Abstract
A method for species-specific detection of orthopoxviruses pathogenic for humans and animals is described. The method is based on hybridization of a fluorescently labeled amplified DNA specimen with the oligonucleotide DNA probes immobilized on a microchip (MAGIChip). The probes identify species-specific sites within the crmB gene encoding the viral analogue of tumor necrosis factor receptor, one of the most important determinants of pathogenicity in this genus of viruses. The diagnostic procedure takes 6 h and does not require any sophisticated equipment (a portable fluorescence reader can be used).
The genus Orthopoxvirus of the family Poxviridae consists of nine species, four of which are able to cause human diseases of various severities (29). The most dangerous member of this taxonomic group is variola virus, the causative agent of smallpox, which has been considered eradicated as a natural cause of disease since 1977. However, other orthopoxviruses (OPVs), such as cowpox virus, buffalopox virus (a subspecies of vaccinia virus), and especially monkeypox virus (1), can also infect humans through contact with wildlife and farm animals, since each of these infectious agents has vast natural reservoirs (7, 31).
Reports of a high frequency of intra- and intermolecular recombination (2, 37, 51), as well as transmission of OPVs between animal species (29), indicate that these pathogenic agents can undergo further evolution. Particularly dangerous is a potential increase in their virulence for humans.
Even the attenuated strains of vaccinia virus that are widely used in genetic engineering and biotechnology can cause severe disease in people with compromised immunity (19, 29). Furthermore, natural or genetically modified OPVs could be used in a bioterrorist attack.
All these facts call for constant monitoring of OPVs that are pathogenic or potentially pathogenic for humans. This requires reliable, inexpensive, and rapid methods of virus detection and species identification.
Several approaches have been proposed for OPV species-level identification in recent years: immunoenzyme analysis of viral antigens (28), immunoblotting (33), PCR followed by electrophoretic analysis of the product length (31, 33), restriction analysis of PCR products (21, 43, 52), and detection of species-specific point substitutions in the hemagglutinin gene (18). All these methods are sensitive and enable one to identify OPVs within 24 h.
Here we describe a method for detection and species identification of OPVs by hybridization on an oligonucleotide MAGIChip (for microarray of gel-immobilized compounds on a chip) that has already been successfully applied to identify the rpoB gene mutations causing resistance to rifampin in Mycobacterium tuberculosis (32).
A fragment of the crmB gene was selected as a target sequence for species-level identification (6, 17). The CrmB protein (alternatively designated the viral tumor necrosis factor receptor [vTNFR]) is a homologue of cellular TNFR. A strong species-specific conservation of the vTNFR (49) offers a convenient way to accurately identify the OPV species (21).
MATERIALS AND METHODS
Oligonucleotides and primers.
Oligonucleotides were synthesized in a model 394 DNA-RNA synthesizer (Applied Biosystems, Foster City, Calif.). Oligonucleotide probes for immobilization on the chip carried a 3′ amino group introduced by 3′-Amino-Modifier C7 CPG 500 (Glen Research, Sterling, Va.). A fragment of the vTNFR gene 245 to 273 bp in length (the length varies from one species to another) was amplified using primers TNFR1f (5′-GCT TCC AGA TTA TGT GAT AGC AAG ACT A-3′) and TNFR3r (5′-TCC GGA TAC TCC GTA TCC TAT TCC-3′). The primer TNFR3r used in the second round of amplification (see below) was 5′ fluorescently labeled with Texas Red (Molecular Probes Inc., Eugene, Oreg.) according to the manufacturer's instructions. Oligonucleotides were purified by high-performance liquid chromatography on a C18 Nucleosil column (Sigma, St. Louis, Mo.).
Virus strains and DNAs.
The OPVs used as sources of DNA in the study are listed in Table 1. A number of OPV DNAs have been received from other laboratories (Table 1). DNAs of myxoma virus strain Lausanne (genus Leporipoxvirus) donated by G. McFadden (5) and fowlpox virus strain FP9 (genus Avipoxvirus) received from M. Skinner (39) were used as negative controls.
TABLE 1.
