Abstract
Despite the development of mutant-selective KRAS inhibitors, colorectal cancer (CRC) responses remain limited, with stable disease and rapid recurrence being common outcomes. The molecular mechanisms enabling CRC cells to tolerate KRAS inhibition and ultimately develop resistance remain poorly understood. Here, we investigated early transcriptional and proteomic responses to KRAS silencing in 3D CRC cell line spheroid models, aiming to identify pathways associated with sensitivity or resistance to KRAS blockade. Cell lines were stratified into KRAS silencing-sensitive (HCT116 and SW480) and -resistant (LS174T and SW837) groups based on spheroid growth, cell cycle progression, and apoptosis induction. Transcriptional profiling revealed the unfolded protein response (UPR) and WNT/β-catenin signaling as pathways specifically upregulated in KRAS silencing-sensitive cells and downregulated in resistant cells. Proteomic analysis of membrane-enriched fractions further supported UPR deregulation, showing a pronounced downregulation of translation-related proteins in sensitive cells. Functional assays validated that the sensitive cell line HCT116 exhibits reduced protein aggregation and lower translational capacity upon KRAS knockdown, consistent with UPR activation. Pharmacological inhibition of IRE1α-mediated UPR signaling did not revert KRAS silencing-induced cell cycle arrest or apoptosis in this cell line. Collectively, our results highlight the UPR activation as an early adaptive response of KRAS-dependent CRC cells to KRAS silencing.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-025-94549-2.
Subject terms: Cancer therapy, Oncogenes, Cell biology, Molecular biology
Introduction
The transforming potential of KRAS-homologous sequences and the identification of KRAS mutations in human cancers date back to the early 1980s1–4. Since then, many studies addressing the relevance of KRAS in human malignancies have established it as the most frequent mutated oncogene in cancer5,6. These studies have highlighted the multifaceted oncogenic functions of KRAS. Mutant KRAS serves not only as a pro-proliferative and pro-survival factor but also as a promoter of stemness, dedifferentiation, migration, invasion, metastasis, therapy resistance, and a modulator of cancer cell-tumor microenvironment interactions7–12. Collectively, these oncogenic functions underscore KRAS as a critical therapeutic target in several cancers.
For a long time, targeting mutated KRAS was unsuccessful, prompting the idea of KRAS as undruggable13. Additionally, numerous attempts to inhibit KRAS oncogenic activity by targeting its upstream regulators or downstream effectors also proved ineffective14. Despite these setbacks, significant efforts persisted in developing selective inhibitors for mutant KRAS14,15, which became fruitful after Shokat et al.16 made a groundbreaking discovery of small molecules that covalently bind to the acquired cysteine residue within the switch II region of the KRASG12C mutant17. These findings laid the foundational steps to therapeutic KRAS blockade. The first mutant-specific KRAS-targeted therapy was recently approved for the treatment of non-small cell lung cancer patients with G12C mutations18,19.
Although 35 to 40% of CRC have a KRAS mutation20, less than 3% present a KRAS-G12C21. Fortunately, the field is advancing rapidly, with numerous inhibitors targeting other KRAS mutations currently in development22, thus offering renewed hope for better treatment options for a larger number of patients with KRAS-mutant CRC. However, in CRC, stable disease remains the most common outcome among patients treated with KRAS-G12C inhibitors as monotherapy23–25. Nevertheless, combining Cetuximab with KRAS G12C inhibition yielded better responses, albeit all partial, showing a modest median progression-free survival increase from 5.6 to 6.9 months26. Still, in June this year, the Food and Drug Administration granted accelerated approval to KRAS G12C inhibitor Adagrasib plus Cetuximab for adults with KRAS G12C-mutated locally advanced or metastatic CRC.
Disease stabilization and the rapid recurrence suggest that KRAS-inhibited cells can persist in a drug-tolerant state, remaining viable long enough to develop resistance mechanisms that eventually drive tumor relapse. The mechanisms that allow cells to tolerate KRAS inhibition and to develop a fully resistant phenotype are still unknown. Addressing this question will be crucial for designing combinatorial strategies that prevent early adaptation and maximize the effectiveness of KRAS-targeted therapies. Our study aimed to uncover the early molecular responses to KRAS silencing. Using a 3D cell culture system, we stratified the cell lines according to their sensitivity to KRAS silencing to allow the identification of response-specific pathways. Using RNA sequencing, proteomic analysis of cell membranes, and functional validation studies, we identified upregulation of the unfolded protein response (UPR) as an early molecular change in KRAS-dependent cells following KRAS silencing.
Results
Evaluation of KRAS-dependency in 3-dimensional cultures of colorectal cancer cell lines
Four mutant KRAS CRC cell lines were used in this study (HCT116, SW480, LS174T, and SW837). Although all four cell lines harbor KRAS mutations, previous studies using 2-dimensional (2D) cell culture models have demonstrated that they exhibit varying levels of dependency on KRAS oncogenic signaling for survival27. However, 2D cell culture models are being progressively substituted by 3-dimensional (3D) models that better mimic the architecture and cellular interactions that exist in tumors28. Therefore, our studies were performed using CRC cell line spheroids. To validate KRAS dependency in this 3D cell culture model, cells were seeded into agarose molds 24 h after KRAS silencing to form spheroids and cultured for another 48 h. As such, all the analyses were performed 72 h after KRAS silencing (Fig. S1A, B).
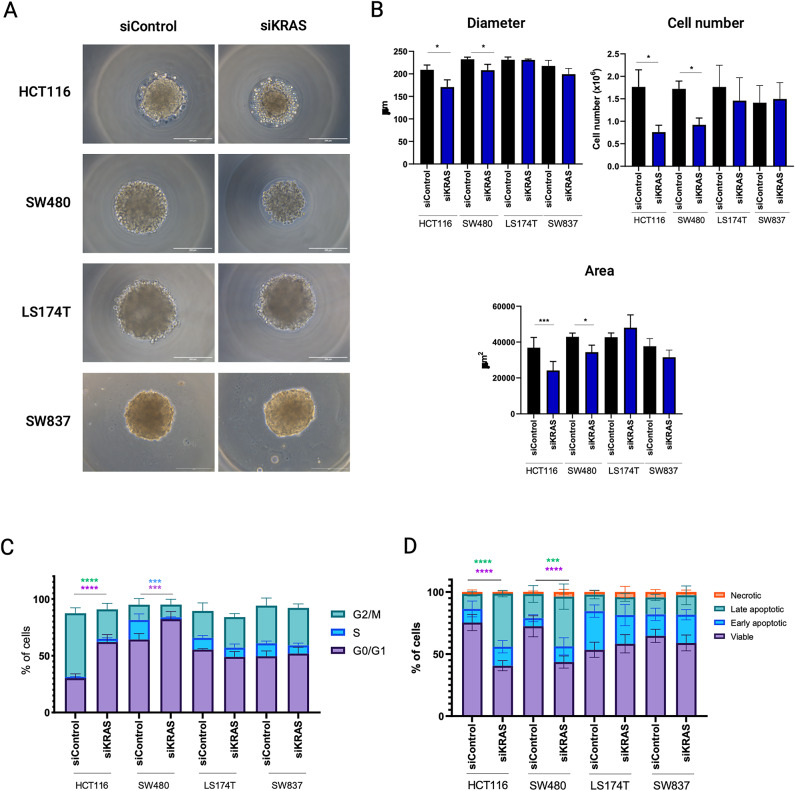
The four cell lines formed spheroids in less than 24 h after seeding, either with a compact (HCT116 and SW837) or loose (SW480 and LS174T) morphology (Fig. 1A). Although all the cell lines exhibited a high degree of KRAS silencing (Figs. S1A–C, and S12–S15), only HCT116 and SW480 cell lines exhibited KRAS-signaling dependency. In both cell lines, spheroids formed by siKRAS cells had a smaller area and diameter compared with spheroids formed by siControl cells, along with a reduction in the number of cells (Fig. 1B). Additionally, HCT116 siKRAS spheroids tended to exhibit decreased circularity and a significant decrease in solidity (Fig. S2). No significant alterations were observed between the siControl and siKRAS conditions in LS174T and SW837 spheroids (Fig. 1A, B and Fig. S2), further corroborating the independence of these cell lines on KRAS signaling.
Fig. 1.
Colorectal cancer cells retain KRAS dependency in 3D cell cultures. HCT116, SW480, LS174T and SW837 cells, both siControl and siKRAS, were grown as spheroids for 48 h. (A) Representative microscopy images of spheroids of siControl and siKRAS cells (scale bar: 200 μm). (B) Quantification of spheroids diameter, area and cell number; paired t-test was used for statistical analysis (*P ≤ 0.05; ***P ≤ 0.001). (C) Cell cycle analysis: bar graphs illustrate the percentage of cells in the different cell cycle phases (G0/G1, S, G2/M); Two-way ANOVA with multiple comparisons was used to compare different populations in siControl with siKRAS conditions (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001). (D) Apoptosis analysis: bar graphs represent the percentage of cells that are in viable, early or late apoptosis, and necrotic states; Two-way ANOVA with multiple comparisons was used to compare different populations in siControl with siKRAS conditions (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 00001). All the bar graphs represent the mean ± SD of at least three independent experiments.
