Abstract
To investigate the distribution of staphylococcal enterotoxin (SE) A to I (SEA to SEI) genes (sea to sei) in Staphylococcus aureus, 146 isolates obtained in Japan from humans involved in and samples from food poisoning outbreaks, healthy humans, cows with mastitis, and bovine raw milk were analyzed by multiplex PCR. One hundred thirteen (77.4%) S. aureus isolates were found to be positive for one or more se genes. The se genotype was classified into 14 genotypes. seg and sei coexisted in the same S. aureus strain. The newly developed sandwich enzyme-linked immunosorbent assay showed that most seh-harboring S. aureus isolates were able to produce a significant amount of SEH. However, most of the S. aureus isolates harboring seg and about 60% of the isolates harboring sei did not produce a detectable level of SEG or SEI, while reverse transcription-PCR analysis proved that the mRNAs of SEG and SEI were transcribed in S. aureus strains harboring seg and sei genes. These results suggest the importance of quantitative assessment of SEG and SEI production in foods in order to clarify the relationship between these new SEs and food poisoning.
Staphylococcal enterotoxins (SEs) are emetic toxins and are one cause of food poisoning in humans. SEs have been classified as members of the pyrogenic toxin superantigen family because of their biological activities and structural relatedness (1, 6). SEs have been divided into five serological types (SEA through SEE) on the basis of their antigenicities (3). In recent years, the existence of new types of SEs (SEG, SEH, SEI, SEJ, SEK, SEL, SEM, SEN, and SEO) has been reported (11, 15, 16, 18, 22, 25). However, the relationship between these new SEs and human food poisoning is not fully understood at present. It is known that about 95% of staphylococcal food poisoning outbreaks are caused by SE types SEA to SEE (3). The remaining 5% of outbreaks may therefore be associated with other newly identified SEs. To clarify the role played by these newly identified SEs in food poisoning, the development of reliable methods of detection of SE proteins is essential. Several reports have described the development of a multiplex PCR for the detection of se genes (2, 10, 12, 13, 14). However, it is noteworthy that the PCR is only able to demonstrate the existence of se genes in Staphylococcus aureus isolates and does not prove that the production of SE proteins occurs. To demonstrate the capability of a strain to produce an amount of SE protein that is sufficient to induce disease, bioassay or immunological methods for the detection of SE protein must be developed. In this study, we used multiplex PCR to analyze the distributions of the sea to sei genes in 146 S. aureus isolates that had been obtained from humans involved in food poisoning outbreaks, healthy humans, cows with mastitis, and bovine raw milk in Japan. Furthermore, the production of SEG, SEH, and SEI by S. aureus isolates harboring the seg, seh, or sei genes was assessed by a newly developed sandwich enzyme-linked immunosorbent assay (ELISA).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids that were used to establish the multiplex PCR and expression constructs for recombinant SEG (rSEG), rSEH, and rSEI are listed in Table 1. The genotypes of the S. aureus type strains shown in Table 1 were confirmed by Southern blot analysis (19) with cloned probes specific for sea to sei and the ECL Direct Nucleic Acid Labeling and Detection system (Amersham Pharmacia Biotech Inc., Piscataway, N.J.). In all, 146 S. aureus isolates (71 isolates from humans involved in 25 outbreaks of food poisoning, 18 isolates from healthy humans, 21 isolates from cows with mastitis, and 36 isolates from bovine raw milk isolated in Japan were analyzed by multiplex PCR. S. aureus cultures were grown in Trypticase soy broth (Nissui, Tokyo, Japan) at 37°C for 18 h with aeration for genomic DNA preparation. S. aureus cultures for SE production were grown in brain heart infusion broth (Difco Laboratories, Detroit, Mich.) supplemented with 1% yeast extract (Difco) at 37°C for 48 h with aeration (4).
TABLE 1.
