Abstract
The tools currently available for genetic subtyping of human immunodeficiency virus type 1 are laborious or can be used only for the analysis of a limited number of samples and/or subtypes. We developed and evaluated a molecular biology-based method using subtype-specific oligonucleotide probes for env genotyping of subtypes A through G, CRF01_AE, and CRF02_AG. DNA enzyme immunoassay (DEIA) genotyping is based on nested PCR amplification of the 5′ end of the env gene (proviral DNA), followed by subtype-specific hybridization and immunoenzymatic detection on microplates. DEIA genotyping was validated with a large number of samples (n = 128) collected in Europe (France; n = 47), West-Central Africa (Cameroon; n = 36), and West Africa (Senegal; n = 45). Three different formats, depending on the distribution of subtypes in the three countries, were developed. The results were compared with those obtained by sequencing of the V3-V5 region and phylogenetic analysis or an env heteroduplex mobility assay. Additional sequencing and phylogenetic analyses of the DEIA region (the first codon of the env coding sequence to the middle of conserved region C1 of gp120) were performed to investigate the reasons for discrepancies. Intense and highly specific reactions between the oligonucleotide probes and the corresponding samples were observed. Overall, correct identification was achieved for 107 of 128 samples (83.6%). One sample was not amplified, 10 (8%) were nontypeable (NT), and 10 (8%) were misidentified. Six of the 10 discordant samples were further investigated by phylogenetic analysis, which indicated that these samples corresponded to recombinants involving the env 5′ end and the V3 and V5 regions of the two parental clades. Sequencing of NT samples showed numerous differences between sample and probe sequences, resulting in a lack of hybridization, and revealed the limitations of the selected probes in terms of specificity and sensitivity. We demonstrated the feasibility of DEIA genotyping: six subtypes plus the two most prevalent circulating recombinant forms were discriminated by using the 5′ end of the env gene. This method can be adapted to the local situation by including only probes that correspond to the prevalent strains.
Phylogenetic analyses have classified human immunodeficiency virus (HIV) type 1 (HIV-1) strains into three groups: M (for major), which is responsible for the HIV-1 pandemic; O (for outlier); and N (for non-M/non-O). O and N strains are mainly isolated from Cameroonian patients (9, 39, 54). Group M isolates are diverse and have been subdivided into genetic subtypes approximately equidistant from each another (29; HIV sequence Database [http://hiv-web.lanl.gov]). The increasing complexity of the sequences of HIV-1 group M isolates, the occurrence of recombinant forms responsible for major epidemics, and the existence of divergent clusters within subtypes have made it necessary to reevaluate the nomenclature. A revised nomenclature system was recently proposed. That system defines 9 “pure” subtypes (subtypes A to D, F to H, J, and K) with no identified recombination and 11 circulating recombinant forms (CRFs), in which members of a lineage that have identical mosaic structures and that play an important role in the pandemic are grouped (49; HIV Sequence Database). Subtypes E and A IbNg in the initial classification are, in fact, two major CRFs now designated CRF01_AE and CRF02_AG, respectively (49). The notion of subsubtypes has also been defined to refer to distinct lineages very closely related to a particular subtype lineage but genetically distant enough to be regarded as a new subtype. Subsubtypes A1 and A2 have been recognized (21), as have subsubtypes F1 and F2, found in Brazil and Europe and in Cameroon, respectively (59). Due to their previously described close relationship, subtypes B and D could be considered subsubtypes B1 and B2, respectively; we refer to them here as subtypes B and D, respectively, to remain consistent with the designations in published studies (49).
This extraordinary degree of diversity complicates diagnosis, disease monitoring, and treatment (1-3, 5, 16-18, 33, 34, 40, 53). It may also affect biological properties such as infectiousness, transmissibility, and immunogenicity, making continuous molecular biology-based epidemiology studies necessary. The diversity of HIV has also been shown to be an important epidemiological marker. Large-scale studies are required to follow the dynamics of the pandemic and are probably also required for vaccine design. HIV-1 strains can be subtyped (i.e., subtypes and CRFs can be identified) directly by genomic analysis (genotyping) or indirectly by analysis of the antibody response of the host by using synthetic peptides corresponding to the V3 loop of gp120 (V3 serotyping) (7, 10, 42, 51). Serological assays have been shown to be useful tools in some geographical areas, but they discriminate only a limited number of antigenically different subtypes (4, 6, 11, 12, 20, 22, 30, 52, 55, 63). Genome analysis based on DNA sequencing followed by phylogenetic analysis is the most informative method for subtyping and is considered the “gold standard.” Although this is becoming easier and is now partly automated, it remains time-consuming and expensive and requires specific equipment, rendering it inappropriate for large-scale studies. The env and, recently, the gag heteroduplex mobility assays (HMAs) have been developed as alternative methods (15, 23); the HMA technique is nonetheless laborious and allows the analysis of only a limited number of samples, and interpretation may be difficult. Other genetic methods can be used in large-scale studies but discriminate fewer subtypes. These methods include subtype A-specific PCR, restriction fragment length polymorphism analysis, combinatorial melting assay, and probe hybridization (19, 26, 28, 35, 38, 41, 43, 47, 48, 56, 57, 60). Probe hybridization has been demonstrated to be effective for rapid screening of the prevalent subtypes in Uganda (subtypes A and D) and Thailand (subtype B and former subtype E) (47, 56). The aim of this study was to develop and to evaluate the feasibility of a similar methodology adapted to the identification of the six major subtypes prevalent worldwide (subtypes A through G), which also allows the discrimination of CRF01_AE and CRF02_AG. We wanted to include CRF discrimination in our objectives because CRF02_AG is now recognized as responsible for the subtype A epidemic in West and West-Central Africa, and no specific tool except gag HMA has been available to track this epidemic (23).
We adapted the DNA enzyme immunoassay (DEIA) technique previously described as relevant for hepatitis C virus (HCV) genotyping (25, 31, 61). This technique is based on the hybridization of amplified proviral DNA with subtype-specific probes used to coat microtiter plate wells, followed by detection with a mouse monoclonal anti-double-stranded DNA antibody. We validated DEIA genotyping using a large number of samples collected in Europe (France), West-Central Africa (Cameroon), and West Africa (Senegal).
MATERIALS AND METHODS
Sample collection.
A total of 131 HIV-1 samples whose envelopes (V3-V5) were genetically characterized were analyzed. Fifty samples were obtained from in- or outpatients, mainly African, attending the Bichat Hospital in northern Paris (European samples). The genetic subtypes of these samples were identified by HMA (n = 47) or sequencing (n = 3). Among these 50 samples, the genetic subtype distribution was as follows: 15 were subtype A-like, 10 were subtype B, 6 were subtype C, 5 were subtype D, 4 were subtype F, 4 were subtype G, 3 were subtype H, and 3 were CRF01_AE. Discrimination between subtype A and CRF02_AG in the envelope was not possible since the genetic subtypes were identified by HMA.
An additional 81 samples (African samples) were obtained from patients attending the Military Hospital in Yaounde, Cameroon (n = 36), and patients attending one of the three major hospitals in Dakar, Senegal (n = 45). For all these samples the genetic subtype was identified by sequence analysis; and the subtype distribution was as follows: 19 were subtype A, 3 were subtype B, 3 were subtype C, 9 were subtype D, 7 were subtype F, 12 were subtype G, and 28 were CRF02_AG.
Peripheral blood mononuclear cell isolation, DNA extraction, and reference subtyping.
Peripheral blood mononuclear cells were separated on a Ficoll gradient. They were washed twice in phosphate-buffered saline (PBS), pelleted, and stored at −80°C. DNA was extracted as described previously (32).
The env subtypes of 47 European samples were determined by env HMA as follows: 1μg of DNA was used for PCR amplification with primers ED3 and ED12. The second round of amplification was performed with 2 μl of the first-round product and primers ES7 and ES8, as described by Delwart et al. (15). Subtype reference samples were amplified with second-round primers and 10 ng of plasmids A to H as templates. HMA was performed as described previously (14).
Three European samples and all African samples were genotyped by sequencing of the V3-V5 region of the env gene (700 bp, amplified with HMA primers ED5 and ED12 as outer primers and primers ES7 and ES8 as inner primers), followed by phylogenetic analysis, under the conditions described previously (62).
DEIA genotyping. (i) Choice of subtyping region and selection of identification probes.
The subtype-specific probes were selected by analyzing the entire consensus sequences of the env genes of pure subtypes A to D and F to G and the two CRFs, CRFs AE and AG (HIV Sequence Database). The criteria for probe specificity were set on the basis of previous experience with the DEIA genotyping of HCV (25, 31, 61). Probes were 20 to 30 bases long and were designed such that the melting temperatures (Tms) of all probes were close, with intersubtype differences of at least 20% (about 4 different bases), and the region corresponding to the probes was conserved for a given subtype. The most suitable region presenting these characteristics comprised the first codon of the env coding sequence (env cds) to the middle of conserved region C1 of gp120 (DEIA region).
