Abstract
The COBAS Amplicor Hepatitis C Virus (HCV) Monitor assay, version 2.0, which reports in international units per milliliter, was compared to the assay reported in copies per milliliter by analyzing dilution series and clinical plasma samples by both methods. In addition, the Amplicor international unit assay was compared to the National Genetics Institute HCV Superquant assay. The dilution series ranged from <100 to 5,000,000 HCV RNA copies/ml and consisted of 32 points, assayed in triplicate in each assay. Thirty clinical samples ranging from 1,000 to 1,000,000 HCV RNA copies/ml were assayed in duplicate. Deming regression analysis comparing the Amplicor HCV RNA international units-per-milliliter and copies-per-milliliter assays was calculated as follows: (Amplicor international units per milliliter) = 1.030(Amplicor copies per milliliter) − 0.392; R2 = 0.981; n = 28; Sy/x (standard error of the estimate) = 0.129. The linearity of the Amplicor international units-per-milliliter assay was as follows: observed = 0.886(expected) + 0.437; R2 = 0.983; n = 30. The linearity of the Superquant assay was as follows: observed= 0.918 (expected) + 0.436; R2 = 0.986; n = 32. Deming regression analysis comparing the Amplicor and Superquant assays was calculated as follows: Superquant = 1.066(Amplicor) − 0.0197; R2 = 0.908; Sy/x = 0.308; n = 28. The Amplicor and Superquant assays were linear through the range of 600 to 600,000 IU of HCV RNA/ml and ∼300 to 5,000,000 HCV RNA copies/ml, respectively. The narrow range of the Amplicor assay means that some samples will require dilution and retesting for accurate quantification above 600,000 IU of HCV RNA/ml. The Amplicor and Superquant assays agreed well within the range of 600 to 600,000 IU of HCV RNA/ml (∼1,000 to ∼1,000,000 HCV RNA copies/ml). Overall, the Amplicor and Superquant assays agree well, and results obtained in one assay could be expected to compare well with results from the other when reported in copies per milliliter.
Current treatments for hepatitis C virus (HCV) infection are limited to alfa interferon alone or in combination therapy with ribavirin for either 24 or 48 weeks (11). Quantitation of HCV RNA in patient samples is becoming a valuable tool in the management of patients undergoing treatment. Indications for test ordering discussed in the literature are used to help predict the likelihood of response to therapy and as a monitor of therapy (2). Both uses are controversial, and interpretation of results and algorithms for clinical decision making are far from standardized. The use of HCV RNA quantitative (viral-load) testing for any patient monitoring is complicated by the fact that several tests are available and none have been thoroughly standardized.
At present, there are several commercially available methods used to measure HCV RNA levels in patients, including the Versant HCV RNA (bDNA) (Bayer Diagnostics, Tarrytown, N.Y.) and COBAS Amplicor HCV Monitor (Roche Diagnostics Corporation, Indianapolis, Ind.) tests, in addition to proprietary, in-house-developed modalities, such as the HCV Superquant (National Genetics Institute [NGI], Los Angeles, Calif.). Since these assays were developed without reference to widely accepted external material, results reported by each method have been shown not to be equivalent (6, 7, 10, 14). Most strikingly, the form of testing recently used by many investigators to evaluate patients undergoing interferon-ribavirin combination therapy (1, 11, 13), the Superquant assay, only available through NGI, is not accessible to other laboratories and has only recently been evaluated and compared to the most commonly used assay, the Amplicor HCV Monitor assay (12). One early study comparing these assays concluded that the Amplicor HCV Monitor version 1.0 and HCV Superquant assays were not interchangeable for monitoring individual patients over time (14), while a recent study (12) found that the results from the COBAS Amplicor HCV Monitor version 2.0 and HCV Superquant assays were interchangeable. The three therapeutic studies previously mentioned used the Superquant assay and presented outcomes for patients based on a cutoff for HCV RNA levels greater than or less than 2,000,000 HCV RNA copies/ml. It is unclear if the treatment recommendations established using one assay method can be extended to all assay methods. Standardization is vital to provide reliable information for good patient management using any commercially available kit or method.
With the introduction of the first World Health Organization (WHO) Nucleic Acid Testing International Standard for HCV RNA (15), testing can be standardized between methods and results from one method should be comparable with results obtained from a different method. The availability of the HCV RNA international unit standard will provide a useful tool for better addressing interassay reproducibility and assessment of the value of HCV RNA quantitative testing for patients undergoing therapy.
MATERIALS AND METHODS
Samples.