Bacterial strains and plasmids used in the study
| Strain or plasmid | Genotype | Origin |
|---|---|---|
| S. aureus | ||
| Type strains | ||
| FRI722 | sea sed | M. S. Bergdoll |
| S6 | sea seb | M. S. Bergdoll |
| FRI361 | sec2 sed seg sei | M. S. Bergdoll |
| 1151-7NG | sed | M. S. Bergdoll |
| FRI326 | see | M. S. Bergdoll |
| FRI569 | seh | Y.-C. Su |
| Isolates | ||
| Nagasaki 1 | sea seb seh | Food poisoning outbreak |
| Fukuoka 2 | seg sei | Food poisoning outbreak |
| E. coli strains | ||
| DH5α | Toyobo | |
| KGX1 | Aprseg, carrying pKGX1 | This work |
| KHX1 | Aprseh, carrying pKHX1 | This work |
| KIX1 | Aprsei, carrying pKIX1 | This work |
| Plasmids | ||
| pGEM3Zf (+) | Apr | Promega |
| pGEX-6P-1 | Apr | Pharmacia |
| pKOG4 | Apr, pGEM3Zf(+) with seg | This work |
| pKOH1 | Apr, pGEM3Zf(+) with seh | This work |
| pKOI6 | Apr, pGEM3Zf(+) with sei | This work |
| pKGX1 | Apr, pGEX-6P-1 with seg | This work |
| pKHX1 | Apr, pGEX-6P-1 with seh | This work |
| pKIX1 | Apr, pGEX-6P-1 with sei | This work |
Chemicals.
Chemicals were obtained from Nakalai Co. Ltd. (Kyoto, Japan), Takara Syuzo Co. Ltd. (Kyoto, Japan), Sigma Chemical Co. (St. Louis, Mo.), and Amersham Pharmacia Biotech Inc.
DNA preparations.
Genomic DNA of S. aureus was purified by the method of Betley and Mekalanos (5) or with the QIAamp DNA purification kit (Qiagen GmbH, Hilden, Germany). Escherichia coli plasmid DNA was purified by the alkaline lysis procedure used by the kit from Qiagen GmbH or the GFX plasmid purification kit (Amersham Pharmacia Biotech Inc.).
PCR.
The nucleotide sequences of all PCR primers and their respective amplified products are listed in Table 2. The primers used to detect sea to see were those described by Becker et al. (2). The primers used to detect seg, seh, and sei and those used for the construction of expression plasmids for rSEG, rSEH, and rSEI were designed according to the published nucleotide sequences of seg, seh, and sei, respectively (15, 18). The amplification was performed in an automated thermal cycler with a hot bonnet (Takara PCR Thermal Cycler MP) and by using TaKaRa EX Taq DNA polymerase (Takara Syuzo Co.). The reaction mixture (20 μl) for the multiplex PCR contained each primer at a concentration of 0.4 μM; 2 mM MgCl2; dGTP, dATP, dTTP, and dCTP (Takara Syuzo Co.) at a concentration of 200 μM each; 0.5 U of Taq polymerase; and 2 μl of 10× buffer (Takara Syuzo Co.). Thermal cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 60 s were repeated 30 times. For the purpose of cloning, TaKaRa Pyrobest polymerase was used under the same conditions. Multiplex PCR was performed with the sea to see primer set and the seg to sei primer set. The specificity and sensitivity of each multiplex PCR were confirmed with genomic DNA purified from the S. aureus type strains or the isolates listed in Table 1.
TABLE 2.
Nucleotide sequences and predicted sizes of PCR products for SE-specific oligonucleotide primers
| Primer set and gene | Primer | Oligonucleotide sequence (5′-3′) | Size (bp) of PCR product | Reference |
|---|---|---|---|---|
| Primer for se gene detection | ||||
| sea | SEA-3 | CCTTTGGAAACGGTTAAAACG | 127 | Becker et al. (2) |
| SEA-4 | TCTGAACCTTCCCATCAAAAAC | |||
| seb | SEB-1 | TCGCATCAAACTGACAAACG | 477 | Becker et al. (2) |
| SEB-4 | GCAGGTACTCTATAAGTGCCTGC | |||
| sec | SEC-3 | CTCAAGAACTAGACATAAAAGCTAGG | 271 | Becker et al. (2) |
| SEC-4 | TCAAAATCGGATTAACATTATCC | |||
| sed | SED-3 | CTAGTTTGGTAATATCTCCTTTAAACG | 319 | Becker et al. (2) |
| SED-4 | TTAATGCTATATCTTATAGGGTAAACATC | |||
| see | SEE-3 | CAGTACCTATAGATAAAGTTAAAACAAGC | 178 | Becker et al. (2) |
| SEE-2 | TAACTTACCGTGGACCCTTC | |||
| seg | SEG-1 | AAGTAGACATTTTTGGCGTTCC | 287 | This study |
| SEG-2 | AGAACCATCAAACTCGTATAGC | |||
| seh | SEH-1 | GTCTATATGGAGGTACAACACT | 213 | This study |
| SEH-2 | GACCTTTACTTATTTCGCTGTC | |||
| sei | SEI-1 | GGTGATATTGGTGTAGGTAAC | 454 | This study |
| SEI-2 | ATCCATATTCTTTGCCTTTACCAG | |||
| Primer for SE protein expression | ||||
| SEG | SEGF1 | CCCCGGATCCCAACCCGATCCTAAATTAGACGAAC | 722 | This study |
| SEGR1 | CCCCGAATTCTCAGTGAGTATTAAGAAATACTTCC | |||
| SEH | SEHF1 | CCCCGGATCCGAAGATTTACACGATAAAAGTGAGTT | 676 | This study |
| SEHR1 | CCCCGAATTCGATTATACTTTTTTCTTAGTATATAG | |||
| SEI | SEIF1 | CCCCGGATCCCAAGGTGATATTGGTGTAGGTAACT | 677 | This study |
| SEIR1 | CCCCGAATTCTTAGTTACTATCTACATATGATATTTCG |
Cloning of seg, seh, and sei and nucleotide sequencing.
