Abstract
A rapid quantitative real-time PCR method was employed to quantify the copy number of E2 and E6 genes for analysis of the physical status of human papillomavirus type 16 (HPV-16) DNA. Significant differences with respect to both copy numbers were found when more than 40% of HPV-16 DNA was integrated with disruption of the E2 gene in an experimental model. The physical status of HPV-16 DNA in 50 clinical samples was exclusively episomal in 21 cases (42%), concomitant in 14 cases (28%), and integrated in 15 cases (30%). The prevalence of integrated and/or concomitant forms of HPV-16 DNA increased with progression of cervical disease. Four of 11 cervical intraepithelial neoplasia involved integrated forms of HPV-16 DNA partially or exclusively. This rapid, sensitive technique is useful in the analysis of the physical status of HPV DNA.
Certain types of human papillomavirus (HPV) play a pivotal role in the carcinogenesis of cervical cancer; moreover, high-level expression of HPV E6 and E7 oncoproteins is required for progression of cancer cell replication (7, 9, 17, 21). Three alternative processes could increase the expression of these proteins. A high copy number of HPV DNA would directly contribute to increased expression of E6 and E7 (15). In fact, numerous cervical cancer cell lines harbor hundreds of copies of HPV DNA per cell. Furthermore, the amount of HPV-16 DNA increased by orders of magnitude with increasing disease grade (22). Secondly, mutations affecting YY1 motifs in the long control region can enhance the viral oncogene expression (5, 15, 16, 18). Finally, integration of HPV DNA into the cellular genome also leads to overexpression of E6 and E7 oncoproteins, since either the E1 or E2 open reading frame (ORF), the product of which represses activity from the promoter P97 for the E6 and E7 genes, is preferentially disrupted or deleted following integration (1, 4, 8, 20, 23).
The present study describes a rapid method of quantitative real-time PCR to quantify copy numbers of E2 and E6 genes for analysis of the physical status of HPV type 16 (HPV-16) DNA. This technique was developed based on the following assumptions: (i) preferential disruption of E2 genes will cause the absence of E2 gene sequences in the PCR product following integration; (ii) copy numbers of both genes should be equivalent in episomal forms; and (iii) E2 gene copy number will be smaller than that for E6 in concomitant forms.
MATERIALS AND METHODS
Specimens and DNA extraction.
Twenty-two invasive cervical carcinoma specimens analyzed in a prior study were evaluated (25). Additionally, cytobrush specimens were collected from outpatients with a confirmed histological diagnosis of CIN or invasive carcinoma at the Department of Obstetrics and Gynecology, Okayama University Medical School Hospital, Okayama, Japan. All specimens were screened for the presence of HPV DNA as described previously (14). Twenty-eight specimens positive for HPV-16 DNA were included. DNA was extracted from these samples via a routine procedure consisting of proteinase K digestion and phenol extraction. Approximately 100 ng of total DNA was obtained per μl of cytobrush specimens. Fifty samples, including 11 cervical intraepithelial neoplasia (CINs) (dysplasia or carcinoma in situ), 11 microinvasive cancers, and 28 invasive cancers, were analyzed by quantitative real-time PCR.
Quantitative real-time PCR.
All samples were tested for disruption of the HPV-16 E2 gene by PCR for the E2 gene (19). Subsequently, samples were subjected to quantitative real-time PCR. E2 and E6 genes of HPV-16 DNA were severally quantified with a GeneAmp 5700 Sequence Detection System (Applied Biosystems, Foster City, Calif.). The PCR mixture consisted of 1 μl of template DNA and 5.0 μl of 10× PCR buffer (supplied by the manufacturer), containing 15 mM MgCl2 (Perkin-Elmer Biosystems, Norwalk, Conn.), a 200 μM concentration of each deoxynucleoside triphosphate, 1.25 U of Mix Ampli Taq Gold (Perkin-Elmer Biosystems), a 1 μM concentration of each oligonucleotide primer, and 1.25 μl of 1:10,000 SYBR Green I (Molecular Probes, Eugene, Oreg.). Total volume was 50 μl.