Next, we evaluated the impact of KRAS silencing on cell cycle progression and apoptosis by flow cytometry. The gating strategy used for cell cycle and apoptosis analysis can be found as supplementary data (Figs. S3 and S4). Compared with siControl cells, siKRAS HCT116 cells exhibited a significant increase in the G0/G1 phase and a decrease in the G2/M phase. siKRAS SW480 cells exhibited a significant increase in the G0/G1 population and a decrease in the S phase population (Fig. 1C) compared with their respective siControl. Regarding apoptosis, statistically significant differences were only detected in siKRAS HCT116 and siKRAS SW480 cells, which showed an increase in late apoptotic cells compared with siControl cells (Fig. 1D). No differences were observed for KRAS-silencing resistant LS174T and SW837 cell lines, either in terms of cell cycle or apoptosis (Fig. 1C, D).
In summary, our results confirm that the SW480 CRC cell line is sensitive to KRAS silencing and the LS174T and SW837 cell lines are resistant. Although HCT116 cells were originally classified as KRAS-independent based on survival analysis in 2D cell cultures, they are classified as sensitive to KRAS silencing in our 3D model based on the proliferation and apoptosis analysis.
The unfolded protein response and WNT pathways are specifically upregulated in KRAS Silencing sensitive cells
To identify pathways of early response to KRAS silencing, we performed RNA-Seq in the four different cell lines in siKRAS and siControl conditions. The analysis revealed that, independently of the sensitivity of the cell lines, KRAS silencing induces a transcriptional profile distinct from siControl cells. Differential expression analysis performed using DESeq2 revealed that a total of 761 genes were found to be differentially expressed between siControl and siKRAS HCT116 cells, with 267 genes up-regulated and 494 down-regulated in siKRAS cells (Table S1). siKRAS SW480 cells presented 1104 differentially expressed genes (DEGs), having 548 up-regulated and 539 down-regulated (Table S2). In LS174T, there was a total of 457 DEGs in siKRAS versus siControl cells, in which 374 were up-regulated, and 83 were down-regulated (Table S3). In siKRAS SW837 cells, a total of 1225 genes were differentially expressed; from these, 415 were up-regulated and 710 were down-regulated (Table S4).
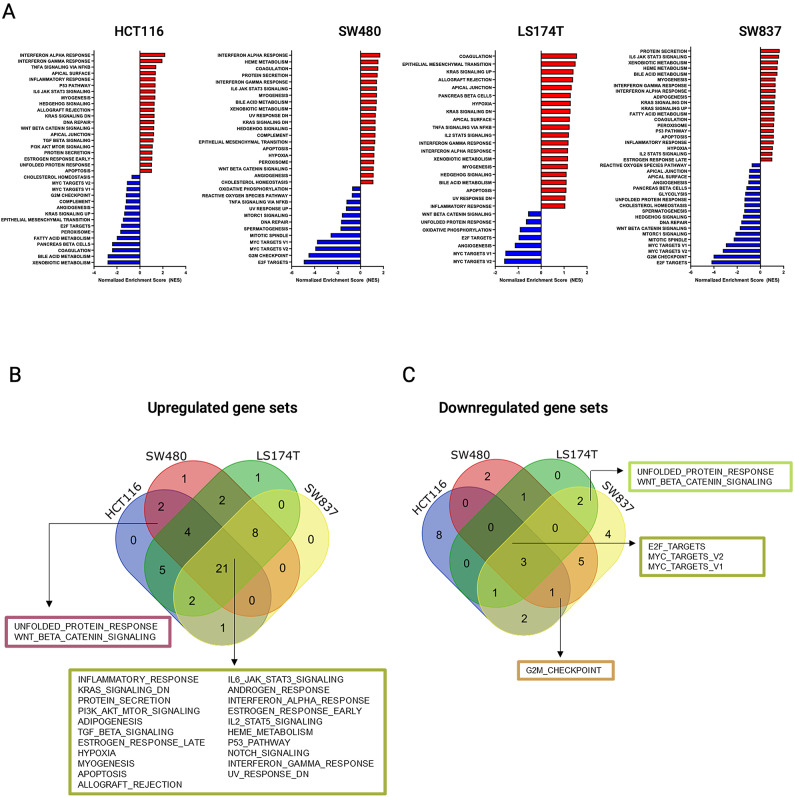
To identify sets of genes with coordinated enrichment or depletion for siKRAS compared to siControl, we performed GSEA for the Hallmarks gene set. In the HCT116 cell line, enrichment was observed in 35 gene sets, and 15 gene sets were depleted in siKRAS cells (Fig. 2A and Table S5). None of the positively enriched gene sets were HCT116-specific, as opposed to 8 out of the 15 negatively enriched gene sets (Fig. 2B, C and Table S5). SW480 cell line showed enrichment in 38 gene sets and depletion in 12 gene sets in siKRAS cells (Fig. 2A and Table S6). One positively enriched gene set and two depleted gene sets were SW480-specific (Fig. 2B, C and Table S6). The LS174T cell line exhibited enrichment in 43 gene sets and depletion in 7 gene sets in siKRAS cells (Fig. 2A and Table S7). Only one of the enriched and none of the depleted gene sets were specific from LS174T (Fig. 2B, C and Table S7). In the SW837, enrichment in 32 gene sets and depletion in 18 gene sets were found for siKRAS cells (Fig. 2A and Table S8); none of the positively enriched were specific, and four of the depleted were exclusive from this cell line (Fig. 2A, B and Table S8). Afterwards, we aimed to distinguish between a general KRAS silencing-related signature (gene sets differentially altered upon KRAS silencing regardless of the sensitivity of the cell lines) from a KRAS silencing- and response-specific signature (gene sets specifically altered in sensitive versus resistant cell lines), which could indicate molecular mediators of response. Most of the gene sets (21 gene sets) were commonly enriched across all cell lines following KRAS silencing, including those associated with inflammatory response, KRAS signaling, and PI3K/AKT/mTOR signaling, among others (Fig. 2B). Only three gene sets were commonly depleted in all cell lines: E2F targets and MYC targets (V2 and V1) (Fig. 2C). Regarding gene sets unique to KRAS-silencing sensitive and KRAS-silencing resistant cell lines, only two gene sets were differentially and oppositely expressed between the two groups of cell lines. Specifically, the unfolded protein response (UPR) and WNT/β-catenin signaling gene sets were exclusively enriched in KRAS-silencing sensitive cell lines (Fig. 2B) and downregulated in KRAS-silencing resistant cell lines (Fig. 2C).
Fig. 2.
Impact of KRAS inhibition on the transcriptomic profile of cells. (A) Results of GSEA Hallmark analysis showing enriched (top 20) and depleted gene sets in HCT116, SW480, LS174T and SW487 cells. A positive Normalized Enrichment Score (NES) value indicates enrichment in the siKRAS phenotype, and a negative NES indicates enrichment in the siControl phenotype. (B) Venn diagram illustrating the unique and shared gene sets enriched in siKRAS cells in HCT116, SW480, LS174T and SW837 cell lines. (C) Venn diagram illustrating the unique and shared gene sets depleted in siKRAS cells in HCT116, SW480, LS174T and SW837 cell lines.
Membrane proteome analysis identifies translation downregulation as a specific feature of KRAS-silencing sensitive cells
The involvement of the UPR in resistance to KRAS inhibition was previously documented in pancreatic and lung cancer models. UPR pathways are mainly initiated by three protein sensor signaling cascades, PERK, IRE1A and ATF6, at the endoplasmic reticulum (ER) membrane29, all of them found upregulated in the RNAseq data of KRAS silencing sensitive cells (Table S9). Therefore, we decided to perform membrane protein enrichment followed by proteomic analysis in the four different cell lines to validate, at the protein level, the UPR alterations found in RNAseq data and to further identify other associated signaling pathways. Two technical replicates and three independent biological replicates were performed for each condition and cell line.
No significant differences were observed among the number of identified proteins between siControl or siKRAS conditions for all cell lines (Fig. S5). Collapsing the protein isoforms into their encoding genes identified in all independent biological and technical replicates resulted in 1015 proteins identified in siControl and 1007 identified in siKRAS of HCT116 cells; 902 proteins in siControl and 960 in siKRAS of SW480 cells; 944 proteins siControl and 942 in siKRAS of LS174T cells; 874 proteins in siControl and 1127 in siKRAS of SW837 cells. Principal component analysis using all quantitative data without any bias filtering of quantitative values showed a complete separation of siControl and siKRAS conditions in HCT116 cells (Fig. S6). The same was not observed for the other cell lines, especially for LS174T and SW837 cells.