S. aureus isolates Fukuoka 2 (seg sei) and Nagasaki 1 (sea seb seh) were selected as the sources for se gene cloning. DNA fragments amplified from the genomic DNA of Fukuoka 2 or Nagasaki 1 with the primers shown in Table 2 were cloned into plasmid vector pGEM3Zf(+). The nucleotide sequences of both strands of three to four independent clones were determined with the following automatic DNA sequencers: ABI 373A (Perkin-Elmer Applied Biosystems, Foster City, Calif.) and DSQ-1000L (Shimazu, Tokyo, Japan). The nucleotide sequences of the seg and seh clones were completely identical to the published seg and seh sequences (15, 18). However, although the nucleotide sequences of the sei clones were almost identical to the published sei sequence (15), the clones had three substitutions, at positions 156 (C to T), 308 (A to T), and 461 (A to G). The substitution at position 308 (A to T) was nonsynonymous, and the deduced amino acid sequence of the cloned mature SEI protein contained a substitution at position 79 (Y to F). The plasmids containing seg, seh, and sei were named pKOG4, pKOH1, and pKOI6, respectively.
Expression of rSEG, rSEH, or rSEI in a GST fusion system.
To construct the rSEG, rSEH, and rSEI expression plasmids, the DNA fragments that code for the mature forms of SEG, SEH, and SEI were digested from pKOG4, pKOH1, and pKOI6, respectively, with BamHI and EcoRI. The fragments were subcloned into the pGEX-6P-1 (Amersham Pharmacia Biotech Inc.) glutathione S-transferase (GST) fusion expression vector and then transformed into E. coli DH5α cells. The resultant plasmids containing seg, seh, and sei were named pKGX1, pKHX1, and pKIX1, respectively. Expression, purification of GST-fused rSEs, cleavage, and removal of the GST tag from rSEs were performed according to the manufacturer's instruction. The resulting mature rSEG, rSEH, and rSEI proteins have five additional amino acid residues, GPLGS, at the N terminus.
Preparation of anti-rSEG, anti-rSEH, and anti-rSEI sera.
Anti-rSEG, anti-rSEH, and anti-rSEI sera were prepared by immunizing rabbits with purified rSEG, rSEH, or rSEI, as described by Shinagawa et al. (20). The titers of the antisera were monitored by gel double diffusion and Western blotting.
Antibody preparation.
Rabbit anti-SEG, anti-SEH, and anti-SEI immunoglobulin G antibodies were purified from hyperimmune sera with an immobilized protein G-Sepharose column (Amersham Pharmacia Biotech Inc.). Monospecific rabbit anti-SEG, anti-SEH, and anti-SEI antibodies were affinity purified from hyperimmune sera with an SEG-, SEH-, or SEI-coupled Sepharose column. One milligram of monospecific antibodies was conjugated to EZ-link plus horseradish peroxidase (Pierce, Rockford, Ill.), according to the manufacturer's instructions.
Sandwich ELISA.
Culture supernatants from the growth of S. aureus isolates were preincubated with 20% (vol/vol) normal rabbit serum at 4°C overnight and were then diluted 10- to 100-fold in phosphate-buffered saline-Tween 20 (0.01 M phosphate buffer and 0.15 M NaCl containing 0.05% Tween 20) to avoid any nonspecific reaction caused by protein A (8). ELISAs were performed in 96-well microplates (Nalge Nunc International, Rochester, N.Y.) by the method of Freed et al. (8), with a slight modification. As a substrate solution, 0.4 mg of ortho-phenylenediamine hydrochloride per ml in 0.1 M citrate buffer (pH 5.0) was used, and the absorbance at 492 nm was read with a 550 Microplate Reader (Bio-Rad, Richmond, Calif.). The concentration of each toxin in the culture supernatants was determined by converting the absorbance values to the corresponding concentrations by use of the standard curve.