A set of different probes specific for this region was defined. Single subtype-specific probes for subtypes C, F, and G were easily identified (Table 1). Unique probes for subtypes B and D could not be found. The lack of specificity of the subtype D probe, which also detected non-B, non-D subtypes, was associated in part with a high level of sequence similarity between subtypes B and D. Therefore, samples of subtypes B and D (called subtype BD) were first distinguished with a common subtype BD-specific probe (probe SBD3), followed, if a positive result was obtained, by specific identification with subtype B- and subtype D-specific probes (Table 1).
TABLE 1.
Characteristics of subtype-specific probes
| Probe | Subtype(s) identified | Oligonucleotide sequencea | Tm (°C) |
|---|---|---|---|
| SAG1 | CRF02_AG | 5′-ATA TTT TGG ATA ATG ATA ATT TGT AAT GC-3′ | 53.7 |
| SA2 | CRF02_AG, A, CRF01_AE | 5′-AAA GAY GCA GAG ACC ACC C-3′ | 55.2 |
| SAE1 | CRF01_AE | 5′-AGA CAC AGA TGA ATT GGC CAA-3′ | 53.9 |
| SB2 | B | 5′-GAM GGR GAY CAG GAA GAA TT-3′ | 51.2 |
| SBD3 | B, BD, D | 5′-ATG CTC CTT GGG ATR TTR ATG-3′ | 52 |
| SD1.1 | D | 5′-AGA GTG AGG GRG ATA GAG AGG AAT T-3′ | 59.5 |
| SC1 | C | 5′-ATC TTA GGC TTT TGG ATG ITA ATG-3′ | 54 |
| SF6 | F | 5′-GGG AAA TGG GGC CTT TTA TT-3′ | 53.2 |
| SG1 | G | 5′-CAT ATA GTA CTG AAA GCC ATA ATG T-3′ | 54.6 |
Degenerate bases (in boldface)are defined as follows: Y, C + T; R, = A + G; M, A + C; I, inosine.
It was difficult to find a specific probe for subtype A due to the close relationship between this subtype, CRF01_AE, and CRF02_AG. The 5′ end of the env gene of the two CRFs is indeed partly subtype A-like (HIV Sequence Database). A common probe (probe SA2) that identifies subtype A, CRF01_AE, and CRF02_AG was therefore identified, with discrimination between subtype A and the two CRFs being achieved with additional CRF-specific probes, designed on the basis of CRF-specific signature sequences (Table 1).
After preliminary assays, nine probes (probes SA2, SAE1, SAG1, SBD3, SB2, SD1.1, SC1, SF6, and SG1) were selected for the detection and identification of samples of subtype A, CRF01_AE, and CRF02_AG; samples belonging to either subtype B or D; samples of subtype B; samples of subtype D; and samples of subtypes C, F, and G (Table 1). They were synthesized, 5′ biotinylated, and purified by high-pressure liquid chromatography (Eurogentec, Seraing, Belgium). All probes were 20- to 29-base oligonucleotide probes. The probe sequences and Tms are presented in Table 1. Nine oligonucleotides with sequences complementary to those of the nine probes were used as positive controls for hybridization.
(ii) Subtype-nonspecific PCR.
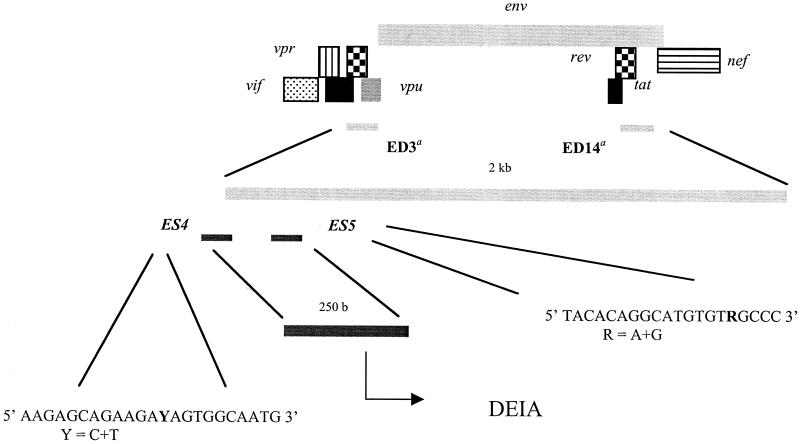
A nested PCR for the region that spans the 5′ end of the env gene (DEIA region) of all genetic subtypes A through H and the two CRFs (CRFs AE and AG) was performed to amplify a 250-bp fragment encompassing the sequences of the identification probes (Fig. 1) (HIV Sequence Database). The primer sets used were primers ED3 and ED14 as outer primers (as described for HMA) and primers ES4 and ES5 as inner primers (designed for DEIA), (see the sequences in Fig. 1). For the first round, 10 μl of DNA template was added to a 90-μl reaction mixture consisting of 10 mM Tris-HCl (pH 9), 50 mM KCl, 0.1% Triton X-100, 1.5 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 0.2 mM, 20 pmol of each primer, and 3.5 U of Expand high-fidelity Taq polymerase (Roche Diagnostics, Mannheim, Germany). Ten microliters of product from the first round was used for the second round in 100 μl of the same mixture used for the first-round PCR and 2.5 U of Taq polymerase (Promega, Madison, Wis). The first-round PCR was performed as previously described by Delwart et al. (14); a first denaturation step for 2 min at 94°C was followed by 3 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min and 32 cycles of 94°C for 15 s, 55°C for 45 s, and 72°C for 1 min, with a final extension of 5 min at 72°C. The conditions for the second round were a denaturation step of 2 min at 94°C, followed by 35 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min, with a final extension of 7 min at 72°C.
FIG. 1.
Principle of genotyping of HIV-1 by DEIA. An env fragment of 250 bp is amplified by nested PCR and is then used for subtype-specific detection and identification by DEIA. a, primers ED3 and ED14 corresponded to those used in the env HMA (15).
(iii) DEIA procedure.
The DEIA was based on the hybridization of amplified single-stranded DNA (ssDNA) with oligonucleotide subtype-specific probes attached to the surfaces of microtiter plate wells by a streptavidin-biotin bond. Hybrids of the probe and the ssDNA were detected with a mouse monoclonal anti-DNA antibody. Antibody binding was detected by means of a colorimetric immunoenzymatic reaction.
The DEIA was performed with a GEN-ETI-K PS0001 DEIA kit, according to the manufacturer's instructions (DiaSorin, Saluggia, Italy). Three parameters had to be defined: the concentration of the identification probe used to coat the microplates, the concentration of the complementary oligonucleotides, and the hybridization temperature. After calibration, the coating concentrations of the identification probes were defined according to the probes used. The concentrations were 0.25 ng/μl for probes SA2, SAE1, SBD3, SD1.1, SC1, and SG1; 0.75 ng/μl for probe SB2; and 0.05 ng/μl for probes SAG1 and SF6. The optimal concentration of the complementary oligonucleotides was defined as 2.5 ng/μl, and the hybridization temperature was 45°C.
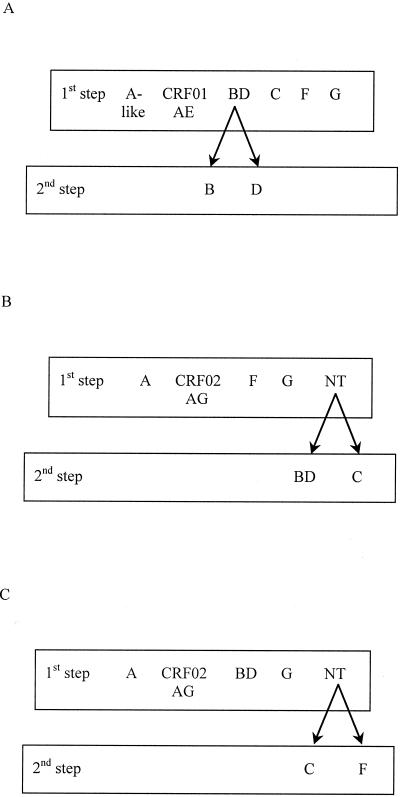
Three formats of the assay, one for each sample collection (i.e., a Europe, a Cameroon, and a Senegal format) were evaluated (Fig. 2). The number and type of probes were adapted to the predominant circulating subtypes.
FIG. 2.
Schematic diagram of the three DEIA formats developed for subtyping of samples from Europe (France; A), West-Central Africa (Cameroon; B) and West Africa (Senegal; C).
(a) Europe format.