Primer sequences were selected utilizing Primer Express software (version 1.0; Applied Biosystems). The primers for each sequence were 5′-TTATTAGGCAGCACTTGGCCA-3′ (nucleotides 3383 to 3403) and 5′-GTGAGGATTGGAGCACTGTCC-3′ (nucleotides 3540 to 3560) for the E2 gene and 5′-AAGGGCGTAACCGAAATCGT-3′ (nucleotides 26 to 46) and 5′-CATATACCTCACGTCGCAG-3′ (nucleotides 215 to 233) for the E6 gene. All oligodeoxynucleotides were synthesized on a model 394 DNA synthesizer (Applied Biosystems). Amplification was effected with initial denaturation for 10 min at 95°C, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 57°C for 30 s, and extension at 72°C for 1 min.
Standardization.
Full-length HPV-16 plasmids were grown and purified by standard methods. DNA solution was diluted with TE buffer (10 mM Tris-Cl, 1 mM EDTA [pH 8.0]) in adequate proportions, and DNA standards containing a series of 104 to 107 copies of HPV-16 genome were prepared. Linear plots of the log of the copy number versus number of cycles at which the fluorescent signals enter the log-linear phase were consistently obtained for both genes. Triplicate dilution series were included as standards in each run. HPV copy number in samples was evaluated using regression analysis. All assays were performed at least in duplicate.
Cutoff value determination.
Full-length HPV-16 plasmids contain equivalent numbers of E2 and E6 ORFs, whereas eucaryotic expression plasmids pSRαE6 (E6 plasmids) carry the entire E6 but not the E2 ORF (11). E6 plasmids were purified and DNA was mixed with full-length HPV-16 DNA in order to prepare a concomitant status of HPV-16 DNA. Full-length HPV-16 plasmid DNA was replaced by an equivalent copy number of E6 plasmid DNA in increments of 20%. Thus, DNA solutions representing episomal (full-length exclusively), integrated (E6 exclusively), and concomitant (20 to 80% integrated) forms of HPV-16 DNA were prepared. Quantification of copy number for E2 and E6 was conducted six times by real-time PCR assay. Differences in the copy number for E2 and E6 genes were determined by unpaired t test. Probability values of less than 0.01 were considered statistically significant.
2D gel electrophoresis.
Four specimens in which a mixed form of HPV-16 DNA was suggested by real-time PCR assay were subjected to two-dimensional (2D) gel electrophoresis. Total DNA (10 μg) was incubated with a no-cut enzyme, HindIII (New England Biolabs, Inc., Beverly, Mass.), and electrophoresed in a 0.4% agarose gel. The resulting lane was excised from the gel and recast within a 0.8% gel, followed by electrophoresis in the second dimension (6, 24). A 2D blot was prepared and hybridized with 32P-labeled HPV-16 as described previously (25).
RESULTS
Cutoff value determination.
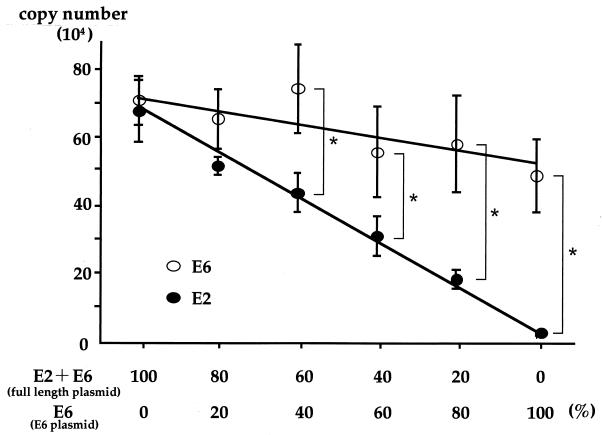
As the concentration of E6 plasmid increased, the copy number for E2 decreased in linear fashion; in contrast the copy number of the E6 gene was largely unchanged (Fig. 1). Statistical analysis demonstrated the existence of significant differences between copy numbers for E2 and E6 when sample DNA contained in excess of 40% E6 plasmid DNA in solution. This finding implies that real-time PCR can reliably distinguish the concomitant presence of episomal and integration forms from a pure episomal form when more than 40% HPV-16 DNA is integrated with disruption of the E2 gene. Hence, the cutoff value was set at 0.6 for the ratio of E2 to E6 copy numbers to distinguish the concomitant form from the pure episomal form.
FIG. 1.
Quantification of E2 and E6 copy number by real-time PCR assay for establishment of a cutoff value. E6 plasmid DNA was mixed with full-length HPV-16 DNA to prepare a concomitant status of HPV-16 DNA. These DNA solutions represent episomal (full-length exclusively), integrated (E6 exclusively), and concomitant (20 to 80% integrated) forms. Quantification of copy numbers for E2 and E6 was conducted six times by real-time PCR assay. Differences in the copy number for E2 and E6 genes were determined by unpaired t test. Error bars, standard deviations; ∗, P < 0.01.