RT-PCR.
Total RNA was extracted from S. aureus cultures by using an RNeasy spin column (Qiagen). Purified total RNA was treated with DNase I to degrade contaminating genomic DNA. cDNA was synthesized with SuperScript II reverse transcriptase (GIBCO-BRL, Grand Island, N.Y.) by incubating 100 ng of total RNA with a random primer (GIBCO-BRL). As a control for genomic DNA contamination, total RNA was also subjected to reverse transcription (RT)-PCR but without the RT step. The seg and sei cDNAs were detected with the same primer sets shown in Table 2 and by the same multiplex PCR performed as described above.
Nucleotide sequence accession numbers.
The nucleotide sequences of pKOG4, pKOH1, and pKOI6 were submitted to the GenBank/EMBL/DDBJ database and have been assigned accession numbers AB060535, AB060536, and AB060537, respectively.
RESULTS
Distributions of sea to sei genes in S. aureus isolates from various sources.
The results of the multiplex PCR analysis for all 146 S. aureus isolates are shown in Table 3. Overall, 113 (77.4%) S. aureus isolates were found to be positive for one or more se genes. The se genotype was classified into 14 genotypes. Fifteen S. aureus isolates possessed only one kind of se gene, and the remaining 98 isolates harbored more than one se gene.
TABLE 3.
se gene distribution among S. aureus isolates
| Genotype | No. (%) of strains from:
|
||||
|---|---|---|---|---|---|
| Humans with food poisoning | Healthy humans | Cows with mastitis | Raw milk | Total | |
| Total | 71 (100) | 18 (100) | 21 (100) | 36 (100) | 146 (100) |
| se negative | 5 (7.0) | 5 (27.8) | 6 (28.6) | 17 (47.2) | 33 (22.6) |
| se positive | 66 (93.0) | 13 (72.2) | 15 (71.4) | 19 (52.8) | 113 (77.4) |
| sea | 4 (5.6) | 4 (2.7) | |||
| sea seb | 2 (2.8) | 2 (1.4) | |||
| sea sed | 2 (2.8) | 2 (1.4) | |||
| sea seg | 2 (2.8) | 2 (1.4) | |||
| sea seh | 6 (8.5) | 1 (5.6) | 7 (4.8) | ||
| sea seb seh | 18 (25.4) | 18 (12.3) | |||
| seb | 3 (4.2) | 1 (5.6) | 1 (2.8) | 5 (3.4) | |
| seb seh | 15 (21.1) | 15 (10.3) | |||
| seb seg sei | 2 (2.8) | 1 (5.6) | 1 (2.8) | 4 (2.7) | |
| sec seg sei | 1 (5.6) | 8 (38.1) | 1 (2.8) | 10 (6.8) | |
| sed seg sei | 3 (16.7) | 3 (2.0) | |||
| seg | 2 (11.1) | 2 (1.4) | |||
| seg sei | 12 (16.9) | 2 (11.1) | 7 (33.3) | 14 (38.9) | 35 (24.0) |
| seh | 2 (11.1) | 2 (5.6) | 4 (2.7) | ||
Among the 71 isolates that originated from individuals and samples with food poisoning, 66 (93.0%) were found to be positive for se genes and 54 (76.1%) possessed the sea, seb, sec, or sed gene. Overall, 10 se genotypes were observed. Thirty-four (47.9%) isolates had the sea gene, and the majority of these isolates possessed other se genes: 2 had seb, 2 had sed, 2 had seg, 6 had seh, and 18 had seb and seh. Isolates that harbored seb and seh were also commonly detected. Twelve (16.9%) isolates from individuals involved in three food poisoning outbreaks possessed only seg and sei genes, and in five (7.0%) isolates no se genes were detected by multiplex PCR.
Among the 18 isolates from healthy humans, 13 (72.2%) were found to be se positive. A total of eight genotypes were observed, and 12 isolates had the seg, seh, or sei genes.
Among the 21 isolates from cows with mastitis, 15 were positive for se genes, and only two genotypes (8 isolates with the sec seg sei genotype and 7 isolates with the seg sei genotype) were observed. On the other hand, 19 of 36 (52.8%) isolates from bovine raw milk were found to be se positive, and five se genotypes were observed among those isolates. The most commonly detected se genotype was seg sei (38.9%).