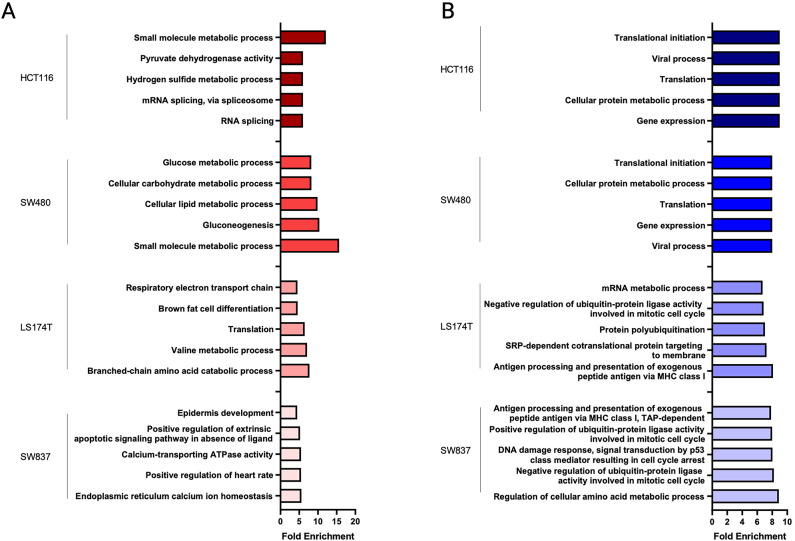
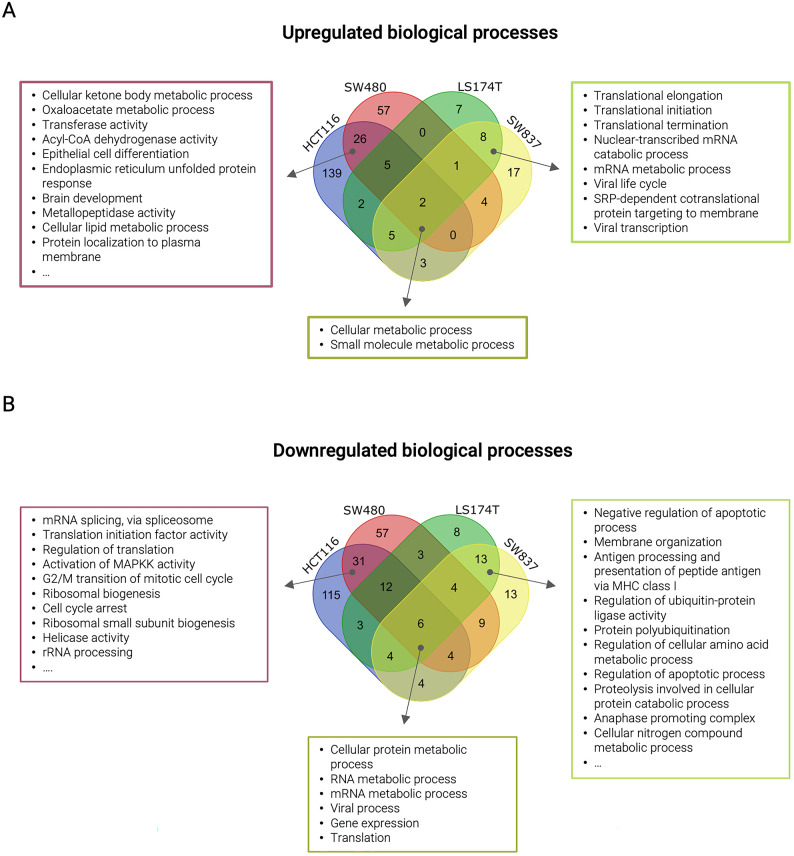
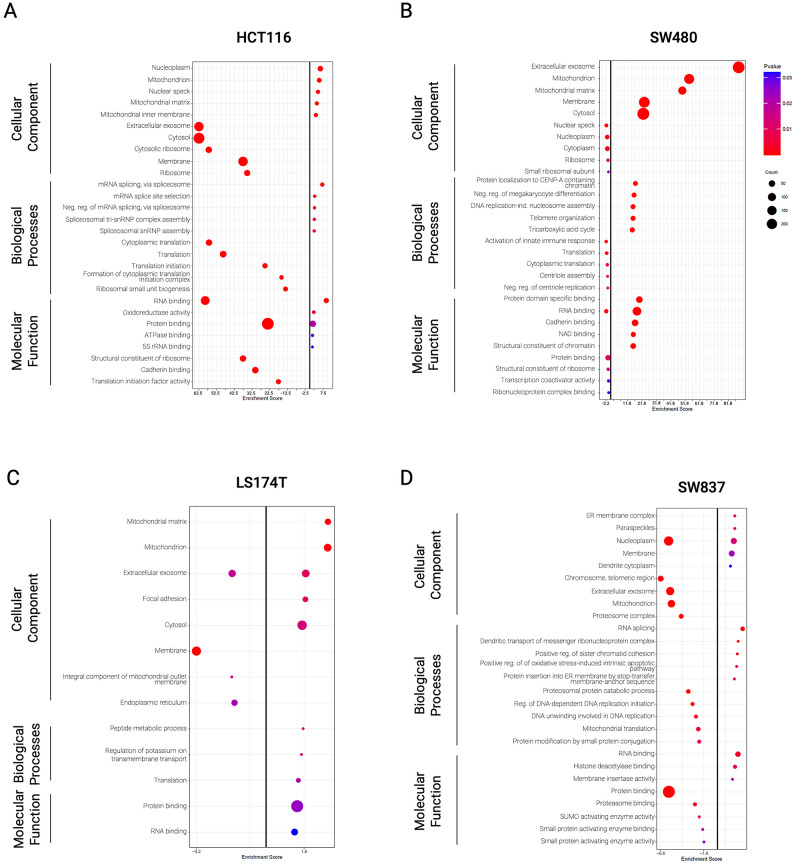
To verify the enrichment efficiency in cell membrane-associated proteins, we performed gene ontology for cellular components with all identified proteins. The analysis revealed an enrichment in proteins associated with Membrane, Extracellular Exosomes, and Mitochondria in all cell lines except for SW837, in which proteins associated with the cellular component Membrane did not figure in the top 5 (Fig. S7 and Table S10). Gene ontology analysis of biological processes using all up- and down-regulated proteins (independently of statistical significance) found in siKRAS cells revealed that up-regulated proteins were mostly associated with metabolic processes in all cell lines (Figs. 3A and 4A). Additionally, in LS174T and SW837 cell lines, up-regulated proteins were associated with translation processes (Figs. 3A and 4A), while in HCT116 and SW480 cell lines were related with the metabolism, epithelial cell differentiation, and the unfolded protein response (Figs. 3A and 4A). Down-regulated proteins were associated with protein and RNA metabolic processes, gene expression and translation in all cell lines (Figs. 3B and 4B). Moreover, translation down-regulation was particularly pronounced in HCT116 and SW480 KRAS-silencing sensitive cell lines in which several of the down-regulated proteins are involved in different translation-related processes (translational elongation, regulation of translational initiation, translation initiation factor activity, translational termination, and negative regulation of translation) (Figs. 3B and 4B). Down-regulated proteins found in LS174T and SW837 KRAS-silencing resistant cell lines were associated with apoptotic and cell cycle processes (Figs. 3B and 4B).
Fig. 3.
Functional enrichment for biological processes using all identified proteins from each cell line. (A) Functional enrichment for biological processes associated with enriched proteins in siKRAS condition. (B) Functional enrichment for biological processes associated with depleted proteins in siKRAS condition. Fold enrichment was calculated by –log(p-value). Data is representative of three biological replicates. Only the top 5 biological processes are represented in the graph.
Fig. 4.
Unique and shared biological processes across cell lines using all identified proteins from each cell line. (A) Venn diagram illustrating the number of unique and shared biological processes from enriched proteins in siKRAS condition in all cell lines. (B) Venn diagram illustrating the number of unique and shared biological processes.