Real-time PCR assay and Southern blot analysis.
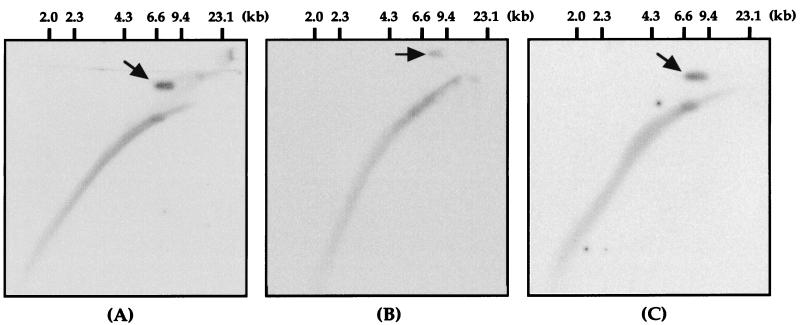
To validate the reliability of the cutoff value, quantification of copy number for E2 and E6 via real-time PCR assay was conducted for 22 invasive cancer specimens, in which confident detection of the physical status of HPV-16 DNA had been effected in a prior study (25). In 10 cases exhibiting integrated HPV-16 DNA, the E2 gene was not amplified by real-time PCR and completely concordant results were obtained (data not shown). It was shown that HPV-16 DNA existed in an episomal or concomitant form in the remaining 12 cases. Table 1 presents final results of the physical status in those cases investigated by four different molecular methods. Four of twelve cases, upon examination of the E2/E6 copy number ratio by real-time PCR assay, displayed values below 0.6; furthermore, mixed forms were suggested. Southern blot analysis and multiplex PCR revealed concomitant forms in two of these four samples. In the remaining cases, E2/E6 copy number ratios calculated from real-time PCR were 0.53 and 0.55 and discordant results were obtained. Subsequently, these four samples were subjected to 2D gel electrophoresis, which clearly demonstrated episomal forms as well as integrated HPV-16 DNA (Fig. 2). In the remaining eight specimens, the E2/E6 copy number ratio ranged from 0.61 to 1.13. Moreover, all testing methods were in complete agreement.
TABLE 1.
Comparison of physical status of HPV-16 DNA in invasive cervical cancers analyzed by four different molecular methods
| Sample no. | E2/E6 ratioa | Result of:
|
|
|---|---|---|---|
| Real-time PCR | Southern hybridization and multiplex PCR | ||
| 1 | 0.53 | Mixedb | Episomal |
| 2 | 1.00 | Episomal | Episomal |
| 3 | 0.41 | Mixedb | Mixed |
| 4 | 1.11 | Episomal | Episomal |
| 5 | 0.91 | Episomal | Episomal |
| 6 | 0.94 | Episomal | Episomal |
| 7 | 0.55 | Mixedb | Episomal |
| 8 | 0.77 | Episomal | Episomal |
| 9 | 0.61 | Episomal | Episomal |
| 10 | 0.51 | Mixedb | Mixed |
| 11 | 0.87 | Episomal | Episomal |
| 12 | 1.13 | Episomal | Episomal |
Obtained by comparison of copy numbers of genes.
Concomitant form of HPV-16 DNA was confirmed by two-dimensional gel electrophoresis.
FIG. 2.
2D gel electrophoresis demonstrating episomal forms as well as integrated HPV-16 DNA. Four specimens in which a mixed form of HPV-16 DNA was suggested by real-time PCR assay were subjected to 2D gel electrophoresis. Three representative photographs are shown here. The HPV-16 DNA in these specimens was present as episomal forms (arrow) as well as integrated HPV DNA. The hybridization signal in the lower arc encompasses the entire distribution of human DNA (data not shown). Molecular size standards are shown above each panel. (A) Sample 1; (B) sample 3; (C) sample 7.
Real-time PCR assay for clinical specimens with cervical lesions.