Development of sandwich ELISA for detection of SEG, SEH, and SEI.
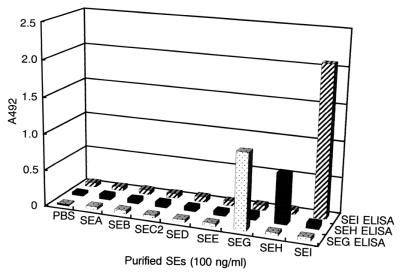
In order to develop immunological methods for the detection of SEG, SEH, and SEI, rSEG, rSEH, and rSEI were expressed in E. coli and were used to immunize rabbits. A sandwich ELISA for the detection of SEG, SEH, and SEI was developed with purified polyclonal antibodies from these antisera. Figure 1 shows the specificities of the sandwich ELISAs for each purified SE. One hundred nanograms each of purified SEA, SEB, SEC, SED, SEE, rSEG, rSEH, and rSEI was subjected to each sandwich ELISA; and no cross-reactivity was observed. The minimum detectable concentrations of SEG, SEH, and SEI by the sandwich ELISA were 0.5, 1.0, and 1.0 ng/ml, respectively. Straight lines were observed at SEG, SEH, and SEI concentrations between 0.5 and 10, 1.0 and 10, and 1.0 and 20 ng/ml, respectively.
FIG. 1.
Specificities of sandwich ELISA systems for detection of SEG, SEH, and SEI. SEA, SEB, SEC2, SED, and SEE (100 ng/ml each) were purified from the culture supernatant of each strain by the method of Shinagawa et al. (21). rSEG, rSEH, and rSEI were each subjected to sandwich ELISA.
Enterotoxin productivity of S. aureus isolates harboring seg, seh, or sei genes.
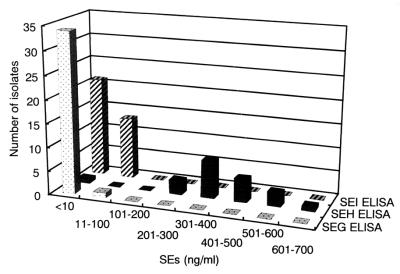
S. aureus isolates harboring seg (n = 35), seh (n = 21), or sei (n = 32) genes were cultured in brain heart infusion broth supplemented with 1% yeast extract, and the culture supernatants were subjected to sandwich ELISA. Among the isolates harboring the seh gene, 17 isolates were from individuals involved in and samples from food poisoning outbreaks, 2 isolates were from healthy humans, and 2 isolates were from bovine raw milk. Among the isolates harboring the seg gene, 15 isolates were from individuals involved in and samples from food poisoning outbreaks, 9 isolates were from healthy humans, 4 isolates were from cows with mastitis, and 7 isolates were from raw milk. Because of the coexistence of the seg and sei genes in most of the isolates, the sei-harboring isolates are included with the seg-harboring isolates with three exceptions: one isolate from a food poisoning outbreak and two isolates from healthy humans. Figure 2 shows the frequency distribution of SEG, SEH, and SEI production by S. aureus isolates. Twenty isolates that harbored the seh gene produced significant levels of SEH (254 to 692 ng/ml), while only one seh-harboring isolate did not produce a detectable level (10 ng/ml) of SEH. In contrast, none of the 34 isolates that harbored the seg gene, including 15 isolates from food poisoning outbreaks, produced a detectable level (5 ng/ml) of SEG. One isolate that harbored the seg gene produced a very small amount of SEG (13.9 ng/ml). Also, 20 isolates that harbored the sei gene produced no detectable level (10 ng/ml) of SEI, while the remaining 12 sei-harboring isolates produced very small amounts of SEI (10.6 to 36 ng/ml). There was no significant correlation between the origins of the isolates and their SEI productivities.
FIG. 2.
Frequency distribution of the productivity of SEG, SEH, and SEI by S. aureus isolates. All S. aureus isolates were grown in brain heart infusion broth-1% yeast extract at 37°C for 48 h. The culture supernatants were preincubated with 20% (vol/vol) normal rabbit serum overnight and were diluted with phosphate-buffered saline-Tween 20 to reduce the background caused by protein A. These pretreated culture supernatants were each subjected to sandwich ELISA, and the concentrations of SEG, SEH, and SEI were determined by use of standard curves.
Detection of seg and sei mRNA by RT-PCR.