Statistical comparison between siKRAS and siControl in HCT116 cells revealed 373 differentially expressed proteins (DEPs): 60 up-regulated and 313 down-regulated in siKRAS (Fig. S8A + Table S11). In SW480 cells, the same analysis revealed 490 DEPs: 469 up-regulated and 21 down-regulated in siKRAS (Fig. S8B + Table S12). For LS174T cells, a total of 50 DEPs were found in siKRAS: 27 up-regulated and 23 down-regulated (Fig. S8C + Table S13). In SW837 cells, the analysis revealed 98 DEPs between siKRAS and siControl cells: 21 up-regulated and 77 down-regulated in siKRAS (Fig. S8D + Table S14). GO enrichment analysis for cellular component, molecular function, and biological process of statistically significant DEPs showed significant enrichment of proteins localized in the nucleoplasm and in the mitochondria, with binding molecular functions (RNA/Protein, ATPase binding), and involved in splicing-related biological processes (mRNA/RNA splicing, spliceosomal assembly) in siKRAS HCT116 cells (Fig. 5A and Table S11). In siKRAS SW480 cells, the same analysis revealed significant enrichment of proteins localized in the extracellular exosome and the mitochondria, with molecular functions related to binding and metabolic activities and associated with several metabolic and chromatin organization biological processes (Fig. 5B and Table S12). An enrichment of metabolic and translation processes, proteins localized in the mitochondrion, and binding functions was observed in siKRAS LS174T cells (Fig. 5C and Table S13). In SW837 cells, a significant enrichment of splicing, metabolism, and chromosome organization processes was observed, along with enrichment of proteins localized in the ER membrane or nucleoplasm and associated with binding molecular functions in siKRAS cells (Fig. 5D and Table S14). GO analysis of significant down-regulated DEPs showed depletion of translation and metabolic processes, proteins localized mainly in extracellular exosome, cytosol, or membranes, and associated with binding functions in siKRAS HCT116 cells (Fig. 5A and Table S11). Innate immune response and translation processes, along with proteins localized in nuclear compartments, cytoplasm and ribosomes and related with binding activities, were found to be depleted in siKRAS SW480 cells (Fig. 5B and Table S12). In LS174T siKRAS cells, down-regulated proteins were primarily localized at membranes, the extracellular exosome, and the endoplasmic reticulum but did not show any association with biological processes or molecular functions (Fig. 5C and Table S13). In siKRAS SW837 cells, the same analysis showed a downregulation of proteins with binding-related molecular functions localized in nuclear compartments, extracellular exosomes, and mitochondrion and associated with metabolic and replication biological processes (Fig. 5D and Table S14).
Fig. 5.
GO terms for cellular component, biological processes and molecular functions using differentially expressed proteins from each cell line. Functional enrichment for cellular components, biological process and molecular functions from up- and down-regulated proteins in siKRAS condition in (A) HCT116 cells, (B) SW480 cells, (C) LS174T cells, (D) SW837 cells. Enrichment was calculated by − log(p-value). Positive values represent up-regulated and negative values represent down-regulated cellular components, biological processes or molecular functions. Only top 5 GO terms are represented in the graphs.
KRAS-silencing sensitive cells increase protein aggregation and lower the translational capacity upon KRAS silencing
RNAseq and proteomic data highlighted the differential regulation of the UPR between KRAS-silencing sensitive and resistant cell lines. UPR activation has been previously implicated in the resistance mechanism to KRAS inhibitors in lung and pancreatic cancers30. Therefore, we sought to validate the differential alteration in UPR found in our study through functional assays.
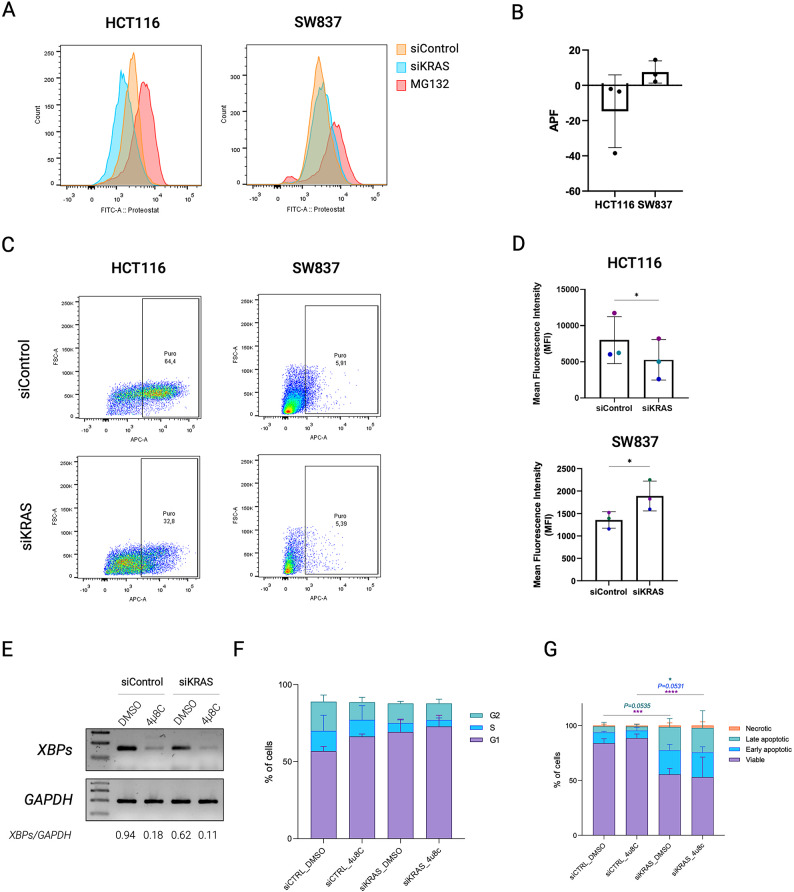
Accumulation of unfolded proteins in the endoplasmic reticulum elicits the UPR, decreasing protein aggregates and enhancing proteostasis31–34. Therefore, we evaluated protein aggregation through PROTEOSTAT assay in HCT116 (sensitive to KRAS silencing) and SW837 cells (resistant to KRAS silencing). In HCT116, PROTEOSTAT staining revealed a decrease in protein aggregates in KRAS-silenced viable cells compared to siControl cells (Fig. 6A, B and Fig. S9), consistent with the UPR upregulation. In contrast, viable siKRAS SW837 cells displayed a slight increase in PROTEOSTAT staining compared to their respective siControl cells (Fig. 6A, B and Fig. S9), in accordance with UPR downregulation. As one of the consequences of UPR activation is the downregulation of protein translation to ensure proper folding and degradation of misfolded proteins, we evaluated the alterations in the translation potential of these cell lines through the SunSET assay. KRAS-silenced HCT116 viable cells presented a decrease in protein translation when compared to siControl cells (Fig. 6C and Fig. S10). In contrast, SW837 cells presented an increase in protein translation upon KRAS silencing (Fig. 6D and Fig. S10). Overall, the PROTEOSTAT and SunSET assays validated the RNASeq and membrane proteomic data, respectively, specifically showing the downregulation of both protein aggregates and translational potential in mutant KRAS CRC cells sensitive to KRAS silencing.
Fig. 6.
Oncogenic KRAS inactivation affects proteostasis. (A) Representative overlaid histograms for the fluorescence intensity of siControl and siKRAS HCT116 and SW837 cells. MG132 was used as positive control. (B) The mean fluorescence intensity values are used to compute the aggresome propensity factor (APF). Data shows the results of three biological replicates and are shown as mean ± SD. (C) Representative flow cytometry profiles for the fluorescence intensity of siControl and siKRAS HCT116 and SW837 cells. (D) The mean fluorescence intensity values of the positive population were used to compute the protein translation potential. Data shows the results of three independent experiments and are shown as mean ± SD. (E) Representative agarose gel of XBPsplicing inhibition after treatment with 4µ8C XBPsplicing and GAPDH PCR products were loaded in different gels. Uncropped versions of the images are provided in Supplementary Fig. 11. (F) Cell cycle analysis in HCT116 cells treated with 4µ8C. Bar graphs illustrate the percentage of cells G0/G1, S, G2/M. (G) Apoptosis analysis of HCT116 cells treated with 4µ8C. Bar graphs represent the percentage of cells that are in viable, early or late apoptosis, and necrotic states. Two-way ANOVA with multiple comparisons was used to compare different populations in siControl with siKRAS conditions (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 00001). All the bar graphs represent the mean ± SD of at least three independent experiments.
Lastly, since UPR activation is often implicated in mediating apoptosis or cell cycle arrest under stress conditions35, we questioned whether it could sensitize cells to KRAS silencing by inducing cell cycle arrest and apoptosis. Since IRE1α-mediated reprogramming of proteostasis was previously implicated in the response of lung and pancreatic cancer cells to KRAS–MAPK inhibition36, we combined KRAS silencing with 4µ8C, an IRE1α pharmacological inhibitor, in the sensitive cell line HCT116. 4µ8C treatment induced a decrease in XBP1 splicing (Fig. 6E; Fig. S11), demonstrating that IRE1α activity was inhibited. Regarding the cell cycle, a slight increase in the percentage of cells in G0/G1 was observed in siControl cells (Fig. 6F); however, this was not statistically significant. Overall, IRE1α inhibition was not able to revert or enhance the cell cycle arrest and apoptosis (Fig. 6F, G) induced by KRAS silencing, suggesting that UPR activation is likely to play a cytoprotective role through alternative pathways.