The samples used in this study were samples submitted to ARUP Laboratories for HCV RNA testing. Serum or EDTA plasma was centrifuged, separated from cells, and frozen at −20°C within 30 min of collection.
Quantitation by COBAS Amplicor HCV Monitor.
The COBAS Amplicor HCV Monitor version 2.0 test utilized a reverse transcription (RT)-PCR approach to quantitation of HCV RNA in patient samples. All amplification and detection steps were performed by the automated COBAS Amplicor instrument. Patient samples were extracted, amplified, and detected as described previously (3, 4, 8). This assay was recently released in a format in which HCV RNA levels are reported in international units as defined by the WHO consensus on nucleic acid testing. Results from the Amplicor assays were reported in HCV RNA copies per milliliter or international units of HCV RNA per milliliter.
Quantitation by NGI HCV Superquant.
The HCV Superquant assay is based on a multicycle RT-PCR method, with an internal control and Southern blot detection for the quantification of HCV RNA. HCV RNA was extracted from the specimens, and a cDNA template was produced. The cDNA was then amplified in four separate PCRs with 25, 30, 35, and 45 cycles. The PCR products were visualized using a Southern blot hybridization, and quantitation was determined through densitometry measurements comparing the wild-type HCV product band intensity to the internal-control product band intensity (17, 18). The Superquant assay utilized robotic instruments for agarose gel electrophoresis, vacuum transfer Southern blot hybridization, and immunostaining (14). Results from the NGI HCV Superquant were reported in HCV RNA copies per milliliter.
Clinical comparisons.
Thirty clinical samples with HCV RNA levels which measured in the COBAS Amplicor HCV Monitor version 2.0 copies-per-milliliter assay between 1,000 and 1,000,000 HCV RNA copies/ml were split into aliquots, stored at −70°C, and submitted to the HCV Superquant, COBAS Amplicor HCV Monitor version 2.0 (copies-per-milliliter), and COBAS Amplicor HCV Monitor version 2.0 (international units-per-milliliter) assays in duplicate. The aliquots of the duplicate specimens were submitted blindly as patient samples in separate weeks. Correlation was determined between the copies- and international units-per-milliliter versions of the Amplicor assays and between the Amplicor international units-per-milliliter assay and the Superquant assay.
Dilution studies.
Four samples containing HCV RNA levels of >1,000,000 HCV RNA copies/ml as measured in the COBAS Amplicor HCV Monitor test version 2.0 were serially diluted in HCV-seronegative defibrinated, delipidated normal human serum (Western States Plasma, Oceanside, Calif.) to obtain a range of HCV RNA levels from <100 to 10,000 HCV RNA copies/ml. An additional four samples with HCV RNA levels of >5,000,000 copies/ml, determined through dilution, were serially diluted in HCV-seronegative normal human serum to obtain a range of HCV RNA levels from 100,000 to 5,000,000 HCV RNA copies/ml. All dilution samples were submitted to the HCV Superquant, COBAS Amplicor HCV Monitor version 2.0 (copies-per-milliliter), and COBAS Amplicor HCV Monitor version 2.0 (international units-per-milliliter) assays in triplicate on the same day.
In addition, 192 samples submitted for HCV RNA quantitative testing and reported as >1,000,000 HCV RNA copies/ml (converted from HCV RNA IU/ml) in the Amplicor international unit-per-milliliter assay were selected. These samples were diluted 1:50 in HCV-seronegative defibrinated, delipidated normal human serum and assayed again. Diluted and undiluted results were compared as to fold difference after the effects of dilution were taken into account.
Lower limit of detection.
Samples with a low HCV RNA concentration were used to examine the lower limits of linearity and lower limits of detection of the Amplicor and Superquant assays. Further evaluation of the lower limit of detection was done by comparison of results from the Amplicor and Superquant assays to those from sensitive qualitative assays. Eight samples reported as less than the lower detection limit but with high background (absorbance > 0.010) in the COBAS Amplicor HCV qualitative assay, were chosen as suspected of containing low levels of HCV RNA. In addition, 23 samples reported as <1,000 HCV RNA copies/ml in the COBAS Amplicor HCV Monitor version 2.0 assay were randomly selected. Samples were split into five aliquots and stored at −70°C. The aliquots were submitted blindly as patient samples to the following assays in singlet: COBAS Amplicor HCV Monitor version 2.0 international unit-per-milliliter assay, COBAS Amplicor HCV Monitor version 2.0 copies-per-milliliter assay, NGI HCV Superquant assay, COBAS Amplicor HCV qualitative assay, and the VERSANT HCV RNA qualitative assay by transcription-mediated amplification (TMA) (Analyte-specific reagent) (Bayer Diagnostics). Samples which were reactive by the TMA assay and were not HCV RNA positive in another assay were repeated in the TMA assay to confirm the initial results. Samples were considered to be positive for HCV RNA if the TMA assay was repeatedly reactive, due to its reported high sensitivity (16).
