Abstract
A lymphotropic papovavirus (LPV) archetypal regulatory region was amplified from DNA from the blood of an immunocompromised rhesus monkey. We believe this is the first nonserological evidence of LPV infection in rhesus monkeys.
Lymphotropic papovavirus (LPV) and simian virus 40 (SV40) are members of the genus Polyomavirus (6). LPV was isolated from a B-lymphoblastic cell line derived from a lymph node of an apparently healthy African green monkey (18). In cell culture, LPV displays a highly restricted host range for continuous lines of B lymphoblasts of monkey or human origin (2, 3). Serological surveys of sera from humans and 11 different species of nonhuman primates, including rhesus monkeys, indicated that, with the exception of baboons, all showed evidence of infection by LPV or an antigenically related virus (14, 19).
Little is known of the natural biology of LPV. Because previous serological data suggested that LPV caused natural infections in different types of primates, we tested whether LPV might be detectable in simian immunodeficiency virus (SIV)-infected rhesus monkeys. We examined DNA that had been extracted for a previous study from brain tissue, peripheral blood mononuclear cells (PBMCs), or lymphoid tissue from SIV-infected rhesus macaques with SV40 disease (5). Due to limited sample availability, few PCR assays were possible, so analyses were designed to detect viral regulatory region sequences. The polyomavirus regulatory regions contain enhancer, promoter, and origin of DNA replication (ori) sequences that are distinct and represent unambiguous genetic markers that characterize each member. Thus, if an LPV regulatory region sequence were detected, there would be a high level of confidence that LPV (or a related virus containing the LPV regulatory region) was present in the sample.
Primer pairs LPR1/LPR2 and LPR3/LPR4 designed to detect LPV regulatory region sequences are given in Table 1. The locations of primer binding sites on the LPV regulatory region are shown (Fig. 1). Virus LPV-K38, from the original LPV isolate passaged in BJAB cells, was the first molecular clone of LPV to be fully sequenced (11). Plasmid pLPV-K38 contains a full-length genome of LPV-K38 and was used as a positive control template for PCR analysis. The oligonucleotides and procedures used for PCR and DNA sequence analysis of SV40 sequences were described previously (5). All oligonucleotides were obtained from Life Technologies, Inc. (Rockville, Md.). PCR amplifications were performed as described previously (5, 7). A second round of PCR was performed for the PBMC DNAs, because polyomavirus-size circular supercoiled DNAs are not efficiently recovered by the spooling method that had been used to prepare PBMC DNA (1). PCR products were cloned and sequenced according to published procedures (5, 7).
TABLE 1.
PCR and DNA sequencing oligonucleotides used for detection and analysis of LPVa
| LPV regulatory region oligo- nucleotide pair | Sequence | Reference positions (nt)b | Size (bp) of PCR product (pLPV-K38) |
|---|---|---|---|
| LPR1 | 5′-GCAACTAGAC- CGCAGAACAGTTG-3′ | 590-567 | 520 |
| LPR2 | 5′-CTCAGGGCAGC- TTACCTAATGAG-3′ | 70-92 | |
| LPR3 | 5′-GGATAGCATCA- AGTGTAAATCCAG-3′ | 790-767 | 819 |
| LPR4 | 5′-CTCTCCTCCTT- AGACAGCGTTTGG-3′ | 5242-5265 |
The temperature used was 65°C.
Reference nucleotide positions in LPV reference strain K38.
FIG. 1.