Discussion
To elucidate the immediate effects of KRAS oncogenic signaling loss on colorectal cancer (CRC) cells, we employed an unbiased RNA sequencing (RNA-seq) approach. This comprehensive transcriptomic analysis allowed us to identify key pathways and gene expression changes that occur in response to the sudden loss of KRAS signaling, providing a snapshot of the early molecular events that shape the cellular response to KRAS inhibition. We identified alterations in the transcriptional profile and proteostasis, which were dependent on the sensitivity of the cell lines to KRAS silencing. By comparing KRAS silencing sensitive cell lines with innate resistant cell lines, only two pathways, the WNT and UPR, revealed opposite deregulation between the two settings. Both pathways were up-regulated in sensitive cell lines and down-regulated in resistant cell lines after KRAS silencing. We explored in more detail the UPR deregulation as this pathway is known to be activated during KRAS oncogenic transformation of normal cells37 and was recently associated with the response and acquisition of resistance to the KRAS inhibitor, being pinpointed as a potential therapeutic target to enhance the response to KRAS inhibitors30. Specifically, UPR activity was downregulated through IRE1α ubiquitination in response to acute KRAS inhibition in lung and pancreatic cancer models, which accords with the well-established UPR activation during KRAS oncogenic transformation. IRE1α activity was selectively reactivated in an ER stress-independent manner in cells that acquired resistance to KRAS inhibitor30. However, nothing was known about UPR activity regarding KRAS inhibition in CRC. Our study also implicates the UPR in the response of CRC cells to KRAS oncogenic signaling loss. However, UPR downregulation and the concomitant increase in protein aggregates and upregulation of translation-associated pathways were only observed in cell lines that were innately resistant to KRAS silencing. Notably, KRAS silencing-sensitive cell lines followed the opposite direction, enhancing the UPR program with a subsequent decrease in protein aggregates and downregulating translation-related pathways. Of note, in the study of Ly et al.36, the lung and pancreatic cell lines used, H358 and Mia-PaCa-2, respectively, were also previously described as KRAS-dependent38. Therefore, the differences observed between our study and the study of Ly et al.36 are likely to highlight differences intrinsic to the cancer models. In addition, in the study of Ly et al., IRE1α inhibition was shown to abrogate resistance to KRAS inhibition, pinpointing it as a therapeutic strategy to improve responses to KRAS inhibitors. Yet, in a previous study from our group using the KRAS silenced HCT116 sensitive cell line in which KRAS-growth independence was induced by treatment with fibroblast-secreted factors, the UPR pathway was one of the most downregulated pathways39. This observation reinforces the opposite trend to what was reported in the lung and pancreatic models. Therefore, considering both of our results showing upregulation of UPR programs shortly after KRAS silencing and downregulation in cells that regained proliferative potential, we suggest that the UPR program activation occurs early, in the tolerant phase to KRAS inhibition and is then lost when cells resume growth.
What triggers UPR upregulation in cells that lost a UPR oncogenic trigger and apparently do not have a high demand for protein synthesis as they are dying or arrested in the G0/G1 cell cycle phase, and which are the functional consequences of UPR upregulation require further studies. The transcriptional and metabolic abnormalities found in the RNAseq and proteomics data can trigger an imbalance in proteostasis, thus increasing ER stress and, consequently, the immediate activation of the UPR40. Regarding the functional effects, it neither seems to be anti-tumorigenic as IRE1α inhibition did not revert the increase in apoptosis, nor does it seem to drive cells toward tolerance to KRAS silencing by endowing a quiescent, cell cycle arrested state. Still, a role for UPR upregulation in supporting the capacity of CRC to withstand KRAS oncogenic signaling loss cannot be excluded. On the one hand, the lack of effect of IRE1α inhibition on cell cycle progression and apoptosis may be due to the compensatory overactivation of other UPR branches, as both PERK and ATF6 were found to be upregulated in KRAS-silencing sensitive cells. On the other hand, UPR activation may not directly regulate cell cycle and survival. It can result from an imbalance in proteostasis due to transcriptional dysregulation, which increases ER stress, thereby triggering immediate UPR activation. For instance, the RNA-seq data and previous findings from our group41, indicate alterations in transcription-related and mRNA splicing pathways alongside a higher transcriptomic instability. These observations suggest that UPR upregulation may be a downstream effect of the transcriptional reprogramming that occurs in sensitive cells as they attempt to adapt to KRAS silencing, ultimately ensuring proteostasis. In addition, UPR activation may have consequences beyond the cancer cell. It has been demonstrated to dynamically reshape the tumor microenvironment and how cancer cells interact with it. For example, the downregulation of protein synthesis resulting from UPR activation may affect what cells present on their surface via the major histocompatibility complex, thus influencing their recognition by the immune system42,43. Moreover, UPR signaling in cancer cells promotes angiogenesis, immune evasion, and extracellular matrix remodeling44–47, likely creating a pro-tumorigenic ecosystem. Thus, further studies are needed to verify whether UPR inhibition can synergize with KRAS inhibitors not by affecting the intrinsic properties of cancer cells but by abrogating pro-tumorigenic interaction with the microenvironment.
Although we use a siRNA strategy to downregulate KRAS oncogenic signaling, the clinical relevance of our model is further supported by the fact that we could recapitulate functional and molecular findings previously described in tumors treated with KRAS inhibitors. For example, cell cycle alterations, namely the G0/G1 arrest found in KRAS-dependent cell lines CRC spheroids, are consistent with the ones described in vivo upon treatment with KRAS inhibitors48. In addition, our transcriptomics data revealed a depletion of E2F and MYC target signatures, and an enrichment in interferon responses (alpha and gamma) upon KRAS silencing. These findings are consistent with previous reports using KRAS inhibitors49–51.
In conclusion, our data collectively implicates UPR in the CRC cell response to KRAS oncogenic signaling loss, highlighting its differential regulation according to the sensitivity of CRC cells. Unlike findings in other cancer types, where UPR activation was associated with acquired resistance, our data suggest that the induction of UPR programs in CRC cells arises early, during the acquisition of tolerance to KRAS inhibition. Further research is needed to elucidate the functional consequences of UPR activation in CRC tolerance and resistance to KRAS inhibition and to inform whether the combined inhibition of UPR and KRAS enhances the anti-tumor responses.
Materials and methods
Cell lines
CRC cell lines HCT116 (RRID: CVCL_0291) and SW480 (RRID: CVCL_0546) were purchased from the American Type Culture Collection (ATCC). LS174T cells were kindly provided by Dr. Ragnhild A. Lothe (Oslo University Hospital, Norway). SW837 cells were kindly provided by Kevin Haigis (Dana-Farber Cancer Institute, USA).
The HCT116 and SW480 cell lines were cultured in RPMI 1640 medium (Gibco, Thermo Fisher Scientific, USA) supplemented with 10% fetal bovine serum (FBS; HyClone, USA) and 1% penicillin–streptomycin (PS; Gibco, Thermo Fisher Scientific, USA). The LS174T and SW837 cell lines were cultured in DMEM supplemented with 10% FBS and 1% PS. All cells were maintained at 37 °C in a humidified atmosphere with 5% CO2.
SiRNA transfection
CRC cell lines were seeded in two-dimensional (2D) conditions in six-well plates, cultured for approximately 16 h, and then transfected using Lipofectamine RNAiMAX (Invitrogen, Thermo Fisher Scientific, USA) in reduced-serum Opti-MEM medium (Gibco, Thermo Fisher Scientific, USA) according to the manufacturer’s guidelines. Gene silencing was performed using ON-TARGETplus SMARTpool small interfering RNA specific for KRAS (siKRAS; L-005069-00-0010) from Dharmacon (USA) at a final concentration of 10 nM. A non-targeting (siControl) siRNA (D-001810-01-50; ON-TARGETplus Non-targeting siRNA #1) was used as a negative control at the same concentration as the siRNA targeting KRAS. Six hours post-transfection, the transfection medium was removed, and the cells were cultured in the respective culture medium.
3D cell culture
Twenty-four hours post-siRNA transfection, the cells were harvested with 0.05% trypsin-EDTA (Gibco, Thermo Fisher Scientific, USA), incubated for 5 min at 37 °C, resuspended in complete medium, and seeded in microwells prepared with micro-molds (3D Petri Dish®, by MicroTissues, Inc., USA). Cell seeding was performed at a density of 1 × 105 cells mL−1, corresponding to 1000 cells per microwell. 3D cultures were incubated for 48 h, after which optical microscopy was used to assess cell growth and spheroid formation. KRAS silencing efficiency was evaluated by Western blotting.