Precision.
Within-run precision was calculated from the triplicate measurements obtained in the dilution studies and was reported as the standard deviation of log10 HCV RNA copies per milliliter or log10 international units of HCV RNA per milliliter. Between-run precision was determined from the duplicate samples used in the correlation studies by analyzing the values for the duplicate determinations of the quantitated samples and comparing the standard deviations of the differences of the duplicates. The between-run precision levels of the Amplicor and Superquant assays were compared through analysis by an F test of log10 HCV RNA copies per milliliter or log10 international units of HCV RNA per milliliter.
Data analysis.
Data were collected on standard report forms, and results were entered into a central database. After being entered into the database, the blinded results were reviewed for data entry errors. The results were converted to log10 units for linearity, comparison, and sensitivity analysis. Observed-versus-expected graphs were analyzed through linear regressions. Correlation between clinical samples was calculated through the use of Deming regression.
RESULTS
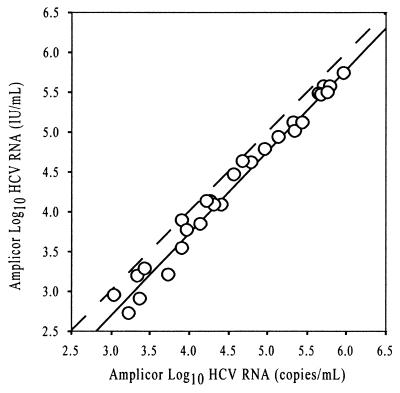
Thirty clinical samples were measured in duplicate in both the Amplicor HCV Monitor version 2.0 copies and international units-per-milliliter assays. Results were obtained for 28 of the 30 samples. Two samples were below the detection limit of the Amplicor assay. The means of the duplicate measurements were used in the calculation of a Deming regression comparing log10 international units of HCV RNA per milliliter to log10 HCV RNA copies per milliliter (the regression analysis is presented in Fig. 1). The analysis yielded the following equation: log international units per milliliter = 1.030(log10 copies per milliliter) − 0.392; R2 = 0.981; Sy/x (standard error of the estimate) = 0.129; n = 28. A high degree of concordance is expected for this comparison because the calibrations for the two assays are linked. These regression results indicate that the two forms of the assay correlate over the range of 600 to 600,000 IU of HCV RNA/ml (∼1,000 to ∼1,000,000 HCV RNA copies/ml) or 2.8 to 5.8 log10 IU of HCV RNA/ml (3.0 to 6.0 log10 HCV RNA copies/ml). The numerical HCV RNA copy-per-milliliter value of a given sample is approximately two times higher than the international units of HCV RNA per-milliliter value.
FIG. 1.
Deming regression comparing log10 international units of HCV RNA per milliliter to log10 HCV RNA copies per milliliter, in the COBAS Amplicor HCV Monitor assays. The means of duplicate measurements are plotted. The equation for the Deming regression is international units = 1.030(copies) − 0.392; R2 = 0.981; Sy/x = 0.129; n = 28. The dashed line represents unity.
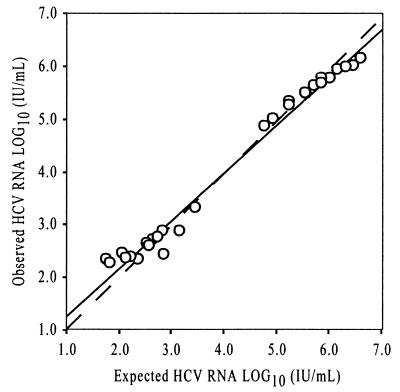
Eight samples were each serially diluted in five dilution steps and assayed in triplicate in the COBAS Amplicor HCV Monitor version 2.0 international units-per-milliliter assay to determine the linearity of the assay across a wide range of HCV RNA levels. The observed versus expected results are presented in Fig. 2. The linear regression equation generated from the dilution study for the assay was as follows: observed = 0.886(expected) + 0.437; R2 = 0.983; n = 30. The linearity of the Amplicor international units-per-milliliter assay is compromised above 5.8 log10 IU of HCV RNA/ml (600,000 IU of HCV RNA/ml) as shown by the decline in observed values above this level, but the assay appears to be sensitive and accurate to the claimed lower limit of 2.8 log10 IU of HCV RNA/ml (600 IU of HCV RNA/ml). From these observations, we conclude that the linear range of the Amplicor international unit assay is 600 to ∼600,000 IU/ml.