OPV DNAs used in microchip analysis
| OPV species | Strain | Reference | Source of strain or DNA |
|---|---|---|---|
| Vaccinia virus | Wyeth | 14 | J. Esposito (DNA) |
| LIVP, clone 2 | 35 | V. Petrov (DNA) | |
| LIVP, clone 4 | 35 | V. Petrov (DNA) | |
| EM-63 | 26 | S. Marennikova | |
| Copenhagen | 29 | J. Esposito (DNA) | |
| Patwadangar | 29 | S. Marennikova | |
| Elstree | 20 | S. Marennikova | |
| CVI-78 | 42 | S. Marennikova | |
| WR | 4 | V. Petrov (DNA) | |
| Rabbitpox Utrechta | 8 | H. Meyer (DNA) | |
| BP-1a | 22 | H. Meyer (DNA) | |
| Elstree/Utrechta | Unpublished | S. Marennikova | |
| Chambon St-Yves | Unpublished | S. Marennikova | |
| Menarda | |||
| Cowpox virus | GRI-90 | 24, 47 | S. Marennikova |
| Hamburg | 38 | S. Marennikova | |
| Turkmenia | 25 | S. Marennikova | |
| EP-1 | 38 | S. Marennikova | |
| EP-2 | 38 | S. Marennikova | |
| EP-3 | 38 | S. Marennikova | |
| EP-4 | 38 | S. Marennikova | |
| EP-5 | 38 | S. Marennikova | |
| EP-6 | 38 | S. Marennikova | |
| EP-7 | 38 | S. Marennikova | |
| EP-8 | 38 | S. Marennikova | |
| EP-267 | 38 | S. Marennikova | |
| RP-1 | 38 | S. Marennikova | |
| RP-9 | 38 | S. Marennikova | |
| Puma M-73 | 27 | S. Marennikova | |
| Brighton | 10 | H. Meyer (DNA) | |
| Monkeypox virus | Congo 8 | 30 | J. Esposito (DNA) |
| Zaire 96-I-16 | 33 | J. Esposito (DNA) | |
| 70-0266 | 13 | J. Esposito (DNA) | |
| 77-0666 | 13 | J. Esposito (DNA) | |
| Variola virus | Garcia-1966 | 48 | J. Esposito (DNA) |
| Ngami | Unpublished | This researchc | |
| Congo-2 | Unpublished | This researchc | |
| Congo-9 | Unpublished | This researchc | |
| Ind-3a | Unpublished | This researchc | |
| Kuw-5 | Unpublished | This researchc | |
| 12-62 | Unpublished | This researchc | |
| 6-58 | Unpublished | This researchc | |
| Butler | Unpublished | This researchc | |
| M-Abr-60 | Unpublished | This researchc | |
| M-Sok-60 | Unpublished | This researchc | |
| M-Sur-60 | Unpublished | This researchc | |
| Aziz | Unpublished | This researchc | |
| Semat | Unpublished | This researchc | |
| Aslamb | Unpublished | This researchc | |
| India 378b | Unpublished | This researchc | |
| Camelpox virus | CP-1 | 41 | H. Meyer (DNA) |
| CP-5 | 36 | H. Meyer (DNA) | |
| CP-1260/95 | Unpublished | H. Meyer (DNA) | |
| CP-1231 | Unpublished | H. Meyer (DNA) | |
| CP Saudi | Unpublished | H. Meyer (DNA) | |
| Ectromelia virus | MP-1 GE | 23 | H. Meyer (DNA) |
| MP-2 GE | 34 | H. Meyer (DNA) | |
| No. 4908 USA | 9 | H. Meyer (DNA) | |
| No. 33221 USA | 9 | H. Meyer (DNA) | |
| K1 | 44 | V. Petrov (DNA) |
Strain produces the rabbitpox virus-like hybridization pattern on the microchip.
Variola virus DNA isolated from human smallpox scabs.
Species identification was performed by conventional methods (host range in animals, morphology of viral pocks produced on CAM, reproductive ceiling temperature in cell culture or on chicken embryo CAM).
Vero cells (American Type Culture Collection) used for virus propagation were grown in Dulbecco's modified Eagle's medium supplemented with 5% fetal calf serum. Monolayer cells were infected and incubated at 35°C for 2 to 4 days. Viral DNA was isolated from the cytoplasm of infected cells as described previously (12). Cultivation of variola virus strains from the Russian collection of variola virus and viral DNA isolation were carried out in the biological safety level 4 (BSL-4) laboratory of the State Research Center of Virology and Biotechnology “Vector” certified by the World Health Organization and the Russian national authorities. Experiments with noninfectious variola virus DNAs were performed under BSL-2 conditions.
The study described in this paper was approved by the World Health Organization as a part of the Russian program on variola virus and other OPV research.
Isolation of variola virus DNA from human scabs.