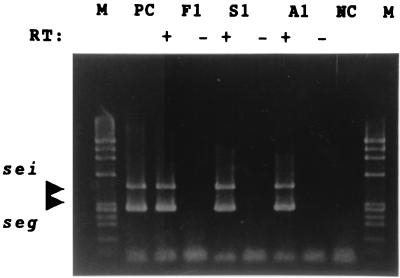
Because most of the isolates harboring seg or sei did not produce a detectable level of SEG or SEI, the transcription of seg and sei mRNAs was examined by RT-PCR. S. aureus isolates Fukuoka 1, Saga 1, and Aomori 1, which were involved in food poisoning outbreaks, were selected for RT-PCR analysis. These isolates harbored both the seg and sei genes but did not produce detectable levels of SEG or SEI. Significant levels of transcription of the mRNAs of the seg and sei genes was detected in all of these isolates, with PCR products of the expected sizes being observed in all samples when RT and both primer sets were used. Because no PCR products were observed in the samples not subjected to RT, it is thought that these PCR products were not amplified from contaminated genomic DNA (Fig. 3).
FIG. 3.
Detection by RT-PCR of SEG and SEI mRNAs in S. aureus isolates harboring seg and sei genes. Total RNAs purified at the mid-log phase from cultures of S. aureus isolates harboring seg and sei were subjected to RT-PCR analysis. Lanes: F1, Fukuoka 1; S1, Saga 1; A1, Aomori 1; M, HaeIII-digested φX174 as a size marker; PC, genomic DNA of Fukuoka 1 as a positive control; NC, Milli-Q water as a negative control; +, with RT; −, without RT.
DISCUSSION
SE genes seg, seh, and sei have been commonly distributed among S. aureus isolates taken from a variety of sources. Such frequent detection of seg, seh, and sei genes in S. aureus has been reported by other researchers as well. McLauchlin et al. (12) collected 129 S. aureus isolates in the United Kingdom and analyzed the distributions of the sea to sei and tsst-1 genes. They showed that sea, seg, seh, and sei were most frequently detected together with other se genes in isolates that originated from food poisoning outbreaks. Jarraud et al. (10) showed that 12 strains of S. aureus isolated from patients with toxic shock syndrome and staphylococcal scarlet fever possessed only the seg and the sei genes. They analyzed the distributions of the seg and the sei genes in 230 S. aureus strains isolated from various clinical sources and showed that seg and sei were most frequently detected both in nasal carriage isolates and in strains from patients with toxemia (11). Moreover, they demonstrated that the seg and sei genes are present in S. aureus in a tandem orientation and that seg and sei belong to an operon of the egc enterotoxin gene cluster, which contains five enterotoxin genes (seg, sei, sem, sen, and seo) and two pseudogenes, in S. aureus strain A900322 (11). The high rate of coexistence of seg and sei in S. aureus isolates suggests that these genes may exist as members of egc in most seg- and sei-positive isolates.
In this study, significant SEH production by most seh-harboring S. aureus isolates was confirmed by a newly developed sandwich ELISA. Su and Wong (23) previously developed a sandwich ELISA for SEH detection using an antibody prepared against native SEH, and they showed that one strain that was involved with food poisoning produced SEH. In addition, Ren et al. (18) purified SEH from a toxin shock syndrome toxin 1-negative S. aureus clinical isolate. Together, these results suggest that most seh-harboring S. aureus isolates would be able to produce SEH and would be capable of causing food poisoning and other staphylococcal superantigen-related diseases. On the other hand, although it has been demonstrated with an animal model that SEG and SEI are emetic toxins, it is not yet clear whether these SEs are responsible for food poisoning in humans. Using a sandwich ELISA, we could not detect significant amounts of SEG production in most S. aureus isolates that harbored seg, including isolates from food poisoning outbreaks, and roughly 60% of sei-harboring isolates did not produce a detectable level of SEI. A sandwich ELISA has been recognized as a good method for the detection of the SEA to SEE proteins (8, 24). It is unlikely that the sandwich ELISA is not suitable for the detection of SEG and SEI. In an epidemiological analysis, it has been reported that a total of 100 to 200 ng of SEA in food was sufficient to cause food poisoning (7). It remains an open question whether these seg- or sei-harboring isolates that were involved in food poisoning have produced SEG or SEI in amounts in foods sufficient to cause food poisoning. The production of SEs is influenced by various factors (pH, water activity, atmosphere, etc.) that cause the amount of SEs produced to vary (3, 4, 9). It is known that the strains that can produce microgram-per-milliliter amounts of SEs in a laboratory medium usually produce only nanogram-per-milliliter amounts of SEs in foods. Hence, it is very difficult to determine what amount of SEs a strain must produce in a laboratory medium in order to qualify as a strain that can cause food poisoning. However, Pereira et al. (17) reported that strains that produce small amounts of SED (nanogram-per-milliliter amounts in laboratory medium) could produce a sufficient amount of SED (nanogram-per-gram amounts) in foods to result in food poisoning. At present, we do not have enough evidence to deny the possibility that strains that produce small amounts of SEG and SEI might produce SEG and SEI in foods in amounts sufficient to cause food poisoning. It is difficult to conclude that SEG and SEI are not responsible for these food poisoning outbreaks. To demonstrate that SEG and SEI have the capability to cause food poisoning, the detection of sufficient amounts of SEG and SEI in foods involved in food poisoning outbreaks must be pursued further.