FIG. 2.
Observed versus expected for a dilution series measured in the COBAS Amplicor HCV Monitor version 2.0 international unit assay. The means of triplicate values are calculated and plotted as log10 international units of HCV RNA per milliliter. The equation for the linear regression analysis is as follows: observed = 0.886(expected) + 0.437; R2 = 0.983; n = 30. The dashed line represents unity.
One hundred ninety-two clinical samples with results of >1,000,000 HCV RNA copies/ml (converted from HCV RNA IU/ml) in the Amplicor HCV Monitor version 2.0 international units-per-milliliter assay were diluted 1:50 and reassayed to determine the accuracy of the initial HCV RNA quantitation. Table 1 presents the results of the samples assayed both diluted and undiluted in the Amplicor international units-per-milliliter assay. Diluted and undiluted results were compared as to fold difference after the effects of dilution were taken into account. These data show that the reliability of results of >600,000 IU of HCV RNA/ml is compromised, as demonstrated by the high proportion (53%) of samples with undiluted HCV RNA levels between 600,000 and 850,000 HCV RNA copies/ml which had 0.5-log10 unit differences when measured undiluted versus diluted. In comparison, in the range of 300,000 to 600,000 IU of HCV RNA/ml, the proportion of samples with undiluted-versus-diluted differences exceeding 0.5 log10 units was 31%. These data suggest that, when tested undiluted, as much as 53% of samples with results of 600,000 IU of HCV RNA/ml may actually contain HCV RNA levels as high as 1,300,000 IU/ml. This would place these patients in a completely different treatment category according to recent recommendations (12). These data indicate that the COBAS Amplicor HCV Monitor version 2.0 international unit assay cannot accurately quantitate above 600,000 IU of HCV RNA/ml, and some samples below this level may be underquantitated as well.
TABLE 1.
Quantitative differences in samples of >300,000 IU/ml assayed diluted and undiluted in the COBAS Amplicor HCV Monitor version 2.0
| HCV RNA levela (IU/ml) | % (no./total) of samples with quantitative differencesb of:
|
||
|---|---|---|---|
| >0.5 log unit | >0.75 log unit | >1.0 log unit | |
| 300,000-600,000 | 31 (15/48) | 8 (4/48) | 0 (0/48) |
| 600,000-850,000 | 53 (18/34) | 18 (6/34) | 12 (4/34) |
| 850,000-12,000,000 | 56 (62/110) | 15 (17/110) | 3 (3/110) |
All measurements were performed using the COBAS Amplicor HCV Monitor version 2.0 assay. Samples were assayed undiluted and diluted 1:50.
log10 international units per milliliter differences determined as log10 international units per milliliter diluted minus log10 international units per milliliter undiluted.
Data from triplicate determinations of eight samples serially diluted and assayed in the COBAS Amplicor HCV Monitor version 2.0 international units-per-milliliter and HCV Superquant assays were used to determine the within-run precision of the assays. The within-run precision of the Amplicor assay, as measured by standard deviation of log10 international units of HCV RNA per milliliter, ranged from 0.004 to 0.388 and from 0.009 to 0.156 log10 HCV RNA copies/ml in the Superquant assay. Higher variations in the standard deviations of the log10 HCV RNA levels were observed near the lower limit of detection of the Amplicor assay. This is expected as the HCV RNA values approach the limit of detection of the assay, but overall, the reproducibility of this assay is comparable to that of the copy/milliliter version of the assay (4). The between-run precision of the assay was determined by analyzing the standard deviation of the differences of the log10 international units of HCV RNA per milliliter values for the duplicate determinations of 28 and 30 samples for the Amplicor and Superquant assays respectively. The standard deviations of the differences of the duplicates were 0.259 log10 IU of HCV RNA/ml in the Amplicor assay and 0.160 log10 HCV RNA copies/ml in the Superquant assay. Overall, the Amplicor and Superquant assays appear to perform well with respect to both within-run and between-run precision. Good precision should be expected from an assay method such as that of the Superquant assay, which uses mostly automated procedures. Amplicor and Superquant between-run precision levels were compared using an F test on the standard deviation of the duplicate log10 determinations for each assay. The F test indicated that the Superquant (standard deviation = 0.160 log10 units) performed significantly better with regard to between-run precision (P = 0.006) than the Amplicor (standard deviation = 0.259 log10 unit) assay.