The Europe format used subtype-specific probes SA2, SAE1, SBD3, SB2, SD1.1, SC1, SF6, and SG1 to identify subtype A-like samples (subtype A and CRF02_AG samples), CRF01_AE samples, and subtype B, D, C, F, and G samples. A first step discriminated six subtypes (subtype A-like, CRF01_AE, and subtypes BD, C, F, and G), as follows (Fig. 2A). Wells of a streptavidin-coated microplate were sensitized with 100 μl of probes appropriately diluted in Tris-EDTA buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8]; one probe per strip for strips 1 to 6 and 7 to 12) by incubation for 20 h at 4°C. The wells were washed five times with PBS containing Tween 20 (PBS-TW; DiaSorin). Before the assay, the products of the nested PCR were denatured for 15 min at 97°C. They were cooled on ice for at least 10 min before DEIA analysis. Hybridization buffer (100 μl, containing Denhardt's solution, SSC [0.15 M NaCl plus 0.015 M sodium citrate], Tris-HCl, and EDTA; DiaSorin) was added to each well. We then added 10 μl of a negative control (NC) consisting of Tris-HCl, MgCl2, and bovine serum albumin (DiaSorin) to wells 1 to 6 of the first line. Ten microliters of each of the six complementary oligonucleotides was added as a positive control to wells 1 to 6 of the second line. Ten microliters of the ssDNA sample (one sample per six wells) was added to each well of the next lines and to each well of all lines on strips 7 to 12. A cardboard lid was placed over the plate to prevent evaporation, and the microplate was incubated for 1 h at 45°C (±1°C). The wells were then washed five times with PBS-TW. One hundred microliters of an anti-double-stranded DNA antibody solution (1:50 dilution in PBS and fetal calf serum; DiaSorin) was added, and the plates were incubated for 30 min at 30°C (±1°C) after they were covered with a cardboard lid. The wells were then washed five times with PBS-TW. One hundred microliters of the enzyme tracer solution (protein A conjugated to horseradish peroxidase [1:50 dilution in PBS-fetal calf serum]; DiaSorin) was added, and the plates were incubated for 30 min at room temperature after they were covered with a cardboard lid. The wells were washed five times with PBS-TW, and the reaction was detected by incubation with 100 μl of a 1:1 chromogen-substrate solution of tetramethylbenzidine and hydrogen peroxide for 30 min at room temperature in the dark. Color development was stopped by adding 1 N H2SO4 (200 μl), and the A450 and A630 were read. After preliminary experiments, the cutoff value was defined as the absorbance for the NC + 0.400.
A second step was performed for the BD-specific probe-reactive samples to distinguish between subtypes B and D. The same conditions used for the first step was applied, except that the cut-off value for the well coated with probe D1.1 was defined as the absorbance for the NC + 0.150.
In this Europe format, we were able to carry out an initial screening of 14 samples per microplate.
(b) Cameroon format.
The Cameroon format involved two steps: the first used probes SA2, SAG1, SF6, and SG1, corresponding to the predominant genotypes in this country. The nontypeable (NT) samples (those with absorbance values below the cutoff) were then analyzed with probes SBD3 and SC1 (Fig. 2B). The same experimental conditions described above for the Europe format were applied for the Cameroon format. In the Cameroon format, we were able to carry out an initial screening of 22 samples per microplate.
(c) Senegal format.
The Senegal format was performed as described above, except that the subtype BD-specific probe replaced the subtype F-specific probe for the first step; the NT samples were further analyzed with subtype C- and F-specific probes (Fig. 2C). In this format, we were able to carry out an initial screening of 22 samples per microplate.
(iv) Probe reactivity.
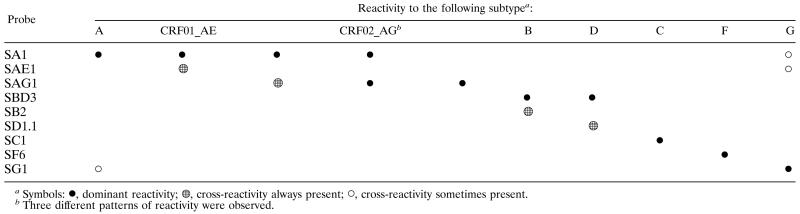
The intersubtype relationships discussed above precluded direct simple subtyping with single reactivity to the specific probe. However, a reproducible profile of probe reactivity including cross-hybridization made it possible to define a chart representing a specific pattern of hybridization for each subtype or CRF (Table 2). A genotype was then assigned according to this chart, with any reactivity profile different from those in this chart being classified as indeterminate.
Table 2.
DNA sequencing of the env cds-C2 region.
DNA sequencing and phylogenetic analysis of the env 5′ end of NT, indeterminate (similar absorbances with two probes), and discordant samples were performed to assign a subtype to the DEIA region. An 820-bp fragment was amplified from proviral DNA by nested PCR with primers ED3 and ED14 as the outer primers and primers ES4 and PSA2 (5′-CCATTTAACAGCAGTTGAGTTG-3′) as the inner primers. The first round was performed with the Expand Long Template PCR system (Roche Diagnostics), according to the manufacturer's instructions. The second round was performed with Taq DNA polymerase (Promega), as follows: 1 to 5 μl from the first-round amplification was used for the second round with the inner primers, the reaction mixture consisting of 50 mM KCl, 10 mM Tris-HCl (pH 9), 0.1% Triton X-100, 1.8 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 0.2 mM, 2.5 U of Taq polymerase, and 20 pmol of each primer. The PCR conditions were 92°C for 5 min, followed by 38 cycles at 92°C for 20 s, 50°C for 30 s, and 72°C for 2 min, with a final extension step at 72°C for 10 min. The amplified fragments were purified with the QIAquick gel extraction kit (QIAGEN, Courtaboeuf, France) and were directly sequenced with the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction kit with AmpliTaq DNA polymerase FS (Perkin-Elmer, Roissy, France) on an Applied Biosystems 373A automatic DNA sequencer (Stretch model).
Phylogenetic analysis.
Nucleotide sequences were aligned with the CLUSTAL W program with minor manual adjustments to take into account the protein sequences (58). Regions that could not be aligned unambiguously due to length or sequence variability were omitted from the analysis. Phylogenetic trees were produced by the neighbor-joining method, and the reliability of the branching orders was assessed by the bootstrap approach with the CLUSTAL W program. Genetic distances were calculated by Kimura's two-parameter method (27).
RESULTS
Probe reactivity.
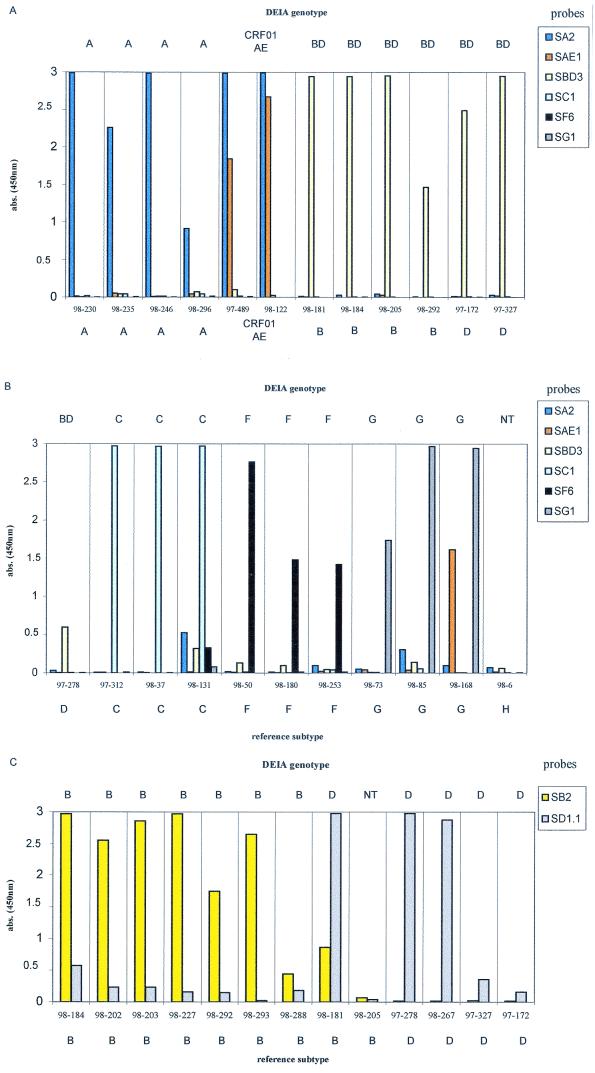
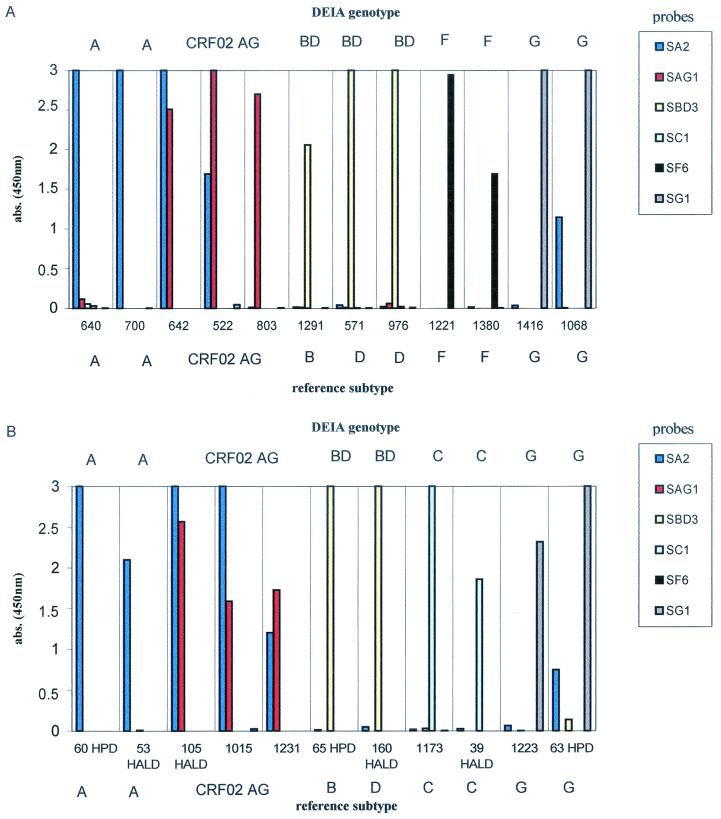
The reactivity profiles of representative European and African samples are presented in Fig. 3 and 4. Subtype BD-, C-, and F-specific probes produced intense and highly specific signals with samples of the subtypes corresponding (Fig. 3A, 3B, and 4). Among the subtype BD-reactive samples from the European collection, the additional identification of subtype B samples was accurate, with high absorbance values, whereas specific identification of subtype D samples was characterized by lower absorbance values (two of four samples had absorbances of <0.4; Fig. 3C). This result was probably due to a higher variability of subtype D sequences and led to a lowering of the cutoff value (absorbance for the NC + 0.150).