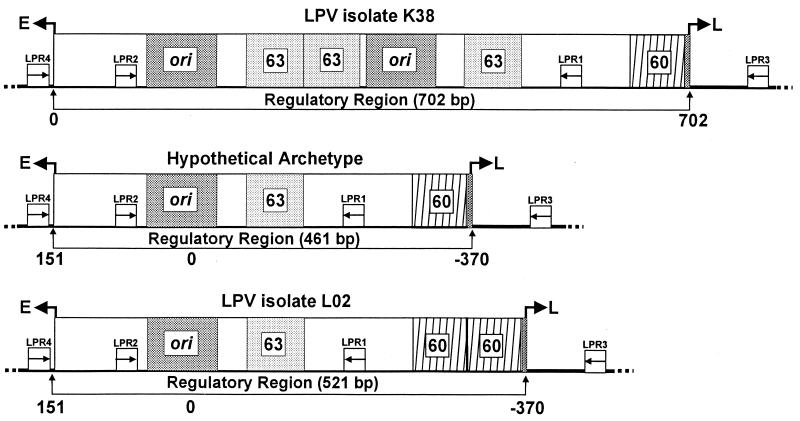
Schematic representation of representative LPV regulatory regions. Structures for LPV isolate K38, isolate L02, and the hypothetical archetype are from Pawlita et al. (11), Furuno et al. (4), and Yoshiike and Takemoto (17). The numbers below the diagrams refer to nucleotide positions within the regulatory region and are from the references cited. The LPV PCR primer binding sites are represented by small boxes inscribed with arrows; arrows indicate the direction of DNA synthesis initiated from those positions. A box inscribed ori represents the origin of DNA replication. Sequences to the right of ori that are within the delimited regulatory region (indicated below each figure) are considered enhancer sequences. The enhancer 63- and 60-bp subgenomic elements are shown as boxes inscribed with the no. 63 or 60. Arrows pointing to the left or right at the borders of each diagram refer to the direction of late (L) or early (E) viral transcription.
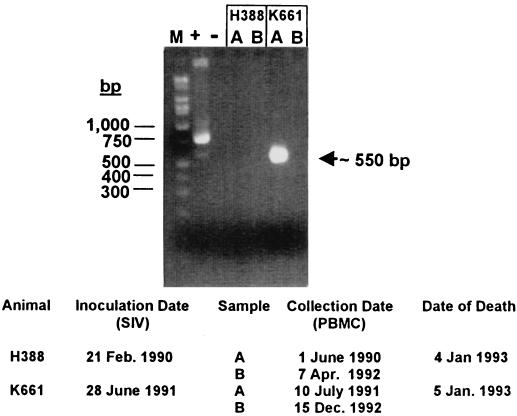
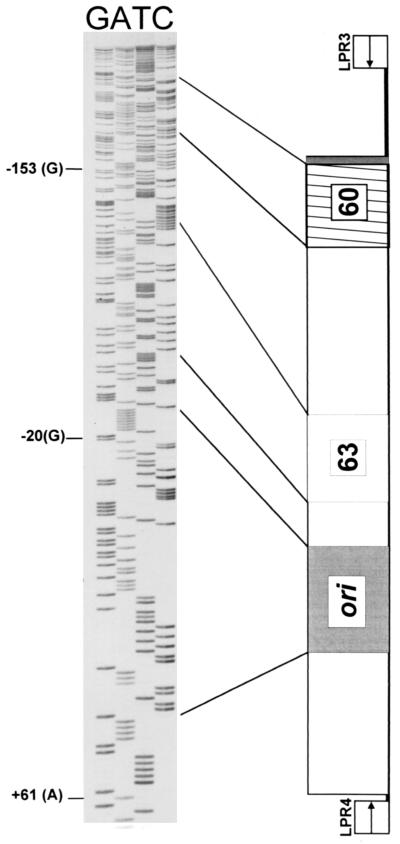
PCR analysis of PBMC DNA with primer pair LPR3/LPR4 yielded a robust PCR-amplified band that was estimated to be about 550 bp in size from the K661 lane A sample, but not from the other samples (Fig. 2). This band was readily distinguished from the positive control PCR, which formed the expected 833-bp product (Fig. 2). Reexamination of a 1:10 dilution of the K661 lane A PCR revealed that two DNA products of similar sizes (approximately 570 and 530 bp) comprised the original PCR product. These PCR products were cloned, and individual clones were analyzed. Sequence analysis of the larger product revealed a 577-bp DNA sequence that matched exactly the hypothetical archetypal regulatory region sequence of LPV (17) (Fig. 3), a sequence readily distinguished from that of LPV-K38, the positive control template. Because we had not worked with LPV prior to this investigation, and because the archetypal LPV sequence was different from that of the control plasmid, we were able to rule out laboratory contamination of the PCR. DNA sequence analysis of the smaller PCR band revealed that it was unrelated to known polyomaviruses.
FIG. 2.
Detection of an LPV regulatory region in a simian PBMC sample. PCR primers LPR3 and LPR4 were used. Molecular weight markers (M) are on the left. + and −, positive and negative controls, respectively. Plasmid pLPV-K38 was used as a positive control template. The band at about 550 bp from animal K661 was shown to contain an LPV-specific 577-bp product as well as a smaller 530-bp nonspecific product. Relevant animal data are listed at the bottom.