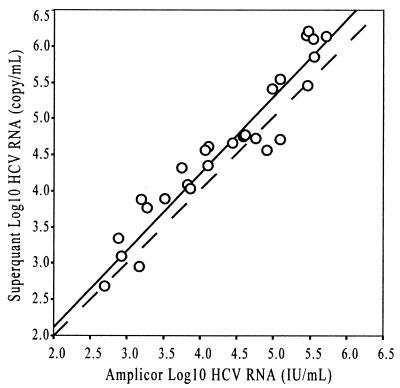
Comparison of the COBAS Amplicor HCV Monitor version 2.0 (international units per milliliter) and HCV Superquant assays was made by testing 30 clinical samples in duplicate in both assays. The Superquant assay reported results in HCV RNA copies/ml, while the Amplicor assay reported in international units of HCV RNA per milliliter. Twenty-eight specimens had results within the dynamic ranges of both assays. The means of the duplicate measurements were used in the analysis presented in Fig. 3. The Deming regression yielded the following equation: Superquant (log10 copies) = 1.066[Amplicor(log10 IU)] − 0.0197; R2 = 0.908; Sy/x = 0.308; n = 28. The Amplicor and Superquant assay results correlated within the range of 600 to 600,000 IU of HCV RNA/ml (∼1,000 to 1,000,000 HCV RNA copies/ml); however, the numerical values for Superquant HCV RNA copies per milliliter averaged ∼2 times greater than the numerical values for the Amplicor international units of HCV RNA per milliliter. This is similar to the proportional relationship between HCV RNA copies per milliliter and international units per milliliter observed in the two Amplicor assays. The Superquant results agreed with the Amplicor results expressed as copies per milliliter. The Deming regression was as follows: log10 Superquant copies per milliliter = 1.073(log10 Amplicor copies per milliliter) − 0.315; Sy/x = 0.293; R2 = 0.926; n = 30. The Superquant results agreed with the Amplicor results expressed as copies per milliliter, converted from international units of HCV RNA per milliliter. The Deming regression was as follows: log10 Superquant copies per milliliter = 1.080(log10 Amplicor copies per milliliter, converted from international units per milliliter) − 0.392; Sy/x = 0.297; R2 = 0.914; n = 28. These data show that there is a proportional relationship between Amplicor international units per milliliter and Superquant copies per milliliter, and therefore, results obtained in the Amplicor assay can be used to monitor patients according to the recommendations made in clinical trials using the Superquant assay. It is important to note that the definition of an HCV RNA copy has not been officially characterized, and as such, the conversion from HCV RNA international units will vary from assay to assay depending on how the manufacturer of a given test defines an HCV RNA copy and the corresponding relationship to international units of HCV RNA. Therefore, it is not possible to directly convert international units of HCV RNA per milliliter to HCV RNA copies per milliliter for all assays or to define how many copies are in 1 IU because HCV RNA copies per milliliter have not been standardized.
FIG. 3.
Deming regression comparing log10 international units of HCV RNA obtained from the COBAS Amplicor HCV Monitor version 2.0 international unit assay and the log10 HCV RNA copies per milliliter from the Superquant assay. The means of the duplicate HCV RNA measurements are plotted. The equation for the Deming regression is as follows: Superquant = 1.0659(Amplicor) − 0.0197; R2 = 0.908; Sy/x = 0.308; n = 28. The dashed line represents unity.
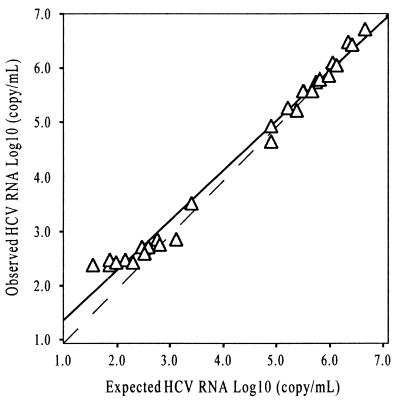
To investigate the linearity of the HCV Superquant assay, eight samples were serially diluted and assayed in triplicate across a wide range of HCV RNA levels. A plot of observed versus expected log10 HCV RNA copies per milliliter is presented in Fig. 4. The linear regression equation for the mean of triplicate determinations was as follows: observed = 0.918(expected) + 0.436; R2 = 0.986; n = 32. The Superquant assay is linear through the range of ∼300 to 5,000,000 HCV RNA copies/ml. The assay appears to be linear to at least 5,000,000 HCV RNA copies/ml, but the linearity of the assay is compromised as expected values decrease below ∼300 HCV RNA copies/ml. This is indicated by the increase of observed versus expected HCV RNA values below this level.