FIG. 3.
Representative profiles of probe reactivities with European samples. The genotype determined by DEIA is given at the top, and the reference subtype is given at the bottom. (A and B) The first screening discriminated subtype A-like, CRF01_AE, and subtype BD, C, F, and G samples. (C) Specific identification of subtypes B and D from the samples reactive with subtype BD-specific probes was performed in a second step with individual B- and D-specific probes. abs., absorbance.
FIG. 4.
Representative profiles of probe reactivity with Cameroonian samples (A) and Senegalese samples (B). The DEIA genotype is given at the top, and the reference subtype is given at the bottom. abs., absorbance.
We analyzed the results for the European and African samples for subtype A, CRF01_AE, and CRF02_AG, separately. According to the interpretation chart, in the absence of the CRF02_AG-specific probe, 11 of the 13 European samples were correctly identified. Subtype A-like samples (subtypes A and CRF02_AG samples) reacted specifically with the subtype A-specific probe, giving intense signals (Fig. 3A). The three CRF01_AE samples were discriminated on the basis of their strong reactivities with the CRF01_AE-specific probe (Fig. 3A). In the African sample collections, 11 of the 13 correctly identified subtype A samples reacted with the subtype A-specific probe, giving an intense signal without cross-hybridization (Fig. 4). For the 26 correctly identified CRF02_AG samples, two major patterns of reactivity were observed (Table 2; Fig. 4): dominant reactivity with the subtype A-specific probe and cross-hybridization to the CRF02_AG-specific probe (21 samples) and dominant reactivity with the CRF02_AG-specific probe and cross-hybridization to the subtype A-specific probe (4 samples). For one sample, only reactivity to the CRF02_AG-specific probe was observed.
Subtype G samples reacted dominantly with the SG1 probe, but various degrees of cross-reactivity to SAE1 and/or SA2 probes were observed (Table 2; Fig. 3B and 4).
Comparison between reference subtyping and DEIA genotyping.
The results of DEIA genotyping were compared to those obtained by env HMA genotyping or env sequencing for the 131 samples.
(i) Subtyping of the European collection.
The Europe format included eight probes for the identification of env subtype A-like samples, CRF01_AE, and the B, C, D, F, and G genotypes (Table 3). We did not distinguish CRF02_AG because it was not distinguished by env HMA. Due to the predominance of subtype B in Europe, specific identification of subtypes B and D with the set of BD-, B-, and D-specific probes was required.
TABLE 3.
Correlation between env HMA or V3-V5 sequencing and DEIA genotyping of the European collection
| Genotype by env HMA or sequencing | No. of samples with the following env genotype by DEIAa:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A-likeb | CRF01_AE | BD | B | D | C | F | G | NT
|
Total (n = 50) | ||
| PCR positive | PCR negative | ||||||||||
| A-likea | 13 | 1 | 1 | 15 | |||||||
| CRF01_AE | 3 | 3 | |||||||||
| B | 9c | 7d | 1d | 1 | 10 | ||||||
| D | 4c | 4d | 1 | 5 | |||||||
| C | 4 | 1 | 1 | 6 | |||||||
| F | 4 | 4 | |||||||||
| G | 4 | 4 | |||||||||
| H | 2 | 1 | 3 | ||||||||
Boldface numbers indicate correct identification.
Genotype A-like samples corresponded to both subtype A and CRF02_AG.
Discrimination of subtype B or D samples from the other subtypes with a common BD-specific probe.
Identification of the subtype BD probe-reactive samples with B- or D-specific probes.
Of the three subtype H samples, one was not amplified and two were nonreactive, providing information about the specificities of the probes. Six of the 47 samples were identified as NT (1 was not amplified and five gave weak signals below the cutoff). Thirty-nine samples (83.0%) were correctly identified. All CRF01_AE, subtype F, and subtype G samples were correctly identified; 13 of 15 (86.6%) subtype A-like samples reacted specifically with the subtype A-specific probe and one was genotyped as subtype G by DEIA. Thirteen of the 15 subtype B and D samples (86.6%) were discriminated with the common BD-specific probe. The subsequent use of probes specific for subtypes B and D correctly identified seven of nine subtype B samples and four of four subtype D samples (Table 3).
(ii) Subtyping of African collections.
For clarity, the results obtained with the Cameroon and Senegal formats are presented in Table 4.
TABLE 4.
Correlation between env V3-V5 genotyping and DEIA genotyping for the African collections
| Genotype by env sequencing | No. of samples with the following env genotype by DEIAa
|
Total (n = 81) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| A | CRF02_AG | BD | C | F | G | Ib | NTc | ||
| A | 13 | 4 | 2 | 19 | |||||
| CRF02_AG | 1 | 26 | 1 | 28 | |||||
| B or D | 11 | 1 | 12 | ||||||
| C | 2 | 1 | 3 | ||||||
| F | 6 | 1 | 7 | ||||||
| G | 10 | 1 | 1 | 12 | |||||
Boldface indicates correct identification.
I, indeterminate, i.e., similar reactivity with subtype A- and subtype G-specific probes.
The NT samples were amplified by nested PCR, but the absorbance value was below the cutoff.
(a) Cameroon format.
Thirty-two of 36 samples (88.9%) were correctly identified by use of the Cameroon format. A first screening of the four predominant subtypes (subtype A, CRF02_AG, and subtypes F and G) with the corresponding probes discriminated 27 samples, with 1 being misidentified as subtype G instead of subtype A. One subtype F, one subtype G, and the seven subtype B or D samples were NT; they were further analyzed in a format containing subtype BD- and C-specific probes. Six of seven subtype B or D samples were correctly identified; the other three samples remained NT.
(b) Senegal format.
Due to the nonnegligible prevalence of subtypes B and D in Senegal, the subtype BD-specific probe instead of the subtype F-specific probe was included in the first step. Thirty-six of 45 samples (80%) were correctly identified. Seven samples were misidentified with the CRF02_AG-specific probe (four samples of genotype A), the subtype G-specific probe (two samples of genotypes A and C, respectively), and the subtype A-specific probe (one sample of genotype CRF02_AG); and three samples (one CRF02_AG sample and two subtype C samples) were NT. The NT samples were further discriminated by an assay with subtype C- and subtype F-specific probes; the two subtype C samples were correctly identified, and the CRF02_AG sample remained NT. One subtype G sample was indeterminate, displaying similar reactivities with subtype A- and subtype G-specific probes.
Molecular basis of DEIA genotyping results.