A scab isolated from a human smallpox patient (as part of the international program for smallpox eradication) and stored in the Russian variola virus collection (State Research Center of Virology and Biotechnology “Vector”) was placed in an Eppendorf tube; supplemented with 200 μl of the lysis buffer containing 100 mM Tris-HCl (pH 8.0), 100 mM EDTA, 100 mM NaCl, and 1% sodium dodecyl sulfate; and incubated for 10 min at 85°C. Twenty microliters of proteinase K solution (10 mg/ml) were added, mixed, and incubated for 1 h at 56°C. Then, 200 μl of the lysis buffer was added and the mixture was kept for 10 min at 56°C. The insoluble debris was removed by centrifugation in an Eppendorf centrifuge for 7 min at 5,000 rpm. DNA was isolated by phenol-chloroform extraction and isopropanol precipitation in the presence of 0.3 M sodium acetate (pH 5.5). After special decontamination, the tubes were transported to the BSL-2 laboratory. The tubes were centrifuged in an Eppendorf centrifuge for 10 min at 14,000 rpm, and the sediment was washed with 70% ethanol. The pellet was dissolved in 50 μl of Tris-EDTA buffer, precipitated with isopropanol, washed with 70% ethanol, air dried, and dissolved in 50 μl of water.
Isolation of vaccinia virus DNA from pocks on CAMs of developing chick embryos.
Pocks isolated from the chorioallantoic membrane (CAM) of developing chick embryos infected with vaccinia virus were transferred to a tube containing 20 μl of 10-mg/ml proteinase K solution and 200 μl of the lysis buffer (see above), and the contents were mixed. The mixture was incubated for 10 min at 56°C and centrifuged for 7 min at 14,000 rpm (in an Eppendorf centrifuge) to remove the insoluble debris. The supernatant was transferred to a fresh tube and extracted with 400 μl of phenol-chloroform (1:1) mixture. The aqueous phase was extracted twice with isoamyl alcohol, precipitated with ethanol in the presence of 0.3 M sodium acetate (pH 5.5), washed with 70% ethanol, air dried, and dissolved in water.
PCR amplification.
The viral DNA was amplified by a two-stage PCR. In the first stage, 50 μl of reaction mixture contained 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 0.2 mM (each) deoxynucleoside triphosphate, 2.5 U of AmpliTaq DNA polymerase (Perkin-Elmer Corporation, Norwalk, Conn.), 100 nM (each) the two primers (TNFR1f and TNFR3r), and 2 μl of a viral DNA sample. The reaction was carried out in a GeneAmp PCR System 2400 (Perkin-Elmer, Foster City, Calif.) as follows: 5 min at 95°C; 30 cycles of 35 s at 95°C, 45 s at 64°C, and 45 s at 72°C; and 5 min at 72°C.
In the second stage, an asymmetric PCR was used to produce the predominantly single-stranded fluorescently labeled product for the subsequent hybridization. Two microliters of the first-stage PCR mixture was added as a template, and the reaction was carried out for 35 cycles under the same conditions except for the concentrations of the TNFR1f and fluorescently labeled TNFR3r primers, which were 10 and 100 nM, respectively.
The PCR products were analyzed by electrophoresis in agarose gels.
MAGIChip with immobilized oligonucleotides.
The MAGIChips were manufactured as described earlier (40, 50, 53). Oligonucleotides were immobilized according to a previously published procedure (32).
The sequences of the specific immobilized probes were selected within the sense strand of the crmB gene. Multiple alignments of the published sequences of the crmB gene from 47 isolates of six OPV species (variola, monkeypox, cowpox, vaccinia, camelpox, and taterapox viruses) and two vaccinia subspecies (rabbitpox and buffalopox viruses) from different geographical areas were constructed using BioEdit (16) and proprietary software. Oligonucleotide probes were selected so that each variable site containing a point substitution, short deletion, or insertion was located near the middle of the probe (Fig. 1). The sequences and probe lengths were adjusted in such a way as to bring their calculated melting temperatures for perfect duplexes within a 3 to 4°C difference.
FIG. 1.
Region of the variola virus vTNFR gene used as a DNA target for on-chip hybridization analysis. The sequences of oligonucleotide probes 1 to 15 and their locations relative to the crmB gene sequence are shown. The species-specific nucleotides are underlined. The dashes indicate deletions.
Hybridization on the MAGIChip.