FIG. 4.
Observed versus expected values for a dilution series measured in the Superquant assay. The means of triplicate values were calculated and plotted as log10 HCV RNA copies per milliliter. The equation for the linear regression is as follows: observed = 0.920(expected) + 0.424; R2 = 0.987; n = 32. The dashed line represents unity.
The results of the analysis of low-level samples in various quantitative and qualitative assays were as follows. Of the eight samples with previously high background values in the COBAS Amplicor HCV qualitative assay, four samples were reactive in the TMA and Amplicor COBAS HCV RNA qualitative assays. Of those four samples, three and two were quantitated with low-level HCV RNA in the Superquant and Amplicor assays, respectively. One sample was not quantitated in either the Amplicor or Superquant assay. Of 23 samples previously reported as <1,000 HCV RNA copies/ml in the Amplicor assay, 5 samples were reactive in the TMA assay. Of those five samples, two were positive in the COBAS Amplicor HCV qualitative assay, two samples were quantitated in the Superquant assay, and no samples were quantitated in the Amplicor Monitor assays. Samples reactive in TMA but not in a second method were retested in the TMA assay to confirm the initial results. Repeat testing confirmed the initial results in all cases. In the remaining 22 samples, HCV RNA was not detected by any of the methods used. These data indicate that the Superquant assay is better able to detect low HCV RNA levels than the Amplicor quantitative assays but is not as sensitive as the qualitative tests that are available. The published sensitivities of the assays are as follows: COBAS Amplicor HCV Monitor version 2.0 assay, 600 IU of HCV RNA/ml (∼1,000 HCV RNA copies/ml); NGI HCV Superquant assay, 100 HCV RNA copies/ml; COBAS Amplicor HCV qualitative, 50 IU of HCV RNA/ml (∼100 HCV RNA copies/ml); TMA HCV qualitative, 50 HCV RNA copies/ml (16). These cutoffs agree with the results which we observed.
DISCUSSION
HCV RNA quantitative measurement has become a routinely ordered test (5) for infected patients despite the lack of consensus regarding its role in patient care. Indeed, several hundred thousand assays are performed in the United States each year even though no submission of an HCV RNA quantitative method to the Food and Drug Administration has yet occurred. Lack of standardization of HCV RNA quantitative methods has contributed to confusion regarding the use of these assays. Clinicians and laboratorians have faced an especially difficult problem when implementing recommendations of clinical trials performed using assays which have not been standardized to more commonly used assays. Most recently, several studies examining the efficacy of interferon-ribovirin combination therapy (1, 11, 13) have employed the assay of the parent drug manufacturer, the HCV Superquant test, leading to recommendations based on the results of this test. However, one of the most commonly used methods for HCV RNA quantification is the Amplicor HCV Monitor assay, which has not been adequately compared to the assays used in clinical-trial testing. This test has recently been calibrated to report RNA levels in standardized international units based on WHO reference materials. Although it is expected that future HCV assays will be standardized to the WHO materials, interpretation of studies utilizing the HCV Superquant assay for Amplicor test users requires a systematic comparison of the two methods.
This study documents the performance attributes of the Amplicor HCV Monitor version 2.0 copy and international unit assays, as well as the NGI HCV Superquant test, and examines their performance using shared replicate samples. We find an equivalence of results produced by the current international unit version of the Amplicor HCV Monitor test with those of the previous HCV RNA copies-per-milliliter version of the assay from the perspectives of accuracy, precision, and dynamic range. This result was expected, since the reagents for the international unit assay are unchanged and only normalization to international unit reporting has been added as an enhancement to the version 2.0 test. The upper limit of linearity of the Amplicor assay expressed in international units was found to be approximately 600,000 IU/ml, which is similar to the upper limit of detection noted in a recent study (8). As noted in Table 1, above this limit, significant variation between values from diluted and undiluted samples is observed. In several instances, >10-fold differences, and up to 20-fold differences, were noted between samples run diluted versus undiluted in the range between 600,000 and 850,000 IU/ml.
The numerical relationship between international units per milliliter and copies per milliliter in the Amplicor HCV Monitor version 2.0 test will vary from lot to lot depending on how a given reagent lot calibrates to the WHO reference material. In this study, using several lots of reagents, the log10 numerical value for international units of HCV RNA per milliliter was approximately 0.4 log10 unit less than the log10 numerical value for HCV RNA copies per milliliter. This approximate ratio may provide a useful guide to physicians for comparing HCV RNA results in international units per milliliter to results obtained previously in copies per milliliter using the Amplicor assay.