Spheroid characterization
After 48 h of 3D culture, spheroids were imaged using the IN Cell Analyzer 2000. Spheroids’ area, diameter, circularity, and solidity were quantified using Fiji software.
Cell cycle and apoptosis analysis
The cell cycle was assessed using Click-iT Plus EdU Alexa Fluor 647 Flow Cytometry Assay kit (Invitrogen, ThermoFisher Scientific, USA), following the manufacturer’s instructions. Apoptosis was evaluated using annexin V-FITC and PI double staining followed by flow cytometry (BD Pharmingen™ FITC Annexin V Apoptosis Detection Kit), following the protocol provided by the manufacturer. The results were analyzed with FlowJo software version X and are presented as the mean ± standard deviation of three biological replicates of each condition.
Protein extraction and Western blotting
Cells were washed twice with PBS, followed by slight dissociation with Accutase (GRiSP, Portugal) for 30 min at 37 °C. Spheroids were recovered, pelleted, and then resuspended in cell lysis buffer containing 1% IGEPAL CA-630, 1% Triton X-100 supplemented with a protease inhibitor cocktail (Roche, Switzerland), and a phosphatase inhibitor cocktail (Sigma-Aldrich, USA). Cell lysates were further incubated on ice for 30 min and centrifuged for 20 min (13,300 rpm, at 4 °C) to pellet the insoluble material. Protein concentration was determined using a DCProtein assay kit from BioRad (USA). The extracted proteins were stored at − 20 °C until further analysis.
Fifteen µg of total protein were separated via 4–20% sodium dodecyl sulfate-polyacrylamide gel electrophoresis under denaturing conditions. Membranes were cut before antibody incubation. Primary antibodies for KRAS (LS-Bio) and GAPDH (Santa Cruz Biotechnology) were incubated overnight at 4 °C with agitation, followed by 1-h incubation with anti-mouse horseradish peroxidase-conjugated secondary antibodies (Cytiva). The signal was detected with a Bio-Rad ChemiDoc XRS System. Band intensity was quantified using ImageJ.
RNA extraction and sequencing
For the analysis of RNA expression, total RNA extraction was performed using TRIzol™ Reagent, according to the manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA, USA). Briefly, spheroids were removed from the molds, subjected to centrifugation at 1200 rpm for 5 min, and the supernatant was discarded. TRIzol™ Reagent was added to the spheroids, and the samples were homogenized, allowing them to stand for 5 min at room temperature. Subsequently, 1-Bromo-3-chloropropane (BCP) was introduced, and the samples were shaken before standing at RT for an additional 5 min. The samples were then centrifuged at 13,200 rpm for 15 min at 4 °C. Following centrifugation, the aqueous phase was carefully removed and transferred to a fresh tube. Isopropanol was incorporated to precipitate RNA, with the tubes being inverted several times for thorough mixing, followed by a 10-min incubation at RT. After centrifugation at 13,200 rpm for 10 min at 4 °C, the supernatant was meticulously discarded, and the RNA pellet was washed with 75% EtOH. After another centrifugation at 10,000 rpm for 5 min at 4 °C, the supernatant was removed, and the pellet was air-dried for 5 min. The pellet was then solubilized by adding DEPC-treated water and incubating at 60 °C for 10 min. Purification of RNA was accomplished using the RNeasy MinElute Cleanup Kit (Qiagen) in accordance with the manufacturer’s protocol. Library preparation and sequencing was performed by Northwestern University NUSeq Core Facility. Library construction was performed following TruSeq mRNA-Seq library preparation. Illumina HiSeq 4000 Sequencer was used to sequence the libraries with the production of single-end, 50–base pair reads.
RNA sequencing data analysis
The quality of the reads was evaluated using FASTQC with fastq files. Adapters were trimmed and reads of poor quality or aligning to rRNA sequences were filtered out using Trimmomatic. The cleaned reads were aligned to the human genome (hg38) using STAR. Counts and transcripts per million for each condition were estimated from mapped reads using RSEM. All the data were processed using RStudio version 2023.06.0. Differential expression was determined using DESeq2. The cutoff for determining significantly differentially expressed genes was an FDR-adjusted p-value less than 0.05 and log2 fold-change < = − 1 or = > 1.
Gene Set Enrichment Analysis (GSEA) was performed on raw counts using GSEA software (version 4.2.2). To run the software, permutation type set to gene-set, the number of permutations set to 1000, enrichment statistic defined to weighted, metric for ranking genes set to Signal2Noise for experiments with 3 replicates and to log2_Ratio_of_Classes for experiments with 2 replicates. Gene list sorting mode was defined to real, gene list ordering mode descending, max size of gene sets was 1000, and min size of gene sets was 15. The analysis used h.all.v2023.2.Hs.symbols.gmt (Hallmarks).
Cell membrane protein extraction
Enrichment of integral membrane proteins and membrane-associated proteins from cells cultured in 3D was performed using Thermo Scientific™ Mem-PER™ Plus Membrane Protein Extraction Kit. Initially, cells were removed from microwells by lifting the matrix containing spheroids, flipping it upside-down, and centrifuging at 1200 rpm for 5 min. Spheroids were collected by collecting all the media in the well after centrifugation. Spheroids were then centrifuged to obtain a pellet. After pelleting cells, they were incubated for 30 min at 37 °C with TryPLE™ Express to dissociated spheroids. Cells were counted, and 5 × 106 cells from each condition were centrifuged at 300g for 5 min. The pellet was then washed with cell wash solution and centrifuged again at 300g for 5 min. The supernatant was carefully removed and discarded; cells were resuspended in the cell wash solution, transferred to a 2 mL centrifuge tube, and centrifuged at 300g for 5 min. The supernatant was again discarded, and a permeabilization buffer was added to the cell pellet. Samples were vortexed briefly to obtain a homogeneous cell suspension and then incubated for 10 min at 4 °C with constant mixing. Permeabilized cells were centrifuged for 15 min at 16,000g. The supernatant containing cytosolic proteins was transferred to a new tube. The pellet was resuspended with solubilization buffer, and tubes were incubated at 4 °C for 30 min with constant mixing. Afterward, tubes were centrifuged at 16,000g for 15 min at 4 °C, and the supernatant containing solubilized membrane and membrane-associated proteins was transferred to a new tube and stored at − 80 °C for future use.
Mass spectrometry analysis for membrane proteome
Twenty µg of proteins from each sample were used for the proteomic analysis. Samples were analyzed by mass spectrometry-based proteomics using nano-LC-MSMS equipment (Dionex RSLCnano 3000) coupled to an Exploris 480 Orbitrap mass spectrometer (Thermo Scientific, Hemel Hempstead, UK). In brief, samples were loaded onto a custom-made fused capillary pre-column (2 cm length, 360 μm OD, 75 μm ID, flowrate 5 µL per minute for 6 min) packed with ReproSil Pur C18 5.0 μm resin (Dr. Maisch, Ammerbuch-Entringen, Germany), and separated using a capillary column (25 cm length, 360 μm outer diameter, 75 μm inner diameter) packed with ReproSil Pur C18 1.9-µm resin (Dr. Maisch, Ammerbuch-Entringen, Germany) at a flow of 250 nL per minute. A 30 min linear gradient from 87% A (0.1% formic acid) to 34% B (0.1% formic acid in 80% acetonitrile) was applied. Mass spectra were acquired in positive ion mode in a data-dependent manner by switching between one Orbitrap survey MS scan (mass range m/z 400 to m/z 1200) followed by the sequential isolation and higher-energy collision dissociation (HCD) fragmentation and Orbitrap detection of fragment ions of the most intense ions with a cycle time of 2 s between each MS scan. MS and MSMS settings: maximum injection times were set to “Auto”, normalized collision energy was 30%, ion selection threshold for MSMS analysis was 10,000 counts, dynamic exclusion of sequenced ions was set to 30 s, isolation window for MSMS was 0.7 Da, and resolution for MS and MSMS was 60,000 and 15,000.
Data and bioinformatics analysis
The acquired raw data were analyzed using the MaxQuant search engine 1.5.3.3052 for protein identification and label-free quantification (LFQ) to estimate iBAQ intensity analysis. The data were searched against the UniProt human database (UP000005640, downloaded October 2020). The search parameters were set with the following conditions: two missed cleavage sites for trypsin/P, carbamidomethylation of cysteine as fixed modification, oxidation of methionine and acetylation of N-terminal residue as variable modifications, and precursor ion mass tolerance of 20 ppm. The false discovery rate (FDR) of peptides and proteins was set to 1%.
Software Perseus 1.6.14.053 was used for data processing and statistical analysis. Reverse, contaminant, and only identified by site proteins were excluded, and iBAQ values were log2 transformed. Missing values were imputed by replacing missing values with a small constant (= 1). All datasets were subjected to a student’s t-test and p-values less than 0.05 with fold-change higher than 2 were considered statistically significant.