DNA sequencing followed by phylogenetic analysis of the env cds to the C2 region of gp120 was performed to assign a subtype to the DEIA region and to account for discrepancies. This was possible for 16 of the 21 samples which were NT (n = 6) or misidentified (n = 10) by the DEIA (Table 5; Fig. 5). The subtype deduced from phylogenetic analysis of the env cds to the C2 region correlated with the DEIA genotype for 6 of the 10 misidentified samples (Table 5). This revealed the occurrence of recombination involving the 5′ end of the env gene and the V3 and V5 regions of the two parental subtypes (Fig. 5 and 6). For one sample (sample SN 165HPD), misidentification was due to the absence of the subtype C-specific probe in the format; this sample reacted dominantly to the subtype C-specific probe when that probe was present and cross-hybridized with the subtype G-specific probe (data not shown).
TABLE 5.
Molecular biology-based analysis of NT samples, misidentified samples, and samples with indeterminate resultsa
| Sample collection | Identification | Subtype by:
|
% Differenceb | Note | ||
|---|---|---|---|---|---|---|
| env HMA or V3-V5 sequencing and phylogenetic analysis | env DEIA | env cds-C2 sequencing and phylogenetic analysis | ||||
| Europe | 98 289 | Ac | G | G | ||
| 98 238 | Ac | NT | CRF02_AG | 15 | Reactive with CRF02_AG-specific probe, if present | |
| 98 181 | Bc | BD or D | B | 10 | Lower specificity of subtype D-specific probe | |
| 98 205 | Bc | BD or NT | B | 5 | Lower sensitivity of subtype B-specific probe | |
| 98 210 | Cc | NT | C | 33 | ||
| Cameroon | CMMP 1432 | A | G | G | ||
| CMMP 1367 | D | NT | D | 24 | ||
| Senegal | SN 191HPD | A | CRF02_AG | CRF02_AG | ||
| SN 65HALD | A | CRF02_AG | CRF02_AG | |||
| SN 19HPD | A | CRF02_AG | CRF02_AG | |||
| SN 103HPD | A | CRF02_AG | A | Lower specificity of CRF02_AG-specific probe | ||
| SN 1048SC | A | G | G | |||
| SN 46HALD | CRF02_AG | A | CRF02_AG | Four bases + two insertions different from the CRF02_AG-specific probe | ||
| SN 1128 | CRF02_AG | NT | CRF06_cpx | 28 | ||
| SN 165HPD | C | G | C | Reacted dominantly with subtype C-specific probe, if present | ||
| SN 99HALD | G | I | G | Similar reactivities to subtype A- and subtype G-specific probes | ||
The reference subtype, the DEIA genotype, and the env 5′ end subtype of 16 analyzed samples are reported. Samples for which identical subtypes were found by DEIA genotyping and DEIA region sequencing are indicated in boldface. Further explanations of discrepancies are reported in the last column.
Difference between sample sequence and probe sequence specific for subtype identification.
Reference subtype determined by env HMA.
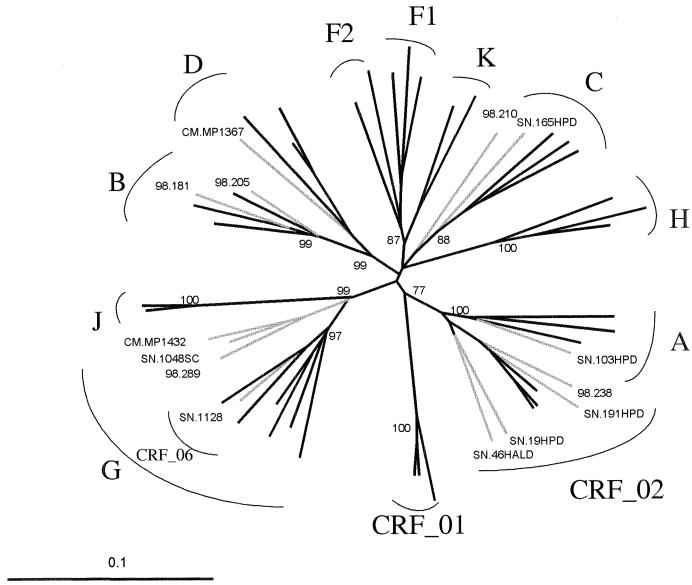
FIG. 5.
Phylogenetic tree based on 750 unambiguously aligned nucleotides from the env cds-C2 region. Fourteen of the 16 discordant samples that could be sequenced are included in the tree. The two remaining samples, SN 65HALD and SN 99HALD, could not be included in the tree because the sequenced fragments were shorter; they were found to be CRF02_AG and subtype G, respectively, on the basis of the sequences of the 618- and 520-bp fragments. The reference strains used to represent the different genetic subtypes were A.UG.92UG037, A.UG.U455, A.SE.SE8131, B.FR.HXB2, B.GA.OYI, B.US.RF, C.BR.92BR025, C.DJ.DJ259A, C.ET.ETH2220, D.CD.ELI, D.CD.NDK, D.UG.94UG1141, F1.BR.93BR020.1, F1.FI.FIN9363, F1.FR.MP411, F2.CM.MP255, F2.CM.MP257, G.BE.DRCBL, G.FI.HH8793, G.NG.92NG083, G.SE.SE6165, H.BE.VI991, H.BE.VI997, H.CF.90CF056, J.SE.SE9173, J.SE.SE9280, K.CD.EQTB11C, K.CM.MP535, CRF01_AE.CF.90CF402, CRF01_AE.TH.93TH253, CRF01_AE.TH.CM240, CRF02_AG.FR.DJ263, CRF02_AG.FR.DJ264, CRF02_AG.NG.IBNG, CRF06_cpx AGJ.ML.95ML84, and AGJ.AU.BFP90. The analysis was performed as described in Materials and Methods.
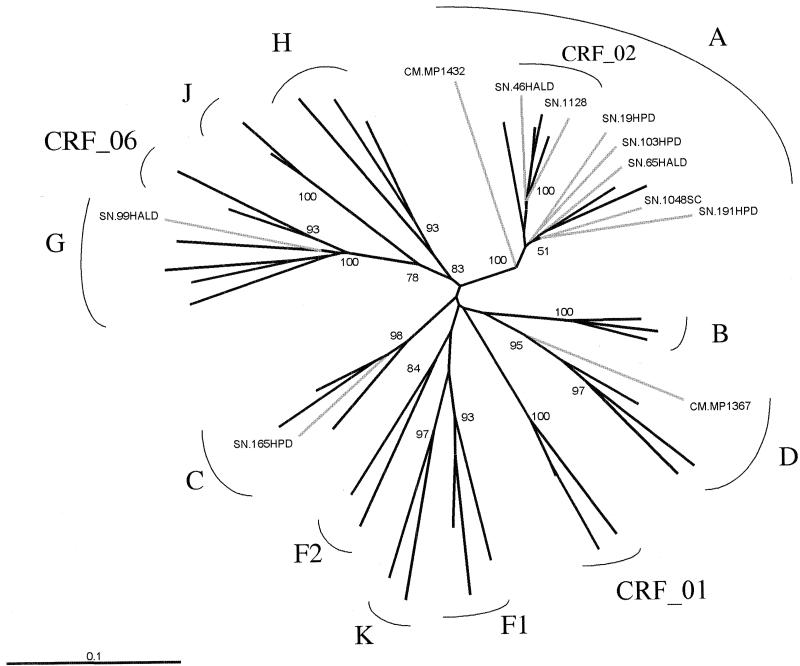
FIG. 6.
Phylogenetic tree based on 670 unambiguously aligned nucleotides from the V3-V5 env region of the 11 discordant samples from the Senegalese and Cameroonian collections (Table 5), for which the reference subtype was determined by env sequencing and phylogenetic analysis.
It was possible to sequence the DNA of the DEIA region for 6 of the 11 NT or indeterminate samples. Three samples (samples 98 210, CMMP 1367, and SN 1128) showed too many differences between sample and complementary probes sequences (more than 20%), probably resulting in a lack of hybridization (Table 5). One of these samples (sample SN 1128) presented a particularly complex env pattern (subtypes A, G, J, and K) involving the V3-V5 region from CRF02_AG associated with the 5′ end of env from the recently described CRF06_cpx (Fig. 5 and 6) (36). For one European sample (sample 98 238) identified as subtype A-like by env HMA, phylogenetic analysis of the 5′ end of env revealed that the DEIA region was related to CRF02_AG (Fig. 5). This sample was initially NT by the European format, but subsequent analysis with the CRF02_AG-specific probe showed that the sample had strong reactivity with this probe. For one Senegalese sample (sample SN 99HALD), the subtype of the DEIA region was identical to that of the V3-V5 region, but the reactivities with the subtype A- and subtype G-specific probes were similar, leading to an indeterminate result by DEIA (Table 5).