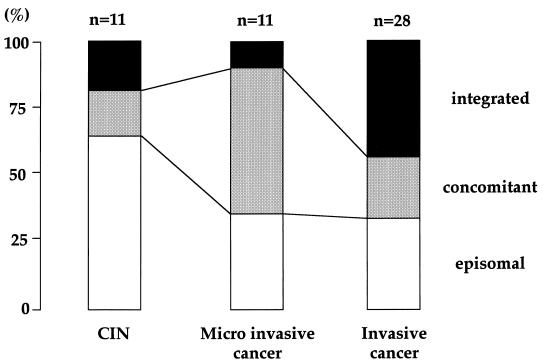
A specific fragment of the E2 gene was not amplified in 15 of 50 cases (30%), suggesting integration of HPV DNA into the host genome. Specimens with intact E2 genes were subjected to real-time PCR assay, and the presence of concomitant forms was considered in cases displaying E2/E6 copy number ratios below 0.6. Figure 3 illustrates the prevalence of the physical status of HPV-16 DNA in dysplastic and neoplastic cervical lesions. The physical status of HPV-16 DNA was exclusively episomal in 21 cases (42%), concomitant in 14 cases (28%), and integrated in 15 cases (30%), respectively. Surprisingly, 4 of 11 CINs (dysplasia or carcinoma in situ) involved integrated forms of HPV-16 DNA partially or exclusively. The prevalence of integrated and/or concomitant forms of HPV-16 DNA increased with progression of cervical disease.
FIG. 3.
Prevalence of the physical status of HPV-16 DNA in cervical lesions. The segments (open, stippled, and black) represent ratios of specimens harboring HPV-16 DNA in pure episomal, concomitant, and pure integrated forms, respectively, according to histopathological diagnosis.
DISCUSSION
The prevalence of “high-risk” HPV, such as HPV-16, -18, and -31, increases with the progression of CIN grade. However, it is possible that viral genomes maintained as episomes would be expelled within several months in most HPV infections, and the vast majority of grade I CINs regress spontaneously (10). Integration of HPV DNA with destruction of the E2 gene is an important mechanism with respect to evaluation of expression of E6 and E7 proteins, since the E2 gene product can repress activities from the HPV promoters that direct expression of these genes (1, 17, 20). In fact, integration of HPV DNA, regardless of type, rarely occurs in grade I CIN in cases where invasive cancers contain integrated HPV DNA (2, 3, 6).
Thus, accurate detection of the physical status of HPV DNA is extremely important for greater understanding of the mechanisms mediating cervical carcinogenesis and cancer progression. A number of Southern blots and, on occasion, 2D gel electrophoresis are essential in order to analyze the physical status of HPV. These procedures require large quantities of high-molecular-weight DNA, which is an obstacle when working with minute intraepithelial lesions. In contrast, a simple PCR technique to amplify the E2 gene can detect integration of HPV DNA even in instances in which the amount of sample DNA is very small (3). However, this method faces limitations related to distinction of a pure episomal form from mixed forms of HPV DNA due to retention of the E2 sequence in both cases.
In a prior study, we developed a new method to solve this problem (25). A cutoff value of the relative ratio of HPV E2 to E6 PCR products (E2/E6 ratio) was determined to discriminate between pure episomal and mixed forms. However, E2/E6 ratios varied extensively depending on viral DNA load, so much so that it was impossible to establish a cutoff value (data not shown). Then, we developed a rapid method of quantitative real-time PCR to quantify the copy number of E2 and E6 genes for analysis of the physical status of HPV-DNA.
This technique can reliably detect concomitant forms with 40% integration accompanied by destruction of target E2 sequences. Southern hybridization may have limitations regarding display of faint signals when the amount of HPV DNA integrated into a random site of the host genome is very small. In fact, our method suggested concomitant form of HPV DNA in two cases that had been misinterpreted by Southern blot hybridization. These results indicated that real-time PCR assay is more sensitive than Southern blot hybridization in the detection of small clusters of integrated HPV DNA.
Disruption outside the E2 ORF is possible; moreover, the target sequences in this real-time PCR assay are extremely small (177 bp in the entire E2 ORF of 1,097 bp). Although the small size of target sequences is preferred for sensitive quantitation by the real-time PCR approach, it will be possible to design several PCR targets contiguously to cover the entire E2 ORF. Several valid primer pairs have been discovered in our laboratory; however, these studies have not been concluded (data not shown). Therefore, the present assay could lead to false results through the misinterpretation of concomitant forms as episomal forms. However, this possibility is not high, as most integration with destruction of the E2 gene involves deletion of a large portion of early genes (12).
