof-of-principle study. Epigenetics. 2021;16(2):196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee E-J, Luo J, Wilson JM, Shi H. Analyzing the cancer methylome through targeted bisulfite sequencing. Cancer Lett. 2013;340(2):171–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu Y, Cheng J, Siejka-Zielińska P, Weldon C, Roberts H, Lopopolo M, et al. Accurate targeted long-read DNA methylation and hydroxymethylation sequencing with TAPS. Genome Biol. 2020;21(1):54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chen J, Cheng J, Chen X, Inoue M, Liu Y, Song C-X. Whole-genome long-read TAPS deciphers DNA methylation patterns at base resolution using PacBio SMRT sequencing technology. Nucleic Acids Res. 2022;50(18): e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Taiwo O, Wilson GA, Morris T, Seisenberger S, Reik W, Pearce D, et al. Methylome analysis using MeDIP-seq with low DNA concentrations. Nat Protoc. 2012;7(4):617–36. [DOI] [PubMed] [Google Scholar]
- 129.Shen SY, Burgener JM, Bratman SV, De Carvalho DD. Preparation of cfMeDIP-seq libraries for methylome profiling of plasma cell-free DNA. Nat Protoc. 2019;14(10):2749–80. [DOI] [PubMed] [Google Scholar]
- 130.De Meyer T, Mampaey E, Vlemmix M, Denil S, Trooskens G, Renard J-P, et al. Quality evaluation of methyl binding domain based kits for enrichment DNA-methylation sequencing. PLoS ONE. 2013;8(3): e59068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Brinkman AB, Simmer F, Ma K, Kaan A, Zhu J, Stunnenberg HG. Whole-genome DNA methylation profiling using MethylCap-seq. Methods. 2010;52(3):232–6. [DOI] [PubMed] [Google Scholar]
- 132.Zhao Y, Xue F, Sun J, Guo S, Zhang H, Qiu B, et al. Genome-wide methylation profiling of the different stages of hepatitis B virus-related hepatocellular carcinoma development in plasma cell-free DNA reveals potential biomarkers for early detection and high-risk monitoring of hepatocellular carcinoma. Clin Epigenet. 2014;6(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2(9):2265–75. [DOI] [PubMed] [Google Scholar]
- 134.Muggerud AA, Rønneberg JA, Wärnberg F, Botling J, Busato F, Jovanovic J, et al. Frequent aberrant DNA methylation of ABCB1, FOXC1, PPP2R2B and PTEN in ductal carcinoma in situ and early invasive breast cancer. Breast Cancer Res. 2010;12(1):R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Higashimoto K, Hara S, Soejima H. DNA methylation analysis using bisulfite pyrosequencing. Methods Mol Biol. 2023;2577:3–20. [DOI] [PubMed] [Google Scholar]
- 136.Estival A, Sanz C, Ramirez J-L, Velarde JM, Domenech M, Carrato C, et al. Pyrosequencing versus methylation-specific PCR for assessment of MGMT methylation in tumor and blood samples of glioblastoma patients. Sci Rep. 2019;9(1):11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Absmaier M, Napieralski R, Schuster T, Aubele M, Walch A, Magdolen V, et al. PITX2 DNA-methylation predicts response to anthracycline-based adjuvant chemotherapy in triple-negative breast cancer patients. Int J Oncol. 2018;52(3):755–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sigalotti L, Covre A, Colizzi F, Fratta E. Quantitative methylation-specific PCR: a simple method for studying epigenetic modifications of cell-free DNA. Methods Mol Biol. 2019;1909:137–62. [DOI] [PubMed] [Google Scholar]
- 139.Oh TJ, Oh HI, Seo YY, Jeong D, Kim C, Kang HW, et al. Feasibility of quantifying SDC2 methylation in stool DNA for early detection of colorectal cancer. Clin Epigenet. 2017;9:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83(22):8604–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Postel M, Roosen A, Laurent-Puig P, Taly V, Wang-Renault S-F. Droplet-based digital PCR and next generation sequencing for monitoring circulating tumor DNA: a cancer diagnostic perspective. Expert Rev Mol Diagn. 2018;18(1):7–17. [DOI] [PubMed] [Google Scholar]
- 142.Neefs I, De Meulenaere N, Vanpoucke T, Vandenhoeck J, Peeters D, Peeters M, et al. Simultaneous detection of eight cancer types using a multiplex droplet digital PCR assay. Mol Oncol. 2025;19(1):188–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Choy LYL, Peng W, Jiang P, Cheng SH, Yu SCY, Shang H, et al. Single-molecule sequencing enables long cell-free DNA detection and direct methylation analysis for cancer patients. Clin Chem. 2022;68(9):1151–63. [DOI] [PubMed] [Google Scholar]
- 144.Yu SCY, Deng J, Qiao R, Cheng SH, Peng W, Lau SL, et al. Comparison of single molecule, real-time sequencing and nanopore sequencing for analysis of the size, end-motif, and tissue-of-origin of long cell-free DNA in plasma. Clin Chem. 2023;69(2):168–79. [DOI] [PubMed] [Google Scholar]
- 145.Sakamoto Y, Zaha S, Nagasawa S, Miyake S, Kojima Y, Suzuki A, et al. Long-read whole-genome methylation patterning using enzymatic base conversion and nanopore sequencing. Nucleic Acids Res. 2021;49(14): e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yang Y, Wu L, Shu X-O, Cai Q, Shu X, Li B, et al. Genetically predicted levels of DNA methylation biomarkers and breast cancer risk: data from 228951 women of european descent. J Natl Cancer Inst. 2020;112(3):295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable. No datasets were generated or analyzed in this study.