DISCUSSION
The aim of this work was to evaluate the feasibility of a simple and rapid env genotyping assay: based on the amplification of proviral DNA by nested PCR, followed by subtype-specific hybridization and immunoenzymatic detection on microplates with DEIA technology. DEIA is used in clinical microbiology laboratories for the diagnosis of viral and bacterial infections (8, 13, 45). We applied this tool to the identification of subtypes A through G, CRF01_AE, and CRF02_AG of HIV-1, which correspond to the eight currently predominant strains of the pandemic (44). HIV-1 subtyping is complex due to the high degree of genetic variability and the similar genomes of viruses belonging to different clades. However, previous studies have shown that DEIA technology can be useful for distinguishing genotypes within a species such as HCV (25, 31, 61).
The subtyping of 131 specimens including all the major pure subtypes and CRF01_AE circulating in France and the predominant pure subtypes and CRF02_AG circulating in Cameroon and Senegal was performed with a set of nine subtype-specific probes. The adaptation of the assay to formats with probes specific for the predominant subtypes or CRFs in a given country made it possible to screen 14 to 22 samples per microplate. The overall data obtained by use of the three formats were compared to those obtained by env HMA or V3-V5 sequencing and indicated the correct identification for 107 of 128 samples (83.6%; range, 80.0 to 88.9%). Sequence analysis of the DEIA region in cases of discordant results partly explained the discrepancies. It revealed the occurrence of recombinations involving the 5′ end of env and the V3 and V5 regions (seven samples) or the presence of too many differences between the sample and the probe sequences, resulting in a lack of hybridization (two samples). For two other samples, the samples were misidentified or NT due to the absence of specific probes in the format.
The data show that DEIA genotyping is a potentially useful tool suitable for rapid and simple env genotyping. Previous studies demonstrated that probe hybridization is an effective technique for molecular biology-based epidemiological analyses (47, 56). These studies were based on the distinction of only two prevalent subtypes, whereas our results indicate that identification of the six major subtypes and discrimination of the two most prevalent CRFs are possible. The adaptation of the assay to give three different formats, corresponding to the subtype distributions in three countries, showed that DEIA genotyping can be modulated as required. This is of value for monitoring the dynamics of the epidemic and the spread of the various subtypes. For instance, HIV is becoming more diverse in Western countries such as France and Sweden, where the prevalences of non-subtype B isolates recently reached 15 and 30%, respectively (6, 55). Different algorithms can be defined for the prescreening of a limited number of predominant subtypes within a large panel of specimens according to the local context or for the screening of a larger number of subtypes within a selected collection. Samples NT or indeterminate by DEIA can be further analyzed by sequencing and phylogenetic analysis.
This methodology has several advantages. It can be used for the direct analysis of the genome, which is more precise than serotyping and which takes advantages of the simplicity of the enzyme-linked immunosorbent assay (ELISA) technique. Microplate detection is less labor-intensive than HMA or sequencing and requires simple ELISA equipment, and the results are based on the simple identification of reactive and nonreactive samples. This would facilitate the screening of numerous samples in large population-based epidemiology studies, especially in developing countries, where sequencing and HMA are difficult to perform and V3 serotyping is limited to a few antigenically different V3 subtypes. In countries where subtypes A, C, F, and G are prevalent, their serologic discrimination is inefficient due to cross-reactivity between subtype-specific peptides (24, 46, 50). Our results suggest that DEIA genotyping could be a valuable tool for distinguishing these subtypes. The identification of specific nucleotide signature sequences made it possible to discriminate associated subtypes such as subtype A, CRF01_AE, and CRF02_AG. The simple and efficient identification of CRF02_AG is particularly important because this form accounts for between 60 and 84% of the subtype A samples identified from West and West-Central Africa (37). CRF02_AG seems to have already spread widely and accounts for a large proportion of the subtype A-like variants found worldwide. The recent development of a gag HMA makes discrimination of such variants possible (23); however, it involves additional gag analysis of subtype A-like samples identified by env HMA. The discrimination of CRF02_AG by DEIA with the env gene may be of value because it is faster and simpler.
As for any subtyping method, DEIA has limitations. DEIA genotyping is based on 20- to 30-base probes and analyzes only a single short region of the env gene. Nonetheless, our results showed that the selected region may provide the same information provided by the longer fragment used in the env HMA technique. The misidentification of 6 of 10 samples was accounted for by recombination events that were detectable only by analysis of the entire env gene. Another limitation is the genetic variability of HIV-1, which increases with the duration of the epidemic; thus, subtype-specific probes should be regularly reevaluated and adapted. The DEIA region was found to be less variable than other parts of the env gene, particularly the V3 region, when the probes were designed. This reduced variability might reduce this limitation.
The occurrence of recombination is a major obstacle to HIV-1 subtyping; it is linked to the prevalence, dynamics, and duration of the epidemic in the region concerned. The description in particular of numerous gag and env recombinants makes subtyping less informative if it is based on only one gene. The development of a rapid and simple method for the identification of gag and env recombinants in one step is necessary. We are therefore developing a gag DEIA genotyping system. However, these data showed that discordant results could be due to intragene recombinations. In countries where the epidemic is relatively old and dynamic, with a high level of diversity and at least 30% recombinant forms, such as in the Democratic Republic of Congo (62), DEIA genotyping may be unsuitable. In such situations, the sequencing of entire genes remains the only suitable technique for correct identification.
In conclusion, this study demonstrated the feasibility and relevance of an HIV-1 DEIA genotyping approach. The primers, probes, and formats developed in this work can be used as they are. However, the high rate of evolution of HIV-1 and its worldwide distribution require the definition of probes adapted to a regional context rather than the unrealistic permanent use of definitively identified universal subtype-specific probes.
Acknowledgments
Funding for the study was obtained from the Agence Nationale de Recherche sur le SIDA (ANRS, Paris, France) and the European commission. J. C. Plantier was supported by a doctoral fellowship from Sidaction.
REFERENCES
- 1.Alaeus, A., K. Lidman, A. Sonnerborg, and J. Albert. 1997. Subtype-specific problems with quantification of plasma HIV-1 RNA. AIDS 11:859-865. [DOI] [PubMed] [Google Scholar]