It is widely believed that integrated sequences rarely occur in CIN (2, 3). We postulate that concomitant forms are likely to be underestimated, as conventional blotting approaches are not applicable to most minute CIN lesions; moreover, concomitant status cannot be detected in some cases. The present investigation, dealing with a relatively small sample size, suggests that some clusters of episomal viral genomes begin to integrate into the host genomes in dysplastic lesion and that this event is an important mechanism of cervical carcinogenesis.
High prevalence of integration in CIN was also detected by a protocol involving amplification of papillomavirus oncogene transcripts (13). This method is able to distinguish episome- from integrate-derived HPV mRNAs. We postulate that the survey including HPV-18-positive samples might produce a slightly lower prevalence of integration as a result of distinct integration profiles of HPV-16 and -18. The majority of HPV-18-containing cancers involve exclusively integrated HPV DNA (2, 25). The amplification of papillomavirus oncogene transcripts assay is a useful technique based on RT-PCR for analysis of the physical status of HPV DNA. However, it is sometimes difficult to obtain high-quality RNAs from clinical samples. In contrast, our methods require a small quantity of DNA. Additionally, the protocol is rapid and quite simple.
In conclusion, quantitative real-time PCR can aid researchers in the analysis of the physical status of HPV DNA. Moreover, this protocol can be applied to the determination of the natural history of HPV as well as the mechanism of cervical carcinogenesis.
Acknowledgments
This work was supported in part by grants-in-aid 13671720 from the Ministry of Education, Sport, Science, Culture, and Technology of Japan.
We thank A. Dusso, Renal Division, Washington University School of Medicine, for assistance in the preparation of the manuscript. We also thank the staff of the 2nd Department of Surgery, Okayama University Medical School, for the use of laboratory equipment.
REFERENCES
- 1.Bernard, B. A., C. Bailly, M. C. Lenoir, M. Darmon, F. Thierry, and M. Yaniv. 1989. The human papillomavirus type 18 (HPV18) E2 gene product is a repressor of the HPV 18 regulatory region in human keratinocytes. J. Virol. 66:4317-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cullen, A. P., R. Reid, M. Campion, and A. T. Lorincz. 1991. Analysis of the physical state of different human papillomavirus DNAs in intraepithelial and invasive cervical neoplasms. J. Virol. 65:606-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Das, B. C., J. K. Sharma, V. Gopalakrishna, and U. K. Luthra. 1992. papillomavirus type 16 DNA in cervical preneoplastic and neoplastic lesions. J. Gen. Virol. 73:2327-2336. [DOI] [PubMed] [Google Scholar]
- 4.Demeret, C., C. Desaintes, M. Yaniv, and F. Thierry. 1997. Different mechanisms contribute to the E2-mediated transcriptional repression of human papillomavirus type 18 viral oncogenes. J. Virol. 71:9343-9349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong, X.-P., F. Stubenrauch, E. Beyer-Finkler, and H. Pfister. 1994. Prevalence of deletions of YY1-binding sites in episomal HPV 16 DNA from cervical cancers. Int. J. Cancer 53:803-808. [DOI] [PubMed] [Google Scholar]
- 6.Dürst, M., A. Kleinheinz, M. Hotz, and L. Gismann. 1985. The physical state of human papillomavirus type 16 DNA in benign and malignant genital tumors. J. Gen. Virol. 66:1515-1522. [DOI] [PubMed] [Google Scholar]
- 7.Dyson, N., P. M. Howley, K. Munger, and E. Harlow. 1989. The human papilloma virus: 16 E7 oncoprotein is able to the retinoblastoma gene product. Science 243:934-937. [DOI] [PubMed] [Google Scholar]
- 8.Francis, D. A., S. I. Schmid, and P. M. Howley. 2000. Repression of the integrated papillomavirus E6/E7 promoter is required for growth suppression of cervical cancer cells. J. Virol. 74:2679-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hausen, H. 1996. Papillomavirus infections: a major cause of human cancers. Biochim. Biophys. Acta 1228:F55-F78. [DOI] [PubMed] [Google Scholar]