FIG. 3.
Sequence analysis of an LPV regulatory region. Primers LPR3 and LPR4 were used for PCR amplification from PBMC DNA from animal K661. Shown are results obtained with LPR4 as the sequencing primer. A schematic of the hypothetical LPV archetypal sequence is shown to the right of the DNA sequence. Reference nucleotides (shown to the left of the DNA sequence) are numbered according to Furuno et al. (4).
LPV was detected in DNA from PBMCs from animal K661 that were drawn in July 1991, but not in a sample drawn in December 1992 (Fig. 2). Both DNA samples had previously tested positive for SV40-K661, ruling out the possibility that PCR inhibitors were present in the latter sample (5). Because the tissue distribution of LPV in natural infections is unknown, PCR assays were performed with available DNA samples extracted from brain tissue from eight animals. All of these DNA samples had previously tested positive for SV40 (5), but all tested negative for LPV sequences.
LPV had originally been isolated from the lymph node of an African green monkey. Lymph node and spleen total DNAs from the previous study for animal T302 were available and had tested positive for SV40 strain T302 (5). Serendipitously, archival autopsy spleen tissue from animal K661 was obtained. The K661 spleen tissue DNA and the T302 spleen and lymph node DNAs were tested by PCR; each was negative for LPV, but positive for SV40.
We attempted to isolate LPV replicating virus by inoculation of Namalwa (12) and BJAB (9) cells with K661 spleen filtrate. Aliquots of the cells were tested for LPV by PCR at each subculture (every 6 days) and at the end of the experiment (after 2 months). LPV-K38 virus, used as a positive control, was prepared by electroporation of linear K38 DNA (obtained from pLPV-K38) into BJAB cells. LPV-K38 could be propagated in the two lymphoid cell lines, although no cytopathic effect was produced. In contrast, both Namalwa and BJAB cell cultures inoculated with K661 spleen filtrate developed a cytopathic effect characterized by swelling of the cells and total cell death within 7 days. PCR and DNA sequence analysis revealed that SV40-K661 was present, but LPV was not. These results were surprising, because SV40 strain 776 and SV40-K661 failed to grow in Namalwa or BJAB cells. Thus, LPV was not detected in spleen cells of animal K661 by PCR or by tissue culture, but some factor in the spleen extract appeared to facilitate the growth of SV40-K661 in Namalwa and BJAB cells.
We report the detection of an archetypal LPV regulatory region sequence in the PBMC DNA of an SIV-infected rhesus monkey that died of SV40 disease. An archetypal LPV regulatory region was detected, rather than the rearranged complex regulatory regions of LPV clones K38 (Fig. 1) (11) and 02 (4, 14), both of which were cloned from virus stocks derived from the original LPV isolate. Based on what is thought to occur with the polyomaviruses BK virus, JC virus, and SV40 (5, 8, 10, 13, 15, 16), we speculate that LPV strains with archetypal regulatory regions are the forms that are transmitted naturally. We suggest that archetypal virus may replicate more abundantly in immunocompromised hosts than in healthy animals.
This report demonstrates that LPV or LPV-like virus can be present in rhesus macaques, validating previous serological determinations (14). As far as we know, this is the first nonserological detection of LPV in a rhesus monkey.
Acknowledgments
We thank Michael Murphey-Corb for providing the specimens from SIV-infected rhesus macaques, S. Kang for the gift of plasmid pLPV-K38, and Paul Ling for Namalwa and BJAB cell cultures. This work was supported in part through Cooperative Agreement NCC 9-58 with the National Space Biomedical Research Institute.