Gene Ontology (GO) pathway analysis was performed using DAVID Bioinformatics (31). For these analyses, a p-value cutoff of 0.05 was applied. All significant pathways were extracted from the GO category, but only the top 5 most significant terms are represented in the graphs.
Protein aggregation detection assay
The PROTEOSTAT Aggresome detection kit (Enzo Life Sciences, ENZ-51035-0025) was used to detect misfolded or aggregated proteins in cells. The PROTEOSTAT aggresome detection assay was performed according to the manufacturer’s instructions. Briefly, spheroids were collected, dissociated for 30 min at 37 °C with TrypLE and then centrifuged at 1200 rpm for 5 min to pellet the cells. Cells were stained with viability dye (1:5000) for 15 min at RT in the dark and then washed with PBS. After, cells were fixed with 4% formaldehyde for 30 min at RT, permeabilized with Permeabilizing Solution (0.5% Triton X-100, 3mM EDTA) for 30 min on ice, and stained with the PROTEOSTAT dye (1:2500 dilution) for 30 min at RT. Cells treated with 5 µM MG132 (provided in the PROTEOSTAT Aggresome detection kit) for 16 h was used as positive control.
Samples were acquired in FACSCanto II cytometer (BD Biosciences). Results were analyzed in FlowJo software version X and are represented by the mean ± standard deviation of three biological replicates of each condition. PROTEOSTAT signal was analyzed considering only live cell population obtained by the viability dye staining. Aggresome propensity factor (APF) was calculated using the mean fluorescence intensity (MFI) values according with the following formula:
 |
1 |
SUrface sensing of translation (SunSET) assay
Protein synthesis was monitored using puromycin labeling and performed as previously described54. Spheroids were incubated with puromycin (Sigma-Aldrich) at 10 µg/ml for 30 min at 37 °C and 5% CO2. After puromycin incorporation the spheroids were washed with cold PBS and incubated for 30 min at 4 °C in the dark with violet fluorescent live/dead fixable cell stain (Invitrogen, Thermo Fisher Scientific, USA) to allow dead cell exclusion. Cells were then fixed in 4% paraformaldehyde for 15 min at RT. For cell permeabilization, cells were incubated for 15 min in FACS buffer (0.5% BSA (NZYTech), 0.01% sodium azide (Sigma-Aldrich), 0.1% saponin (Alfa-Aesar), PBS) at 4 °C. Cells were then stained with a mouse monoclonal anti-puromycin-AF647 (Clone 12D10; Merck Millipore, Burlington, MA, USA) at 4 °C. After 1 h of incubation, cells were washed, resuspended in PBS and dissociated/filtered using a 40 μm cell strainer. Samples were analyzed in a FACSCanto II cytometer (BD Biosciences). Results were analyzed in FlowJo software version.
Combination of KRAS silencing with IRE1α Inhibition
KRAS silencing was performed by siRNA transfection as described above. Six hours post-transfection, the medium was replaced by regular cell culture medium supplemented with 12.2 µM of IRE1α inhibitor 4µ8C (Calbiochem, San Diego, USA). Twenty-four hours post-transfection, cells were seeded in the 3D spheroid molds and kept for an additional 48 h in culture in regular cell culture medium supplemented with of 4µ8C (12,2 µM). Apoptosis and cell cycle analysis were performed 72 h post-transfection (48 h post-3D seeding) following the protocols described above. IRE1α activity inhibition was confirmed by evaluating XBP1 splicing expression levels using the primers Fw: 5′-CTGAGTCCGCAGCAGGTG-3′ and Rv: 5′-CTCTGGGGAAGGGCATTTGA-3′ (Sigma Aldrich, Germany).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors acknowledge the support of Catarina Meireles and Emilia Cardoso from the Translational Cytometry unit (TraCy) i3S Scientific Platform. We acknowledge the COST Action CA20113 “PROTEOCURE” supported by COST (European Cooperation in Science and Technology).
Author contributions
Study conceptualization: FM, RM, VB and SV; Data acquisition: FM, ALM, JC, ASC, and SV; Data analysis: FM, ALM, SV, HCB and RM; Supervision: RM, VB and SV; Funding acquisition: VB, SV, JC and RM; Project administration: RM, VB, and SV; Resources: VB, SV, HCB, RM, and CA; Manuscript writing: FM (original draft) and FM, ALM, JC, CA, HCB, ASC, RM, VB and SV (review and editing).
Funding
The work was funded by internal grants (MSI and Cancer Challenge 2022) provided by the Institute for Molecular Pathology and Immunology of the University of Porto (IPATIMUP) and by national funds through FCT in the scope of the project 2022.05346.PTDC (10.54499/2022.05346.PTDC). This article is also a result of the projects (iNOVA4Health–UIDB/04462/2020 and UIDP/04462/2020, and by the Associated Laboratory LS4FUTURE (LA/P/0087/2020), two programs financially supported by Fundação para a Ciência e Tecnologia / Ministério da Ciência, Tecnologia e Ensino Superior. FM is a Ph.D. student in the Doctoral Program in Biomedicine at the Faculty of Medicine of the University of Porto, and she is funded through a Ph.D. fellowship (SFRH/BD/143669/2019) awarded by the Portuguese Foundation for Science and Technology (FCT). ALM is a Ph.D. student in the Doctoral Program in Biomedicine from the Faculty of Medicine of the University of Porto, and she is funded through a Ph.D. fellowship (2020.08932.BD) awarded by the FCT. JC is hired by IPATIMUP under norma transitória do DL n.° 57/2016 alterada pela lei n.° 57/2017 (10.54499/DL57/2016/CP1363/CT0012). CA was financially supported by national funds (OE), through Fundação para a Ciência e a Tecnologia (FCT)/MCTES, within the scope of iBiMED—Institute of Biomedicine (10.54499/UIDP/04501/2020). ASC is supported by Fundação para a Ciência e a Tecnologia (10.54499/DL57/2016/CP1457/CT0013). VB was supported by NIH grants R01CA228272, R02CA225002, U54CA268084, and U54CA261694, NSF grant EFMA1830961, the Center for Physical Genomics and Engineering at Northwestern University, and philanthropic support from K. Hudson and R. Goldman, S. Brice and J. Esteve, M. E. Holliday and I. Schneider, the Christina Carinato Charitable Foundation, and D. Sachs. RM is supported by Fundação para a Ciência e a Tecnologia (CEEC position, 10.54499/CEECIND/03906/2017/CP1421/CT0004). SV is supported by FCT (10.54499/2021.01550.CEECIND/CP1663/CT0012).