The hybridization mixture was prepared by adding 10 μl of the second-stage asymmetric PCR mixture to 20 μl of 1.5 M NaCl, 75 mM HEPES (pH 7.5), and 8 mM EDTA. Hybridization was performed as described earlier (32) except for the incubation time (2 h). After hybridization, the chip was washed three times at 37°C with 0.8 M NaCl, 50 mM HEPES (pH 7.0), 6 mM EDTA, and 0.5% Tween 20 and air dried. The resulting fluorescence pattern was recorded using a research charge-coupled device camera-equipped setup as described earlier (15) or a portable fluorescence analyzer (3).
The optimal NaCl concentration, temperature, and time of hybridization were determined by analyzing the melting curves and hybridization kinetics of perfect duplexes for all available samples. The melting curves were obtained on a microchip under near-equilibrium conditions as described earlier (11).
RESULTS
The microchip contained 15 oligonucleotide probes directed towards five species-specific segments of the vTNFR gene. The probe locations relative to the sequence of the vTNFR gene of the variola virus (EMBL accession no. X69198 [45, 46]) are shown in Fig. 1.
The microchip contained five gel pad columns (Fig. 2), each column representing a separate interrogated segment of the amplified fragment of viral DNA. Within each column, only one species-specific probe can form a perfect duplex with a viral DNA sample, while all other probes form mismatched duplexes. As a result, the hybridization pattern is unique for every tested DNA sample, thus allowing an accurate species assignment.
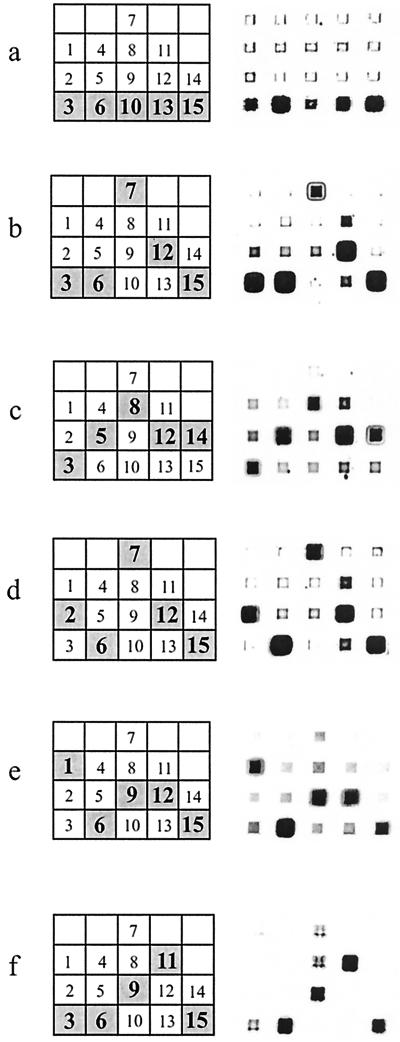
FIG. 2.
Hybridization patterns of six OPV DNAs from variola minor virus strain Garcia-1966 (a), vaccinia virus strain Wyeth (b), monkeypox virus strain Zaire-96 (c), vaccinia virus strain rabbitpox Utrecht (d), cowpox virus strain GRI-90 (e), and camelpox virus strain CP-1 (f). In the graphs on the left, the expected fluorescence patterns are shown. The numbers inside the microchip elements correspond to the probe numbering in Fig. 1.
In Fig. 2, the graphs on the left show the expected hybridization patterns for all tested species. On the right, the actual hybridization images are displayed. The gel pads containing perfect duplexes emit much stronger fluorescence than those containing the corresponding mismatched duplexes. The relative fluorescence intensities of individual gel pads should be considered within each column separately; for example, consider the leftmost column of the microchip after hybridization with the camelpox virus DNA sample (Fig. 2f). A positive signal corresponding to the perfect duplex formed by hybridization of the target DNA with probe 3 can be identified unambiguously within the column, although its absolute fluorescence level is comparable to some of the signals corresponding to the mismatched duplexes in other columns.
Fifty-nine samples of OPV DNA representing six different species were analyzed (Table 1). Different strains of variola, monkeypox, cowpox, vaccinia, and camelpox viruses were successfully identified by hybridization of amplified DNA to the microchip. No discrepancy between hybridization and conventional identification results was observed. No amplification with the chosen specific primers (TNFR1f and TNFR3r) was observed when we used as a template DNA isolated from the related ectromelia virus (Table 1), in whose genome the locus under consideration was lacking. The primers also failed to amplify any PCR product from human genomic DNA, cellular DNA isolated from uninfected Vero cell cultures, and DNAs of unrelated poxviruses—myxoma virus (genus Leporipoxvirus) and fowlpox virus (genus Avipoxvirus) (data not shown).