The low level of production of SEG and SEI by the isolates in this study is not inconsistent with previous studies (11, 15), which have described the transcription of seg and sei mRNAs by some seg- and sei-harboring strains. In this study, the transcription of seg and sei mRNAs in three seg- and sei-harboring S. aureus isolates was confirmed, although these isolates did not produce detectable levels of SEG and SEI. It seems that SEG and SEI production may be regulated at the translational level. As mentioned above, seg and sei may exist as a part of egc in S. aureus. In this study, we could not analyze the levels of production of the newly reported enterotoxins SEK, SEL, SEM, SEN, and SEO. The genes coding for SEM, SEN, and SEO may exist within seg- and sei-harboring S. aureus isolates. Previous studies have shown that the mRNA transcribed from egc is polycistronic and that it can code for five enterotoxin genes (11). Although SEM, SEN, and SEO have not demonstrated emetic activities in animal models, the possibility exists that these enterotoxins may be translated from mRNA in larger amounts than SEG and SEI and that these SEs may cause food poisoning.
The detection of se genes by multiplex PCR is reliable for the se genotyping of S. aureus isolates, although the positive results of PCR only confirm the existence of se genes in isolates. The existence of se genes in S. aureus isolates is necessary for these strains to cause food poisoning or other diseases. However, it is debatable whether all se gene-positive strains, especially strains harboring newly reported se genes, can cause food poisoning or other diseases. To confirm the relationship between these newly reported SEs and food poisoning or other diseases, it is important to demonstrate the production of toxin at levels that are sufficient to cause diseases by strains harboring these se genes. The difficulty of developing immunological methods for the detection of newly reported SEs results from the difficulty of purifying such SEs and preparing specific antibodies. Our strategy of developing sandwich ELISA systems using recombinant SEs will be useful for overcoming these difficulties.
Acknowledgments
We thank Akemi Kai (Tokyo Metropolitan Research Laboratory of Public Health) and Keiko Saito (Azabu University) for kindly providing strains used in this work.
This work was partly supported by grants-in-aid for scientific research from the Japan Society for the Promotion of Science (grants 11760213 and 12660281).
REFERENCES
- 1.Balaban, N., and A. Rasooly. 2000. Staphylococcal enterotoxins. Int. J. Food Microbiol. 61:1-10. [DOI] [PubMed] [Google Scholar]
- 2.Becker, K., R. Roth, and G. Peters. 1998. Rapid and specific detection of toxigenic Staphylococcus aureus: use of two multiplex PCR enzyme immunoassays for amplification and hybridization of staphylococcal enterotoxin genes, exfoliative toxin genes, and toxic shock syndrome toxin 1 gene. J. Clin. Microbiol. 36:2548-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergdoll, M. S. 1983. Enterotoxins, p. 559-598. In C. S. F. Easton and C. Adlam (ed.), Staphylococci and staphylococcal infections. Academic Press, London, United Kingdom.
- 4.Bergdoll, M. S. 1989. Staphylococcus aureus, p. 463-523. In M. P. Doyle (ed.), Foodborne bacterial pathogens. Marcel Dekker, Inc., New York, N.Y.