Data availability
RNA-Seq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE254832, GSE254833, and GSE283752 are available at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE254832, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE254833, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE283752. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE50 partner repository55 with the dataset identifier PXD059013 and 10.6019/PXD059013 (Reviewer access details: Log in to the PRIDE website using the following details: Project accession: PXD059013; Token: mWuPTfVRoRJw. Alternatively, reviewer can access the dataset by logging in to the PRIDE website using the following account details: Username: reviewer_pxd059013@ebi.ac.uk; Password: 5T3sqfr623Xs).The remaining data generated in this work is contained within the article or supplementary material.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Capon, D. J. et al. Activation of Ki-ras2 gene in human colon and lung carcinomas by two different point mutations. Nature304, 507–513 (1983). [DOI] [PubMed] [Google Scholar]
- 2.Shimizu, K. et al. Structure of the Ki-ras gene of the human lung carcinoma cell line Calu-1. Nature304, 497–500 (1983). [DOI] [PubMed] [Google Scholar]
- 3.Pulciani, S. et al. Oncogenes in solid human tumours. Nature300, 539–542 (1982). [DOI] [PubMed] [Google Scholar]
- 4.Der, C. J., Krontiris, T. G. & Cooper, G. M. Transforming genes of human bladder and lung carcinoma cell lines are homologous to the ras genes of Harvey and Kirsten sarcoma viruses. Proc. Natl. Acad. Sci. 79, 3637–3640 (1982). [DOI] [PMC free article] [PubMed]
- 5.Chhichholiya, Y., Singh, H. V., Vashistha, R., Singh, S. & Munshi, A. Deciphering the role of KRAS gene in oncogenesis: focus on signaling pathways, genetic alterations in 3′UTR, KRAS specific MiRNAs and therapeutic interventions. Crit. Rev. Oncol. Hematol.194, 104250 (2024). [DOI] [PubMed] [Google Scholar]
- 6.Haigis, K. M. KRAS alleles: The devil is in the detail. Trends Cancer 3, 686–697 (2017). [DOI] [PMC free article] [PubMed]
- 7.Pylayeva-Gupta, Y. & Grabocka, E. Bar-Sagi, D. RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer11, 761–774 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dias Carvalho, P. et al. Mutant KRAS modulates colorectal cancer cells invasive response to fibroblast-secreted factors through the HGF/C-MET axis. Int. J. Cancer. 151, 1810–1823 (2022). [DOI] [PubMed] [Google Scholar]
- 9.Velho, S. & Haigis, K. M. Regulation of homeostasis and oncogenesis in the intestinal epithelium by Ras. Exp. Cell. Res.317, 2732 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamarsheh, S., Groß, O., Brummer, T. & Zeiser, R. Immune modulatory effects of oncogenic KRAS in cancer. Nat. Commun.11, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carvalho, P. D., Machado, A. L., Martins, F., Seruca, R. & Velho, S. Targeting the tumor microenvironment: an unexplored strategy for mutant KRAS tumors. Cancers (Basel) 11 (2019). [DOI] [PMC free article] [PubMed]
- 12.Carvalho, P. D. et al. KRAS oncogenic signaling extends beyond cancer cells to orchestrate the microenvironment. Cancer Res.78, 7–14 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Huang, L., Guo, Z., Wang, F. & Fu, L. KRAS mutation: from undruggable to druggable in cancer. Signal. Transduct. Target. Ther.6, 1–20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haidar, M. & Jacquemin, P. Past and future strategies to inhibit membrane localization of the KRAS oncogene. Int. J. Mol. Sci.22, 22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merz, V. et al. Targeting KRAS: the elephant in the room of epithelial cancers. Front. Oncol.11 (2021). [DOI] [PMC free article] [PubMed]
- 16.Zhang, Z., Guiley, K. Z. & Shokat, K. M. Chemical acylation of an acquired Serine suppresses oncogenic signaling of K-Ras (G12S). Nat. Chem. Biol.18, 1177–1183 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ostrem, J. M. et al. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature503, 548–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakajima, E. C. et al. FDA approval summary: Sotorasib for KRAS G12C mutated metastatic NSCLC. Clin. Cancer Res.28, 1482 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong, D. S. et al. KRASG12C inhibition with sotorasib in advanced solid tumors. N. Engl. J. Med.383, 1207–1217 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu, G., Pei, L., Xia, H., Tang, Q. & Bi, F. Role of oncogenic KRAS in the prognosis, diagnosis and treatment of colorectal cancer. Mol. Cancer. 20, 1–17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neumann, J., Zeindl-Eberhart, E., Kirchner, T. & Jung, A. Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol. Res. Pract.205, 858–862 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Oya, Y., Imaizumi, K. & Mitsudomi, T. The next-generation KRAS inhibitors… What comes after sotorasib and adagrasib? Lung Cancer 194 (2024). [DOI] [PubMed]
- 23.Johnson, M. L. et al. KRYSTAL-1: activity and safety of adagrasib (MRTX849) in patients with colorectal cancer (CRC) and other solid tumors harboring a KRAS G12C mutation. Eur. J. Cancer138, S2 (2020). [Google Scholar]
- 24.Fakih, M. G. et al. Sotorasib for previously treated colorectal cancers with KRASG12C mutation (CodeBreaK100): A prespecified analysis of a single-arm, phase 2 trial. Lancet Oncol.23, 115–124 (2022). [DOI] [PubMed] [Google Scholar]
- 25.Ciardiello, D., Maiorano, B. A. & Martinelli, E. Targeting KRASG12C in colorectal cancer: the beginning of a new era. ESMO Open8 (2023). [DOI] [PMC free article] [PubMed]
- 26.Rona, Y. et al. Adagrasib with or without cetuximab in colorectal cancer with mutated KRAS G12C. N. Engl. J. Med.388, 44–54 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh, A. et al. TAK1 Inhibition promotes apoptosis in KRAS-dependent colon cancers. Cell148, 639–650 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peirsman, A. et al. MISpheroID: A knowledgebase and transparency tool for minimum information in spheroid identity. Nat. Methods18, 1294–1303 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hetz, C., Zhang, K. & Kaufman, R. J. Mechanism, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell. Biol.21, 421 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lv, X. et al. Modulation of the proteostasis network promotes tumor resistance to oncogenic KRAS inhibitors. Science381, eabn4180 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siwecka, N. et al. Dual role of Endoplasmic reticulum stress-mediated unfolded protein response signaling pathway in carcinogenesis. Int. J. Mol. Sci.20 (2019). [DOI] [PMC free article] [PubMed]
- 32.Gorman, A. M., Healy, S. J. M., Jäger, R. & Samali, A. Stress management at the ER: regulators of ER stress-induced apoptosis. Pharmacol. Ther.134, 306–316 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Wen, J. H. et al. Cellular protein aggregates: formation, biological effects, and ways of elimination. Int. J. Mol. Sci.24 (2023). [DOI] [PMC free article] [PubMed]
- 34.Francisco, S., Ferreira, M., Moura, G., Soares, A. R. & Santos, M. A. Does proteostasis get lost in translation? Implications for protein aggregation across the lifespan. Ageing Res. Rev.62, 101119 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Chen, G., Wei, T., Ju, F. & Li, H. Protein quality control and aggregation in the Endoplasmic reticulum: from basic to bedside. Front. Cell. Dev. Biol.11 (2023). [DOI] [PMC free article] [PubMed]
- 36.Lv, X. et al. Modulation of the proteostasis network promotes tumor resistance to oncogenic KRAS inhibitors. Science (1979)381, eabn4180 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, R. F. et al. RasGRF couples Nox4-dependent Endoplasmic reticulum signaling to Ras. Arterioscler. Thromb. Vasc Biol.37, 98–107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh, A. et al. A gene expression signature associated with ‘K-Ras addiction’ reveals regulators of EMT and tumor cell survival. Cancer Cell15, 489–500 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliveira, S. M. et al. Fibroblasts promote resistance to KRAS Silencing in colorectal cancer cells. Cancers (Basel) 16 (2024). [DOI] [PMC free article] [PubMed]
- 40.Chevet, E., Hetz, C. & Samali, A. Endoplasmic reticulum stress-activated cell reprogramming in oncogenesis. Cancer Discov. 5, 586–597 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Martins, F. et al. KRAS Silencing impacts chromatin organization and transcriptional activity in colorectal cancer cells. Res. Sq. 10.21203/rs.3.rs-3752760/v2 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Granados, D. P. et al. ER stress affects processing of MHC class I-associated peptides. BMC Immunol.10, 10 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mercier, R. & LaPointe, P. The role of cellular proteostasis in antitumor immunity. J. Biol. Chem.298, 101930 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urra, H., Aravena, R., González-Johnson, L. & Hetz, C. The uprising connection between Endoplasmic reticulum stress and the tumor microenvironment. Trends Cancer. 10.1016/j.trecan.2024.09.011 (2024). [DOI] [PubMed] [Google Scholar]
- 45.Cubillos-Ruiz, J. R., Bettigole, S. E. & Glimcher, L. H. Tumorigenic and immunosuppressive effects of endoplasmic reticulum stress in cancer. Cell168, 692–706 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Izadpanah, A., Willingham, K., Chandrasekar, B., Alt, E. U. & Izadpanah, R. Unfolded protein response and angiogenesis in malignancies. Biochim. Biophys. Acta (BBA) Rev. Cancer1878, 188839 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen, X. & Cubillos-Ruiz, J. R. Endoplasmic reticulum stress signals in the tumour and its microenvironment. Nat. Rev. Cancer. 21, 71–88 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu, L. et al. 143D, a novel selective KRASG12C inhibitor exhibits potent antitumor activity in preclinical models. Acta Pharmacol. Sin. 44, 1475–1486 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liao, W. et al. KRAS-IRF2 axis drives immune suppression and immune therapy resistance in colorectal cancer. Cancer Cell35, 559–572e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mugarza, E. et al. Therapeutic KRAS G12C Inhibition drives effective interferon-mediated antitumor immunity in Immunogenic lung cancers. Sci. Adv. 8 (2022). [DOI] [PMC free article] [PubMed]
- 51.Hallin, J. et al. The KRASG12C inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov. 10, 54–71 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol.26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Tyanova, S. et al. The perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Schmidt, E. K., Clavarino, G., Ceppi, M. & Pierre, P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods. 6, 275–277 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Perez-Riverol, Y. et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res.50, D543–D552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE254832, GSE254833, and GSE283752 are available at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE254832, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE254833, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE283752. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE50 partner repository55 with the dataset identifier PXD059013 and 10.6019/PXD059013 (Reviewer access details: Log in to the PRIDE website using the following details: Project accession: PXD059013; Token: mWuPTfVRoRJw. Alternatively, reviewer can access the dataset by logging in to the PRIDE website using the following account details: Username: reviewer_pxd059013@ebi.ac.uk; Password: 5T3sqfr623Xs).The remaining data generated in this work is contained within the article or supplementary material.