Computer analysis showed that the locus under consideration was variable within the vaccinia species (Fig. 1). Accordingly, two types of hybridization patterns of vaccinia virus strains were identified (Fig. 2b and d). Nine of 13 vaccinia virus strains analyzed produced the “vaccinia virus-like” hybridization patterns, while the other 4 produced the “rabbitpox virus-like” patterns (Table 1).
Sixteen of 59 OPV DNA samples were hybridized in quadruplicate to check the reproducibility of the procedure. No false-negative or false-positive results were observed, and the registered deviations were insignificant.
To evaluate the sensitivity of the method, which was primarily determined by the sensitivity of PCR amplification, serial dilutions of vaccinia virus strain LIVP suspension were prepared. The virion concentration was determined by electron microscopy. PCR amplification was performed with DNA samples isolated from each of the serial viral dilutions. The amount of template DNA corresponding to 102 to 103 virions was needed to produce a visible specific amplicon (data not shown).
DNA samples extracted from smallpox scabs (Table 1) and a vaccinia virus strain LIVP pock on the CAM were used to test whether the proposed method can be applied for analysis of clinical specimens. In all cases, the sensitivity of the method was sufficient to identify OPV species accurately.
DISCUSSION
Although a number of PCR-based methods for OPV identification have been described (31, 33, 43), they all suffer from significant shortcomings. PCR analysis that relies solely on the lengths of the amplified products and their restriction fragments is limited to large-scale rearrangements of the viral genome and is unable to detect minor genome polymorphism, e.g., point nucleotide variations, short deletions, insertions, and inversions. The allele-specific PCR in solution can reveal point substitutions, but it requires several reactions to be run in parallel and therefore has limited clinical application.
The method described in the present study resolves most of these difficulties. Among the advantages of the analysis on the MAGIChip are its simplicity and the potential for further extension without technical limitations caused by an excessively high number of detecting oligonucleotides. At the same time, like other PCR-based methods, it dramatically decreases the biological hazard by avoiding large-scale growth and isolation of live virus and can be applied to the analysis of clinical specimens. Using human smallpox scabs collected from 1970 to 1975 and pocks formed by vaccinia virus on the CAM, we have demonstrated that the pathogen can be unambiguously detected and identified to the species level by MAGIChip analysis.
The proposed method introduces further simplification of the hybridization protocol developed in our laboratory (S. Bavykin, unpublished data): the relatively short length of the amplified fragment makes it suitable for immediate hybridization without any fragmentation, the use of a fluorescently labeled primer ensures a highly efficient incorporation of the label in the final PCR product, and the whole procedure can be performed in 6 h. The method is simple enough to be used for rapid OPV species identification in both pure viral cultures and clinical specimens.
Acknowledgments
We are grateful to I. Taran and S. Surzhikov for the synthesis of oligonucleotides; to V. Chupeeva and E. Kreindlin for manufacturing MAGIChips; to A. Chikova for technical assistance; to J. Esposito, H. Meyer, V. Petrov, G. McFadden, and M. Skinner for the samples of viral DNA; to S. Marennikova for OPV strains; to G. Kochneva for cultivation of vaccinia and cowpox viruses; to E. Ryabchikova for electron microscopy analysis of virus samples; and to E. Novikova, A. Krichevskaya, and A. Kolchinsky for editorial help.
This work was supported by ISTC grants 1516 and 1987.
REFERENCES
- 1.Anonymous. 1997. Human monkeypox in Kasai Oriental, Democratic Republic of the Congo (former Zaire). Wkly. Epidemiol. Rec. 72:369-372. [PubMed] [Google Scholar]
- 2.Ball, L. A. 1987. High-frequency homologous recombination in vaccinia virus DNA. J. Virol. 61:1788-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bavykin, S. G., J. P. Akowski, V. M. Zakhariev, V. E. Barsky, and A. D. Mirzabekov. 2001. Portable system for microbial sample preparation and oligonucleotide microarray analysis. Appl. Environ. Microbiol. 67:922-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buller, R. M. L., and G. J. Palumbo. 1992. Safety and attenuation of vaccinia virus, p. 235-267. In M. M. Binns and G. L. Smith (ed.), Recombinant poxviruses. CRC Press, Inc., Boca Raton, Fla.