For physicians using the Amplicor test reporting in international units of HCV RNA per milliliter, interpretation and implementation of much of the recent HCV clinical-trial literature depends on a direct comparison of the Amplicor test with the Superquant assay used in the clinical trials. This study confirms recent observations (12) that results obtained in the Superquant and Amplicor assays correlate well for samples that fall within the dynamic range of the Amplicor assay. Deming regression analysis of 28 clinical samples within the Amplicor HCV Monitor range of 600 to 600,000 IU of HCV RNA/ml supported a close correlation of Superquant HCV RNA copies per milliliter with Amplicor HCV Monitor international units of HCV RNA per milliliter. However, as the discrepant results for diluted samples above 600,000 IU of HCV RNA/ml show, values generated from undiluted samples in the Amplicor assay of >600,000 IU of HCV RNA/ml are unreliable, and comparison to Superquant results above the level is unwarranted and not recommended. On this point we differ from the conclusions (12) which suggest that results from samples assayed undiluted in the Amplicor assay can be reliably used to determine HCV RNA levels up to 2,000,000 to 3,000,000 HCV RNA copies/ml. This study supports an upper limit of the Amplicor assay of no greater than 600,000 IU of HCV RNA/ml. If accurate viral-load measurements above this level are required, samples should be diluted and reassayed. Alternatively, reporting Amplicor results simply as >600,000 IU/ml may be appropriate.
The limitations in the upper range of the Amplicor HCV Monitor version 2.0 assay also raise general questions about the utility of cutoff values suggested for the management of HCV-infected patients. An early study of the efficacy of interferon (19) with a small number of patients established general groupings of less than or greater than 2,000,000 HCV RNA copies/ml as important in predicting response to therapy. This study used the Quantiplex HCV RNA version 1.0 bDNA test for the determination of the HCV load. The cutoff of 2,000,000 HCV RNA copies/ml has persisted in the literature as a benchmark for many outcome studies even though other nonstandardized methods have been used in various clinical trials. The inflection point of 2,000,000 HCV RNA copies/ml may or may not be representative of equivalent HCV RNA levels, depending on which assay is used for quantification. Standardization of HCV RNA quantitative assays has the potential to improve the quality of data obtained from clinical trials for HCV therapy and to provide more reliable reference points for guiding therapy. While the reference point of 2,000,000 HCV RNA copies/ml persists in the literature, it has been noted that treatment response can still occur in patients with viral-load levels above this point (9). Additional studies with interferon-ribavirin combination therapy and pegylated interferon may also yield new information on the value of an HCV RNA level cutoff. Cautious evaluation of study data and careful consideration of the meaning of any cutoff value is needed before a value becomes established as a predictive cutoff. This type of evaluation has not yet been done, and clinicians should be careful of overinterpreting HCV loads and of compounding that error by assuming cutoffs are sharp breakpoints with respect to clinical outcome. In the absence of more complete reporting of data from clinical trials, the significance of HCV values across the spectrum of possible viral loads remains uncertain.
Acknowledgments
This work was supported by a grant from Roche Molecular Systems, Inc.
We give special thanks to Stewart Wood, Amy Stuyvesant, Lisa Bergantz, Ward Willoughby, James Sorensen, Jay Hansen, and Tyler Gerritsen for performing the Amplicor assays.