- 5.Betley, M. J., and J. J. Mekalanos. 1988. Nucleotide sequence of the type A staphylococcal enterotoxin gene. J. Bacteriol. 170:34-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dinges, M. M., P. M. Orwin, and P. M. Schlievert. 2000. Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 13:16-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evenson, M. L., M. W. Hinds, R. S. Bernstein, and M. S. Bergdoll. 1988. Estimation of human dose of staphylococcal enterotoxin A from a large outbreak of staphylococcal food poisoning involving chocolate milk. Int. J. Food Microbiol. 7:311-316. [DOI] [PubMed] [Google Scholar]
- 8.Freed, R. C., M. L. Evenson, R. F. Reiser, and M. S. Bergdoll. 1982. Enzyme-linked immunosorbent assay for detection of staphylococcal enterotoxins in foods. Appl. Environ. Microbiol. 44:1349-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Genigeorgis, C. A. 1989. Present state of knowledge on staphylococcal intoxication. Int. J. Food Microbiol. 9:327-360. [DOI] [PubMed] [Google Scholar]
- 10.Jarraud, S., G. Cozon, F. Vandenesch, M. Bes, J. Etienne, and G. Lina. 1999. Involvement of enterotoxin G and I in staphylococcal toxic shock syndrome and staphylococcal scarlet fever. J. Clin. Microbiol. 37:2446-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jarraud, S., M. A. Peyrat, A. Lim, A. Tristan, M. Bes, C. Mougel, J. Etienne, F. Vandenesch, M. Bonneville, and G. Lina. 2001. egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J. Immunol. 166:669-677. [DOI] [PubMed] [Google Scholar]
- 12.McLauchlin, J., G. L. Narayanan, V. Mithani, and G. O'Neill. 2000. The detection of enterotoxins and toxic shock syndrome toxin genes in Staphylococcus aureus by polymerase chain reaction. J. Food Prot. 63:479-488. [DOI] [PubMed] [Google Scholar]
- 13.Mehrotra, M., G. Wang, and W. M. Johnson. 2000. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J. Clin. Microbiol. 38:1032-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monday, S. R., and G. A. Bohach. 1999. Use of multiplex PCR to detect classical and newly described pyrogenic toxin genes in staphylococcal isolates. J. Clin. Microbiol. 37:3411-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munson, S. H., M. T. Tremaine, M. J. Beteley, and R. A. Welch. 1998. Identification and characterization of staphylococcal enterotoxin type G and I from Staphylococcus aureus. Infect. Immun. 66:3337-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orwin, P. M., D. Y. M. Leung, H. L. Donahue, R. P. Novick, and P. M. Schlievert. 2001. Biochemical and biological properties of staphylococcal enterotoxin K. Infect. Immun. 69:360-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pereira, J. L., S. P. Salzberg, and M. S. Bergdoll. 1991. Production of staphylococcal enterotoxin D in foods by low-enterotoxin-producing staphylococci. Int. J. Food. Microbiol. 14:19-26. [DOI] [PubMed] [Google Scholar]
- 18.Ren, K., J. D. Bannan, V. Pancholi, A. L. Cheung, J. C. Robbins, V. A. Fischetti, and J. B. Zabriskie. 1994. Characterization and biological properties of a new staphylococcal enterotoxin. J. Exp. Med. 180:1675-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Shinagawa, K., M. Ishibashi, H. Yamamoto, N. Kunita, and K. Hisa. 1974. A consideration to immune doses of staphylococcal enterotoxin B to rabbits. Jpn. J. Med. Sci. Biol. 27:309-314. [DOI] [PubMed] [Google Scholar]
- 21.Shinagawa, K., M. Mitsumori, N. Matsusaka, and S. Sugii. 1991. Purification of staphylococcal enterotoxin A and E by immunoaffinity chromatography using a murine monoclonal antibody with dual specificity for both of these toxins. J. Immnol. Methods 139:49-53. [DOI] [PubMed] [Google Scholar]
- 22.Su, Y.-C., and A. C. L. Wong. 1995. Identification and purification of a new staphylococcal enterotoxin, H. Appl. Environ. Microbiol. 61:1438-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su, Y.-C., and A. C. L. Wong. 1996. Detection of staphylococcal enterotoxin H by an enzyme-linked immunosorbent assay. J. Food Prot. 59:327-330. [DOI] [PubMed] [Google Scholar]
- 24.Thompson, N. E., M. Razdan, G. A. Kuntsmann, J. M. Aschenbach, M. L. Evenson, and M. S. Bergdoll. 1986. Detection of staphylococcal enterotoxins by enzyme-linked immunosorbent assays and radioimmunoassays: comparison of monoclonal and polyclonal antibody systems. Appl. Environ. Microbiol. 51:885-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, S., J. J. Iandolo, and G. C. Stewart. 1998. The enterotoxin D plasmid of Staphylococcus aureus encodes a second enterotoxin determinant (sej). FEMS Microbiol. Lett. 168:227-233. [DOI] [PubMed] [Google Scholar]