- 5.Cameron, C., S. Hota-Mitchell, L. Chen, J. Barrett, J. X. Cao, C. Macaulay, D. Willer, D. Evans, and G. McFadden. 1999. The complete DNA sequence of myxoma virus. Virology 264:298-318. [DOI] [PubMed] [Google Scholar]
- 6.Cunnion, K. M. 1999. Tumor necrosis factor receptors encoded by poxviruses. Mol. Genet. Metab. 67:278-282. [DOI] [PubMed] [Google Scholar]
- 7.Czerny, C. P., A. M. Eis-Hubinger, A. Mayr, K. E. Schneweis, and B. Pfeiff. 1991. Animal poxviruses transmitted from cat to man: current event with lethal end. J. Vet. Med. B 38:421-431. [DOI] [PubMed] [Google Scholar]
- 8.Czerny, C. P., and H. Mahnel. 1990. Structural and functional analysis of orthopoxvirus epitopes with neutralizing monoclonal antibodies. J. Gen. Virol. 71:2341-2352. [DOI] [PubMed] [Google Scholar]
- 9.Dick, E. J., Jr., C. L. Kittell, H. Meyer, P. L. Farrar, S. L. Ropp, J. J. Esposito, R. M. Buller, H. Neubauer, Y. H. Kang, and A. E. McKee. 1996. Mousepox outbreak in a laboratory mouse colony. Lab. Anim. Sci. 46:602-611. [PubMed] [Google Scholar]
- 10.Downie, A. W. 1939. A study of the lesions produced experimentally by cowpox virus. J. Pathol. Bacteriol. 48:361-379. [Google Scholar]
- 11.Drobyshev, A., N. Mologina, V. Shick, D. Pobedimskaya, G. Yershov, and A. Mirzabekov. 1997. Sequence analysis by hybridization with oligonucleotide microchip: identification of beta-thalassemia mutations. Gene 188:45-52. [DOI] [PubMed] [Google Scholar]
- 12.Esposito, J., R. Condit, and J. Obijeski. 1981. The preparation of orthopoxvirus DNA. J. Virol. Methods 2:175-179. [DOI] [PubMed] [Google Scholar]
- 13.Esposito, J. J., and J. C. Knight. 1985. Orthopoxvirus DNA: a comparison of restriction profiles and maps. Virology 143:230-251. [DOI] [PubMed] [Google Scholar]
- 14.Fenner, F., D. A. Henderson, I. Arita, Z. Jezek, and I. D. Ladnyi. 1988. Smallpox and its eradication. World Health Organization, Geneva, Switzerland.
- 15.Fotin, A., A. Drobyshev, D. Proudnikov, A. Perov, and A. Mirzabekov. 1998. Parallel thermodynamic analysis of duplexes on oligodeoxyribonucleotide microchips. Nucleic Acids Res. 26:1515-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Res. Symp. Ser. 41:95-98. [Google Scholar]
- 17.Hu, F.-Q., Y. S. Chan, and D. J. Pickup. 1994. Cowpox virus contains two copies of an early gene encoding a soluble secreted form of the type II TNF receptor. Virology 204:343-356. [DOI] [PubMed] [Google Scholar]
- 18.Ibrahim, M. S., J. J. Esposito, P. B. Jahrling, and R. S. Lofts. 1997. The potential of 5′-nuclease PCR for detecting a single-base polymorphism in Orthopoxvirus. Mol. Cell. Probes 11:143-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kempe, C. H., and A. S. Beneson. 1955. Smallpox and vaccinia. Symposium on unusual infections of childhood. Pediatr. Clin. N. Am. 2:19-32. [DOI] [PubMed] [Google Scholar]
- 20.Krag, H., and M. Bentzon. 1963. The international reference preparation of smallpox vaccine: an international collaboration essay. Bull. W. H. O. 29:299-309. [PMC free article] [PubMed] [Google Scholar]
- 21.Loparev, V. N., R. F. Massung, J. J. Esposito, and H. Meyer. 2001. Detection and differentiation of Old World orthopoxviruses: restriction fragment length polymorphism of the crmB gene region. J. Clin. Microbiol. 39:94-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahnel, H. 1974. Laboratory differentiation of orthopox viruses. J. Vet. Med. B 21:242-258. (In German.) [PubMed]
- 23.Mahnel, H. 1983. Attenuation of mouse pox (ectromelia) virus. Zentbl. Veterinarmed. 30:701-707. [PubMed] [Google Scholar]
- 24.Marennikova, S. S., P. V. Gashnikov, O. A. Zhukova, E. I. Ryabchikova, V. V. Streltsov, O. I. Ryazankina, E. V. Chekunova, N. N. Yanova, and S. N. Shchelkunov. 1996. Biotype and genetic characteristics of the cowpox virus strain that caused illness in a child. J. Microbiol. 4:6-10. (In Russian.) [PubMed]
- 25.Marennikova, S. S., I. E. Ladnyj, Z. I. Ogorodnikova, E. M. Shelukhina, and N. N. Maltseva. 1978. Identification and study of a poxvirus isolated from wild rodents in Turkmenia. Arch. Virol. 56:7-14. [DOI] [PubMed] [Google Scholar]
- 26.Marennikova, S. S., and N. N. Maltseva. 1964. Comparative study on some vaccinia virus strains. Peculiarities of growth on chicken embryo, hemagglutinin activity and thermal resistance. Vopr. Virusol. 3:280-286. (In Russian.)