- 2.Apetrei, C., D. Descamps, G. Collin, I. Loussert-Ajaka, F. Damond, M. Duca, F. Simon, and F. Brun-Vezinet. 1998. Human immunodeficiency virus type 1 subtype F reverse transcriptase sequence and drug susceptibility. J. Virol. 72:3534-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apetrei, C., I. Loussert-Ajaka, D. Descamps, F. Damond, S. Saragosti, F. Brun-Vezinet, and F. Simon. 1996. Lack of screening test sensitivity during HIV-1 non-subtype B seroconversions. AIDS 10:F57-F60. [DOI] [PubMed] [Google Scholar]
- 4.Apetrei, C., A. Necula, C. Holm-Hansen, I. Loussert-Ajaka, I. Pandrea, C. Cozmei, A. Streinu-Cercel, F. R. Pascu, E. Negut, G. Molnar, M. Duca, M. Pecec, F. Brun-Vezinet, and F. Simon. 1998. HIV-1 diversity in Romania. AIDS 12:1079-1085. [PubMed] [Google Scholar]
- 5.Arnold, C., K. L. Barlow, S. Kaye, C. Loveday, P. Balfe, and J. P. Clewley. 1995. HIV type 1 sequence subtype G transmission from mother to infant: failure of variant sequence species to amplify in the Roche Amplicor Test. AIDS Res. Hum. Retrovirs. 11:999-1001. [DOI] [PubMed] [Google Scholar]
- 6.Barin, F., A. M. Courouce, J. Pillonel, L. Buzelay, et al. 1997. Increasing diversity of HIV-1M serotypes in French blood donors over a 10-year period (1985-1995). AIDS 11:1503-1508. [DOI] [PubMed] [Google Scholar]
- 7.Barin, F., Y. Lahbabi, L. Buzelay, B. Lejeune, A. Baillou-Beaufils, F. Denis, C. Mathiot, S. M'Boup, V. Vithayasai, U. Dietrich, and A. Goudeau. 1996. Diversity of antibody binding to V3 peptides representing consensus sequences of HIV type 1 genotypes A to E: an approach for HIV type 1 serological subtyping. AIDS Res. Hum. Retrovir. 12:1279-1289. [DOI] [PubMed] [Google Scholar]
- 8.Cassinotti, P., H. Mietz, and G. Siegl. 1996. Suitability and clinical application of a multiplex nested PCR assay for the diagnosis of herpes simplex virus infections. J. Med. Virol. 50:75-81. [DOI] [PubMed] [Google Scholar]
- 9.Charneau, P., A. M. Borman, C. Quillent, D. Guetard, S. Chamaret, J. Cohen, G. Remy, L. Montagnier, and F. Clavel. 1994. Isolation and envelope sequence of a highly divergent HIV-1 isolate: definition of a new HIV-1 group. Virology 205:247-253. [DOI] [PubMed] [Google Scholar]
- 10.Cheingsong-Popov, R., S. Lister, D. Callow, P. Kaleebu, S. Beddows, J. Weber, et al. 1994. Serotyping HIV type I by antibody binding to the V3 loop: relationship to viral genotype. AIDS Res. Hum. Retrovir. 10:1379-1386. [DOI] [PubMed] [Google Scholar]
- 11.Cheingsong-Popov, R., C. Williamson, S. Lister, L. Morris, J. van Harmelen, H. Bredell, R. Wood, P. Sonnenberg, E. van der Ryst, D. Martin, and J. Weber. 1998. Usefulness of HIV-1 V3 serotyping in studying the HIV-1 epidemic in South Africa. AIDS 12:949-950. [PubMed] [Google Scholar]
- 12.Couturier, E., F. Damond, P. Roques, H. Fleury, F. Barin, J. B. Brunet, F. Brun-Vezinet, F. Simon, et al. 2000. HIV-1 diversity in France, 1996-1998. AIDS 14:289-296. [DOI] [PubMed] [Google Scholar]
- 13.Del Prete, R., A. Mosca, M. D'Alagni, R. Sabato, V. Picca, and G. Miragliotta. 1997. Detection of Mycobacterium tuberculosis DNA in blood of patients with acute pulmonary tuberculosis by polymerase chain reaction and non-isotopic hybridisation assay. J. Med. Microbiol. 46:495-500. [DOI] [PubMed] [Google Scholar]
- 14.Delwart, E. L., M. P. Busch, M. L. Kalish, J. W. Mosley, and J. I. Mullins. 1995. Rapid molecular epidemiology of human immunodeficiency virus transmission. AIDS Res. Hum. Retrovir. 11:1081-1093. [DOI] [PubMed] [Google Scholar]
- 15.Delwart, E. L., E. G. Shpaer, J. Louwagie, F. E. McCutchan, M. Grez, H. Rubsamen-Waigmann, and J. I. Mullins. 1993. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science 262:1257-1261. [DOI] [PubMed] [Google Scholar]
- 16.Descamps, D., C. Apetrei, G. Collin, F. Damond, F. Simon, and F. Brun-Vezinet. 1998. Naturally occurring decreased susceptibility of HIV-1 subtype G to protease inhibitors. AIDS 12:1109-1111. [PubMed] [Google Scholar]
- 17.Descamps, D., G. Collin, F. Letourneur, C. Apetrei, F. Damond, I. Loussert-Ajaka, F. Simon, S. Saragosti, and F. Brun-Vezinet. 1997. Susceptibility of human immunodeficiency virus type 1 group O isolates to antiretroviral agents: in vitro phenotypic and genotypic analyses. J. Virol. 71:8893-8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Descamps, D., G. Collin, I. Loussert-Ajaka, S. Saragosti, F. Simon, and F. Brun-Vezinet. 1995. HIV-1 group O sensitivity to antiretroviral drugs. AIDS 9:977-978. [PubMed] [Google Scholar]
- 19.Ellenberger, D. L., D. Pieniazek, J. Nkengasong, C. C. Luo, S. Devare, C. Maurice, M. Janini, A. Ramos, C. Fridlund, D. J. Hu, I. M. Coulibaly, E. Ekpini, S. Z. Wiktor, A. E. Greenberg, G. Schochetman, and M. A. Rayfield. 1999. Genetic analysis of human immunodeficiency virus in Abidjan, Ivory Coast reveals predominance of HIV type 1 subtype A and introduction of subtype G. AIDS Res. Hum. Retrovir. 15:3-9. [DOI] [PubMed] [Google Scholar]
- 20.Engelbrecht, S., T. L. Smith, P. Kasper, E. Faatz, M. Zeier, D. Moodley, C. G. Clay, and E. J. van Rensburg. 1999. HIV type 1 V3 domain serotyping and genotyping in Gauteng, Mpumalanga, KwaZulu-Natal, and Western Cape Provinces of South Africa. AIDS Res. Hum. Retrovir. 15:325-328. [DOI] [PubMed] [Google Scholar]
- 21.Gao, F., N. Vidal, Y. Li, S. A. Trask, Y. Chen, L. G. Kostrikis, D. D. Ho, M. Oh, M. Salminen, D. Robertson, G. M. Shaw, B. Hahn, and M. Peeters. 2001. Evidence for two distinct sub-subtypes within the HIV-1 subtype A radiation. AIDS Res. Hum. Retrovir. 17:675-688. [DOI] [PubMed]
- 22.Gaywee, J., A. W. Artenstein, T. C. VanCott, R. Trichavaroj, A. Sukchamnong, P. Amlee, M. de Souza, F. E. McCutchan, J. K. Carr, L. E. Markowitz, R. Michael, and S. Nittayaphan. 1996. Correlation of genetic and serologic approaches to HIV-1 subtyping in Thailand. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 13:392-396. [DOI] [PubMed] [Google Scholar]
- 23.Heyndrickx, L., W. Janssens, L. Zekeng, R. Musonda, S. Anagonou, G. Van der Auwera, S. Coppens, K. Vereecken, K. De Witte, R. Van Rampelbergh, M. Kahindo, L. Morison, F. E. McCutchan, J. K. Carr, J. Albert, M. Essex, J. Goudsmit, B. Asjo, M. Salminen, A. Buve, Study Group on the Heterogeneity of HIV Epidemics in African Cities, and G. van Der Groen. 2000. Simplified strategy for detection of recombinant human immunodeficiency virus type 1 group M isolates by gag/env heteroduplex mobility assay. J. Virol. 74:363-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoelscher, M., S. Hanker, F. Barin, R. Cheingsong-Popov, U. Dietrich, B. Jordan-Harder, D. Olaleye, E. Nagele, A. Markuzzi, D. Mwakagile, F. Minja, J. Weber, L. Gurtler, and F. Von Sonnenburg. 1998. HIV type 1 V3 serotyping of Tanzanian samples: probable reasons for mismatching with genetic subtyping. AIDS Res. Hum. Retrovir. 14:139-149. [DOI] [PubMed] [Google Scholar]
- 25.Imberti, L., E. Cariani, A. Bettinardi, A. Zonaro, A. Albertini, and D. Primi. 1991. An immunoassay for specific amplified HCV sequences. J. Virol. Methods 34:233-243. [DOI] [PubMed] [Google Scholar]
- 26.Janini, L. M., D. Pieniazek, J. M. Peralta, M. Schechter, A. Tanuri, A. C. Vicente, N. dela Torre, N. J. Pieniazek, C. C. Luo, M. L. Kalish, G. Schochetman, and M. A. Rayfield. 1996. Identification of single and dual infections with distinct subtypes of human immunodeficiency virus type 1 by using restriction fragment length polymorphism analysis. Virus Genes 13:69-81. [DOI] [PubMed] [Google Scholar]
- 27.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 28.Kostrikis, L. G., S. Shin, and D. D. Ho. 1998. Genotyping HIV-1 and HCV strains by a combinatorial DNA melting assay (COMA). Mol. Med. 4:443-453. [PMC free article] [PubMed] [Google Scholar]
- 29.Kuiken, C. L., B. Foley, B. Hahn, B. Korber, F. McCutchan, P. A. Marx, J. W. Mellors, J. I. Mullins, J. Sodroski, and S. Wolinksy (eds.). 1999. Human retroviruses and AIDS 1999: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.M.