REFERENCES
- 1.Amedee, A. M., N. Lacour, J. L. Gierman, L. N. Martin, J. E. Clements, R. Bohm, Jr., R. M. Harrison, and M. Murphey-Corb. 1995. Genotypic selection of simian immunodeficiency virus in macaque infants infected transplacentally. J. Virol. 69:7982-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brade, L., N. Muller-Lantzsch, and H. zur Hausen. 1981. B-lymphotropic papovavirus and possibility of infections in humans. J. Med. Virol. 6:301-308. [DOI] [PubMed] [Google Scholar]
- 3.Brade, L., W. Vogl, L. Gissman, and H. zur Hausen. 1981. Propagation of B-lymphotropic papovavirus (LPV) in human B-lymphoma cells and characterization of its DNA. Virology 114:228-235. [DOI] [PubMed] [Google Scholar]
- 4.Furuno, A., T. Miyamura, and K. Yoshiike. 1984. Monkey B-lymphotropic papovavirus DNA: nucleotide sequence of the region around the origin of replication. J. Virol. 50:451-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lednicky, J. A., A. S. Arrington, A. R. Stewart, X. M. Dai, C. Wong, S. Jafar, M. Murphey-Corb, and J. S. Butel. 1998. Natural isolates of simian virus 40 from immunocompromised monkeys display extensive genetic heterogeneity: new implications for polyomavirus disease. J. Virol. 72:3980-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lednicky, J. A., and J. S. Butel. 1999. Polyomaviruses and human tumors: a brief review of current concepts and interpretations. Frontiers Biosci. 4:153-164. [DOI] [PubMed] [Google Scholar]
- 7.Lednicky, J. A., A. R. Stewart, J. J. Jenkins III, M. J. Finegold, and J. S. Butel. 1997. SV40 DNA in human osteosarcomas shows sequence variation among T-antigen genes. Int. J. Cancer 72:791-800. [DOI] [PubMed] [Google Scholar]
- 8.Loeber, G., and K. Dörries. 1988. DNA rearrangements in organ-specific variants of polyomavirus JC strain GS. J. Virol. 62:1730-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menezes, J., W. Leibold, G. Klein, and G. Clements. 1975. Establishment and characterization of an Epstein-Barr virus (EBV)-negative lymphoblastoid B cell line (BJA-B) from an exceptional, EBV-genome-negative African Burkitt's lymphoma. Biomedicine 22:276-284. [PubMed] [Google Scholar]
- 10.Newman, J. T., and R. J. Frisque. 1997. Detection of archetype and rearranged variants of JC virus in multiple tissues from a pediatric PML patient. J. Med. Virol. 52:243-252. [DOI] [PubMed] [Google Scholar]
- 11.Pawlita, M., A. Clad, and H. zur Hausen. 1985. Complete DNA sequence of lymphotropic papovavirus: prototype of a new species of the polyomavirus genus. Virology 143:196-211. [DOI] [PubMed] [Google Scholar]
- 12.Reedman, B. M., and G. Klein. 1973. Cellular localization of an Epstein-Barr virus (EBV)-associated complement-fixing antigen in producer and non-producer lymphoblastoid cell lines. Int. J. Cancer 11:499-520. [DOI] [PubMed] [Google Scholar]
- 13.Rubinstein, R., B. C. A. Schoonakker, and E. H. Harley. 1991. Recurring theme of changes in the transcriptional control region of BK virus during adaptation to cell culture. J. Virol. 65:1600-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takemoto, K. K., A. Furuno, K. Kato, and K. Yoshiike. 1982. Biological and biochemical studies of African green monkey lymphotropic papovavirus. J. Virol. 42:502-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White, F. A., III, M. Ishaq, G. L. Stoner, and R. J. Frisque. 1992. JC virus DNA is present in many human brain samples from patients without progressive multifocal leukoencephalopathy. J. Virol. 66:5726-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yogo, Y., T. Kitamura, C. Sugimoto, T. Ueki, Y. Aso, K. Hara, and F. Taguchi. 1990. Isolation of a possible archetypal JC virus DNA sequence from nonimmunocompromised individuals. J. Virol. 64:3139-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshiike, K., and K. K. Takemoto. 1986. Studies with BK virus and monkey lymphotropic papovavirus, p. 295-326. In N. P. Salzman (ed.), The Papovaviridae, . The polyomavirusesvol. 1. Plenum Press, New York, N.Y. [Google Scholar]
- 18.zur Hausen, H., and L. Gissmann. 1979. Lymphotropic papovaviruses isolated from African green monkey and human cells. Med. Microbiol. Immunol. 167:137-153. [DOI] [PubMed] [Google Scholar]
- 19.zur Hausen, H., L. Gissmann, A. Mincheva, and J. F. Böcker. 1980. Characterization of a lymphotropic papovavirus, p. 365-372. In M. Essex, G. Todaro, and H. zur Hausen (ed.), Viruses in naturally occurring cancers. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.