- 27.Marennikova, S. S., N. N. Maltseva, V. I. Korneeva, and N. M. Garanina. 1977. Outbreak of pox disease among Carnivora (Felidae) and Edentata. J. Infect. Dis. 135:358-366. [DOI] [PubMed] [Google Scholar]
- 28.Marennikova, S. S., F. G. Nagieva, G. R. Matsevich, E. M. Shelukhina, N. A. Khabahpasheva, and G. M. Platonova. 1988. Monoclonal antibodies to monkeypox virus: preparation and application. Acta Virol. 32:19-26. [PubMed] [Google Scholar]
- 29.Marennikova, S. S., and S. N. Shchelkunov. 1998. Orthopoxviruses pathogenic for humans. KMK Scientific Press Ltd., Moscow, Russia.
- 30.Marennikova, S. S., E. M. Shelukhina, N. N. Maltseva, K. L. Cimiskjan, and G. R. Matsevic. 1972. Isolation and properties of the causal agent of the new variola-like disease (monkeypox). Bull. W. H. O. 46:599-611. [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer, H., M. Pfeffer, and H.-J. Rziha. 1994. Sequence alterations within and downstream of the A-type inclusion protein genes allow differentiation of Orthopoxvirus species by polymerase chain reaction. J. Gen. Virol. 75:1975-1981. [DOI] [PubMed] [Google Scholar]
- 32.Mikhailovich, V., S. Lapa, D. Gryadunov, A. Sobolev, B. Strizhkov, N. Chernyh, O. Skotnikova, O. Irtuganova, A. Moroz, V. Litvinov, M. Vladimirskii, M. Perelman, L. Chernousova, V. Erokhin, A. Zasedatelev, and A. Mirzabekov. 2001. Identification of Mycobacterium tuberculosis strains resistant to rifampin by hybridization, PCR, and ligase detection reaction on oligonucleotide microchips. J. Clin. Microbiol. 39:2531-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukinda, V. B., G. Mweta, M. Kilundu, D. L. Heymann, A. S. Khan, J. J. Esposito, et al. 1997. Re-emergence of human monkeypox in Zaire in 1996. Lancet 349:1449-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osterrieder, N., H. Meyer, and M. Pfeffer. 1994. Characterization of the gene encoding for the A-type inclusion body protein of mousepox virus. Virus Genes 8:125-135. [DOI] [PubMed] [Google Scholar]
- 35.Petrov, V. S., V. V. Raspopin, V. N. Pak, I. H. Urmanov, N. B. Cherny, and A. G. Bachinsky. 1991. Study on the clonal variability in vaccinia virus strain L-IVP. Vopr. Virusol. 3:235-237. (In Russian.) [PubMed]
- 36.Pfeffer, M., H. Meyer, U. Wernery, and O.-R. Kaaden. 1996. Comparison of camelpox viruses isolated in Dubai. Vet. Microbiol. 49:135-146. [DOI] [PubMed] [Google Scholar]
- 37.Pickup, D. J., B. S. Ink, B. L. Parsons, W. Hu, and W. K. Joklik. 1984. Spontaneous deletions and duplications of sequences in the genome of cowpox virus. Proc. Natl. Acad. Sci. USA 81:6817-6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pilaski, J., A. Rosen, and G. Darai. 1986. Comparative analysis of the genomes of orthopoxviruses isolated from elephant, rhinoceros and okapi by restriction enzymes. Arch. Virol. 88:135-142. [DOI] [PubMed] [Google Scholar]
- 39.Pollitt, E., M. A. Skinner, and S. Heaphy. 1998. Nucleotide sequence of the 4.3 kbp BamHI-N fragment of fowlpox virus FP9. Virus Genes 17:5-9. [DOI] [PubMed] [Google Scholar]
- 40.Proudnikov, D., E. Timofeev, and A. D. Mirzabekov. 1998. Immobilization of DNA in polyacrylamide gel for the manufacture of DNA and DNA-oligonucleotide microchips. Anal. Biochem. 259:34-41. [DOI] [PubMed] [Google Scholar]