REFERENCES
- 1.Davis, G. L., R. Esteban-Mur, V. Rustgi, J. Hoefs, S. C. Gordon, C. Trepo, M. L. Shiffman, S. Zeuzem, A. Craxi, M. H. Ling, J. Albrecht, et al. 1998. Interferon alfa-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. N. Engl. J. Med. 339:1493-1499. [DOI] [PubMed] [Google Scholar]
- 2.Davis, G. L., and J. Y. Lau. 1997. Factors predictive of a beneficial response to therapy of hepatitis C. Hepatology 26:122S-127S. [DOI] [PubMed]
- 3.DiDomenico, N., H. Link, R. Knobel, T. Caratsch, W. Weschler, Z. G. Loewy, and M. Rosenstraus. 1996. COBAS AMPLICOR: fully automated RNA and DNA amplification and detection system for routine diagnostic PCR. Clin. Chem. 42:1915-1923. [PubMed] [Google Scholar]
- 4.Erali, M., E. R. Ashwood, and D. R. Hillyard. 2000. Performance characteristics of the COBAS AMPLICOR hepatitis C virus MONITOR Test, version 2.0. Am. J. Clin. Pathol. 114:180-187. [DOI] [PubMed] [Google Scholar]
- 5.Gish, R. G. 1999. Standards of treatment in chronic hepatitis C. Semin. Liver Dis. 19:35-47. [PubMed] [Google Scholar]
- 6.Gretch, D. R., C. dela Rosa, R. L. Carithers, Jr., R. A. Willson, B. Williams, and L. Corey. 1995. Assessment of hepatitis C viremia using molecular amplification technologies: correlations and clinical implications. Ann. Intern. Med. 123:321-329. [DOI] [PubMed] [Google Scholar]
- 7.Jacob, S., D. Baudy, E. Jones, L. Xu, A. Mason, F. Regenstein, and R. P. Perrillo. 1997. Comparison of quantitative HCV RNA assays in chronic hepatitis C. Am. J. Clin. Pathol. 107:362-367. [DOI] [PubMed] [Google Scholar]
- 8.Lee, S. C., A. Antony, N. Lee, J. Leibow, J. Q. Yang, S. Soviero, K. Gutekunst, and M. Rosenstraus. 2000. Improved version 2.0 qualitative and quantitative AMPLICOR reverse transcription-PCR tests for hepatitis C virus RNA: calibration to international units, enhanced genotype reactivity, and performance characteristics. J. Clin. Microbiol. 38:4171-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lok, A. S., and N. T. Gunaratnam. 1997. Diagnosis of hepatitis C. Hepatology 26:48S-56S. [DOI] [PubMed]
- 10.Lunel, F., P. Cresta, D. Vitour, C. Payan, B. Dumont, L. Frangeul, D. Reboul, C. Brault, J. C. Piette, and J. M. Huraux. 1999. Comparative evaluation of hepatitis C virus RNA quantitation by branched DNA, NASBA, and monitor assays. Hepatology 29:528-535. [DOI] [PubMed] [Google Scholar]
- 11.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, J. K. Albrecht, et al. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 12.Pawlotsky, J. M., M. Bouvier-Alias, C. Hezode, F. Darthuy, J. Remire, and D. Dhumeaux. 2000. Standardization of hepatitis C virus RNA quantification. Hepatology 32:654-659. [DOI] [PubMed] [Google Scholar]
- 13.Poynard, T., P. Marcellin, S. S. Lee, C. Niederau, G. S. Minuk, G. Ideo, V. Bain, J. Heathcote, S. Zeuzem, C. Trepo, J. Albrecht, et al. 1998. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. Lancet 352:1426-1432. [DOI] [PubMed] [Google Scholar]
- 14.Reichard, O., G. Norkrans, A. Fryden, J. H. Braconier, A. Sonnerborg, and O. Weiland. 1998. Comparison of 3 quantitative HCV RNA assays—accuracy of baseline viral load to predict treatment outcome in chronic hepatitis C. Scand. J. Infect. Dis. 30:441-446. [DOI] [PubMed] [Google Scholar]
- 15.Saldanha, J., N. Lelie, A. Heath, et al. 1999. Establishment of the first international standard for nucleic acid amplification technology (NAT) assays for HCV RNA. Vox Sang. 76:149-158. [DOI] [PubMed] [Google Scholar]
- 16.Sarrazin, C., G. Teuber, R. Kokka, H. Rabenau, and S. Zeuzem. 2000. Detection of residual hepatitis C virus RNA by transcription-mediated amplification in patients with complete virologic response according to polymerase chain reaction-based assays. Hepatology 32:818-823. [DOI] [PubMed] [Google Scholar]
- 17.Tong, M. J., L. M. Blatt, J. G. McHutchison, R. L. Co, and A. Conrad. 1997. Prediction of response during interferon alfa 2b therapy in chronic hepatitis C patients using viral and biochemical characteristics: a comparison. Hepatology 26:1640-1645. [DOI] [PubMed] [Google Scholar]
- 18.Tong, M. J., S. J. Hwang, M. Lefkowitz, S. D. Lee, R. L. Co, A. Conrad, P. Schmid, and K. J. Lo. 1994. Correlation of serum HCV RNA and alanine aminotransferase levels in chronic hepatitis C patients during treatment with ribavirin. J. Gastroenterol. Hepatol. 9:587-591. [DOI] [PubMed] [Google Scholar]
- 19.Yamada, G., M. Takatani, F. Kishi, M. Takahashi, T. Doi, T. Tsuji, S. Shin, M. Tanno, M. S. Urdea, and J. A. Kolberg. 1995. Efficacy of interferon alfa therapy in chronic hepatitis C patients depends primarily on hepatitis C virus RNA level. Hepatology 22:1351-1354. [PubMed] [Google Scholar]