- 10.Ho, G. Y., R. Bierman, L. Beardsley, C. J. Chang, and R. D. Burk. 1998. Natural history of cervicovaginal papillomavirus infection in young women. N. Engl. J. Med. 338:423-428. [DOI] [PubMed] [Google Scholar]
- 11.Kanda, T., S. Watanabe, S. Zanma, H. Sato., A. Furuno, and K. Yoshiike. 1991. Human papillomavirus type 16 E6 protein with glycine substitution for cysteine in the metal-binding motif. Virology 185:536-543. [DOI] [PubMed] [Google Scholar]
- 12.Kitagawa, K., H. Yoshikawa, T. Onda, T. Kawana, Y. Taketani, H. Yoshikawa, and A. Iwamoto. 1996. Genomic organization of human papillomavirus type 18 in cervical cancer specimens. Jpn. J. Cancer Res. 87:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klaes, R., S. M. Woerner, R. Ridder, N. Wentzensen, M. Duerst, A. Schneider, B. Lotz, P. Melsheimer, and M. K. Doeberitz. 1999. Detection of high-risk cervical intraepithelial neoplasia and cervical cancer by amplification of transcripts derived from integrated papillomavirus oncogenes. Cancer Res. 59:6132-6136. [PubMed] [Google Scholar]
- 14.Kobayashi, Y., M. Yoshinouchi, G. Tianqi, K. Nakamura, A. Hongo, S. Kamimura, Y. Mizutani, J. Kodama, Y. Miyagi, and T. Kudo. 1998. Presence of human papilloma virus DNA in pelvic lymph nodes can predict unexpected recurrence of cervical cancer in patients with histologically negative lymph nodes. Clin. Cancer Res. 4:979-983. [PubMed]
- 15.Kozuka, T., Y. Aoki, K. Nakagawa, K. Ohtomo, H. Yoshikawa, K. Matsumoto, K. Yoshiike, and T. Kanda. 2000. Enhancer-promoter activity of human papillomavirus type16 long control regions isolated from cell lines SiHa and CaSki and cervical cancer biopsies. Jpn. J. Cancer Res. 91:271-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laimins, L. A. 1998. Regulation of transcription and replication by human papillomaviruses. p. 201-223. In D. J. McCance (ed.), Human tumor viruses. American Society for Microbiology, Washington, D.C.
- 17.Lorincz, A. T., R. Reid, A. Bennett, M. D. Greenberg, W. Lanster, and R. J. Kurman. 1992. Human papillomavirus infection of the cervix: relative risk associations of 15 common anogenital types. Obstet. Gynecol. 79:328-337. [DOI] [PubMed] [Google Scholar]
- 18.May, M., X-P. Dong., E. Beyer-Finkler, F. Stubenrauch, P. G. Funchs, and H. Pfister. 1994. The E6/E7 promoter of extrachromosomal HPV 16 DNA in cervical cancers escapes from cellular repression by mutation of target sequences for YY-1. EMBO J. 13:1460-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park, J. S., E. S. Hwang, S. N. Park, H. K. Ahn, S. J. Um, C. J. Kim, and S. J. Kim. 1997. Physical status and expression of HPV genes in cervical cancers. Gynecol. Oncol. 65:121-129. [DOI] [PubMed] [Google Scholar]
- 20.Romanczuk, H., F. Thierry, and P. M. Howley. 1990. Mutational analysis of cis elements involved in E2 modulation of human papillomavirus type 16 P97 and type 18 P105 promoters. J. Virol. 64:2849-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75:495-505. [DOI] [PubMed] [Google Scholar]
- 22.Swan, D. C., R. A. Tucker, G. Tortolero-Luna, M. F. Mitchell, L. Wideroff, E. R. Unger, R. A. Nisenbaum, W. C. Reeves, and J. P. Icenogle. 1999. Human papillomavirus (HPV) DNA copy number is dependent on grade of cervical disease and HPV type. J. Clin. Microbiol. 37:1030-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thierry, F., and P. M. Howley. 1991. Functional analysis of E2 mediated repression of the HPV 18 P105 promoter. New Biol. 3:90-100. [PubMed] [Google Scholar]
- 24.Wettstein, F. O., and J. G. Stevens. 1982. Variable-sized free episomes of Shope papilloma virus DNA are present in all non-virus producing neoplasms and integrated episomes are detected in some. Proc. Natl. Acad. Sci. USA 79:790-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshinouchi, M., A. Hongo, K. Nakamura, J. Kodama, S. Itoh, H. Sakai, and T. Kudo. 1999. Analysis by multiplex PCR of the physical status of human papillomavirus type 16 DNA in cervical cancers. J. Clin. Microbiol. 37:3514-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]