- 41.Ramyar, H., and M. Hessami. 1972. Isolation, cultivation and characterization of camel pox virus. Zentbl. Veterinarmed. 19:182-189. [DOI] [PubMed] [Google Scholar]
- 42.Rivers, T. M., S. M. Ward, and R. D. Baird. 1939. Amount and duration of immunity induced by intradermal inoculation of cultured vaccine virus. J. Exp. Med. 69:857-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ropp, S. L., Q. Jin, J. C. Knight, R. F. Massung, and J. J. Esposito. 1995. PCR strategy for identification and differentiation of smallpox and other orthopoxviruses. J. Clin. Microbiol. 33:2069-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryazankina, O. I., F. I. Muravlev, V. V. Gutorov, N. N. Mikrjukov, I. O. Cheshenko, and S. N. Shchelkunov. 1993. Comparative analysis of the conserved region of the orthopoxvirus genome encoding the 36K and 12K proteins. Virus Res. 29:281-303. [DOI] [PubMed] [Google Scholar]
- 45.Shchelkunov, S. N., V. M. Blinov, and L. S. Sandakhchiev. 1993. Genes of variola and vaccinia viruses necessary to overcome the host protective mechanisms. FEBS Lett. 319:80-83. [DOI] [PubMed] [Google Scholar]
- 46.Shchelkunov, S. N., S. M. Resenchuk, A. V. Totmenin, V. M. Blinov, S. S. Marennikova, and L. S. Sandakhchiev. 1993. Comparison of the genetic maps of variola and vaccinia viruses. FEBS Lett. 327:321-324. [DOI] [PubMed] [Google Scholar]
- 47.Shchelkunov, S. N., P. F. Safronov, A. V. Totmenin, N. A. Petrov, O. I. Ryazankina, V. V. Gutorov, and G. J. Kotwal. 1998. The genomic sequence analysis of the left and right species-specific terminal region of a cowpox virus strain reveals unique sequences and a cluster of intact ORFs for immunomodulatory and host range proteins. Virology 243:432-460. [DOI] [PubMed] [Google Scholar]
- 48.Shchelkunov, S. N., A. V. Totmenin, V. N. Loparev, P. F. Safronov, V. V. Gutorov, V. E. Chizhikov, J. C. Knight, J. M. Parsons, R. F. Massung, and J. J. Esposito. 2000. Alastrim smallpox variola minor virus genome DNA sequences. Virology 266:361-386. [DOI] [PubMed] [Google Scholar]
- 49.Smith, T. D., P. J. Smolak, D. Friend, H. Hagen, M. Gerhart, L. Park, D. J. Pickup, D. Torrance, K. Mohler, K. Schooley, and R. G. Goodwin. 1997. Poxvirus genomes encode a secreted, soluble protein that preferentially inhibits b-chemokine activity yet lacks sequence homology to known chemokine receptors. Virology 236:316-327. [DOI] [PubMed] [Google Scholar]
- 50.Timofeev, E., S. Kochetkova, A. Mirzabekov, and V. Florentiev. 1996. Regioselective immobilization of short oligonucleotides to acrylic copolymer gels. Nucleic Acids Res. 24:3142-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turner, P. C., and R. W. Moyer. 1990. The molecular pathogenesis of poxviruses. Curr. Top. Microbiol. Immunol. 163:125-151. [DOI] [PubMed] [Google Scholar]
- 52.Wienecke, R., H. Wolff, M. Schaller, H. Meyer, and G. Plewig. 2000. Cowpox virus infection in an 11-year-old girl. J. Am. Acad. Dermatol. 42:892-894. [DOI] [PubMed] [Google Scholar]
- 53.Yershov, G., V. Barsky, A. Belgovskiy, E. Kirillov, E. Kreindlin, I. Ivanov, S. Parinov, D. Guschin, A. Drobyshev, S. Dubiley, and A. Mirzabekov. 1996. DNA analysis and diagnostics on oligonucleotide microchips. Proc. Natl. Acad. Sci. USA 93:4913-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]