- 30.Lara, C., M. Sallberg, B. Johansson, I. L. de Rivera, and A. Sonnerborg. 1997. The Honduran human immunodeficiency virus type 1 (HIV-1) epidemic is dominated by HIV-1 subtype B as determined by V3 domain sero- and genotyping. J. Clin. Microbiol. 35:783-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Pogam, S., F. Dubois, R. Christen, C. Raby, A. Cavicchini, and A. Goudeau. 1998. Comparison of DNA enzyme immunoassay and line probe assays (Inno-LiPA HCV I and II) for hepatitis C virus genotyping. J. Clin. Microbiol. 36:1461-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loussert-Ajaka, I., M. L. Chaix, B. Korber, F. Letourneur, E. Gomas, E. Allen, T. D. Ly, F. Brun-Vezinet, F. Simon, and S. Saragosti. 1995. Variability of human immunodeficiency virus type 1 group O strains isolated from Cameroonian patients living in France. J. Virol. 69:5640-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loussert-Ajaka, I., D. Descamps, F. Simon, F. Brun-Vezinet, M. Ekwalanga, and S. Saragosti. 1995. Genetic diversity and HIV detection by polymerase chain reaction. Lancet 346:912-913. [DOI] [PubMed] [Google Scholar]
- 34.Loussert-Ajaka, I., T. D. Ly, M. L. Chaix, D. Ingrand, S. Saragosti, A. M. Courouce, F. Brun-Vezinet, and F. Simon. 1994. HIV-1/HIV-2 seronegativity in HIV-1 subtype O infected patients. Lancet 343:1393-1394. [DOI] [PubMed] [Google Scholar]
- 35.Luo, C. C., R. G. Downing, N. Dela Torre, J. Baggs, D. J. Hu, R. A. Respess, D. Candal, L. Carr, J. R. George, T. J. Dondero, B. Biryahwaho, and M. A. Rayfield. 1998. The development and evaluation of a probe hybridization method for subtyping HIV type 1 infection in Uganda. AIDS Res. Hum. Retrovir. 14:691-694. [DOI] [PubMed] [Google Scholar]
- 36.Montavon, C., F. Bibollet-Ruche, D. Robertson, B. Koumare, C. Mulanga, E. Esu-Williams, C. Toure, S. Mboup, E. Saman, E. Delaporte, and M. Peeters. 1999. The identification of a complex A/G/I/J recombinant HIV type 1 virus in various West African countries. AIDS Res. Hum. Retrovir. 15:1707-1712. [DOI] [PubMed] [Google Scholar]
- 37.Montavon, C., C. Toure-Kane, F. Liegeois, E. Mpoudi, A. Bourgeois, L. Vergne, J. L. Perret, A. Boumah, E. Saman, S. Mboup, E. Delaporte, and M. Peeters. 2000. Most env and gag subtype A HIV-1 viruses circulating in West and West Central Africa are similar to the prototype AG recombinant virus IBNG. J. Acquir. Immune Defic. Syndr. 23:363-374. [DOI] [PubMed] [Google Scholar]
- 38.Morgado, M. G., M. L. Guimaraes, C. B. Gripp, C. I. Costa, I. Neves, V. G. Veloso, M. I. Linhares-Carvalho, L. R. Castello-Branco, F. I. Bastos, C. Kuiken, E. A. Castilho, B. Galvao-Castro, V. Bongertz, et al. 1998. Molecular epidemiology of HIV-1 in Brazil: high prevalence of HIV-1 subtype B and identification of an HIV-1 subtype D infection in the city of Rio de Janeiro, Brazil. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 18:488-494. [DOI] [PubMed] [Google Scholar]
- 39.Myers, G., B. Korber, B. H. Hahn, K. T. Jeang, J. W. Mellors, F. E. McCutchan, L. E. Henderson, and G. N. Pavlakis (eds.). 1995. Human retroviruses and AIDS 1995: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.M.
- 40.Nkengasong, J. N., M. Kalou, C. Maurice, C. Bile, M. Y. Borget, S. Koblavi, E. Boateng, M. Sassan-Morokro, E. Anatole-Ehounou, P. Ghys, A. E. Greenberg, and S. Z. Wiktor. 1998. Comparison of NucliSens and Amplicor monitor assays for quantification of human immunodeficiency virus type 1 (HIV-1) RNA in plasma of persons with HIV-1 subtype A infection in Abidjan, Côte d'Ivoire. J. Clin. Microbiol. 36:2495-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nkengasong, J. N., C. C. Luo, L. Abouya, D. Pieniazek, C. Maurice, M. Sassan-Morokro, D. Ellenberger, D. J. Hu, C. P. Pau, T. Dobbs, R. Respess, D. Coulibaly, I. M. Coulibaly, S. Z. Wiktor, A. E. Greenberg, and M. Rayfield. 2000. Distribution of HIV-1 subtypes among HIV-seropositive patients in the interior of Côte d'Ivoire. J. Acquir. Immune Defic. Syndr. 23:430-436. [DOI] [PubMed] [Google Scholar]
- 42.Pau, C. P., M. Kai, D. L. Holloman-Candal, C. C. Luo, M. L. Kalish, G. Schochetman, B. Byers, J. R. George, et al. 1994. Antigenic variation and serotyping of HIV type 1 from four World Health Organization-sponsored HIV vaccine sites. AIDS Res. Hum. Retrovir. 10:1369-1377. [DOI] [PubMed] [Google Scholar]
- 43.Peeters, M., F. Liegeois, F. Bibollet-Ruche, D. Patrel, N. Vidal, E. Esu-Wiliams, S. Mboup, E. Mpoudi Ngole, B. Koumare, N. Nzila, J. L. Perret, and E. Delaporte. 1998. Subtype-specific polymerase chain reaction for the identification of HIV-1 genetic subtypes circulating in Africa. AIDS 12:671-673. [PubMed] [Google Scholar]
- 44.Peeters, M., and P. M. Sharp. 2000. Genetic diversity of HIV-1: the moving target. AIDS 14:S129-S140. [PubMed] [Google Scholar]
- 45.Petitjean, J., F. Vincent, M. Fretigny, A. Vabret, J. D. Poveda, J. Brun, and F. Freymuth. 1998. Comparison of two serological methods and a polymerase chain reaction-enzyme immunoassay for the diagnosis of acute respiratory infections with Chlamydia pneumoniae in adults. J. Med. Microbiol. 47:615-621. [DOI] [PubMed] [Google Scholar]
- 46.Plantier, J. C., F. Damond, M. Lasky, J. L. Sankale, C. Apetrei, M. Peeters, L. Buzelay, S. M'Boup, P. Kanki, E. Delaporte, F. Simon, and F. Barin. 1999. V3 serotyping of HIV-1 infection: correlation with genotyping and limitations. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 20:432-441. [DOI] [PubMed] [Google Scholar]
- 47.Rayfield, M. A., R. G. Downing, J. Baggs, D. J. Hu, D. Pieniazek, C. C. Luo, B. Biryahwaho, R. A. Otten, S. D. Sempala, T. J. Dondero, et al. 1998. A molecular epidemiologic survey of HIV in Uganda. AIDS 12:521-527. [DOI] [PubMed] [Google Scholar]
- 48.Robbins, K. E., L. G. Kostrikis, T. M. Brown, O. Anzala, S. Shin, F. A. Plummer, and M. L. Kalish. 1999. Genetic analysis of human immunodeficiency virus type 1 strains in Kenya: a comparison using phylogenetic analysis and a combinatorial melting assay. AIDS Res. Hum. Retrovir. 15:329-335. [DOI] [PubMed] [Google Scholar]
- 49.Robertson, D. L., J. P. Anderson, J. A. Bradac, J. K. Carr, B. Foley, R. K. Funkhouser, F. Gao, B. H. Hahn, M. L. Kalish, C. Kuiken, G. H. Learn, T. Leitner, F. McCutchan, S. Osmanov, M. Peeters, D. Pieniazek, M. Salminen, P. M. Sharp, S. Wolinsky, and B. Korber. 2000. HIV-1 nomenclature proposal. Science 288:55-56. [DOI] [PubMed] [Google Scholar]
- 50.Sherefa, K., M. Sallberg, B. Johansson, M. Salminen, and A. Sonnerborg. 1997. Subtyping of human immunodeficiency virus type 1 strains by using antibodies specific for the third variable domain (V3) of gp120: results may be affected by divergent V3 sequences. J. Clin. Microbiol. 35:2419-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sherefa, K., A. Sonnerborg, J. Steinbergs, and M. Sallberg. 1994. Rapid grouping of HIV-1 infection in subtypes A to E by V3 peptide serotyping and its relation to sequence analysis. Biochem. Biophys. Res. Commun. 205:1658-1664. [DOI] [PubMed] [Google Scholar]
- 52.Simon, F., I. Loussert-Ajaka, F. Damond, S. Saragosti, F. Barin, and F. Brun-Vezinet. 1996. HIV type 1 diversity in northern Paris, France. AIDS Res. Hum. Retrovir. 12:1427-1433. [DOI] [PubMed] [Google Scholar]
- 53.Simon, F., T. D. Ly, A. Baillou-Beaufils, V. Fauveau, J. De Saint-Martin, I. Loussert-Ajaka, M. L. Chaix, S. Saragosti, A. M. Courouce, D. Ingrand, et al. 1994. Sensitivity of screening kits for anti-HIV-1 subtype O antibodies. AIDS 8:1628-1629. [DOI] [PubMed] [Google Scholar]
- 54.Simon, F., P. Mauclere, P. Roques, I. Loussert-Ajaka, M. C. Muller-Trutwin, S. Saragosti, M. C. Georges-Courbot, F. Barre-Sinoussi, and F. Brun-Vezinet. 1998. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat. Med. 4:1032-1037. [DOI] [PubMed] [Google Scholar]
- 55.Sonnerborg, A., S. Durdevic, J. Giesecke, and M. Sallberg. 1997. Dynamics of the HIV-1 subtype distribution in the Swedish HIV-1 epidemic during the period 1980 to 1993. AIDS Res. Hum. Retrovir. 13:343-345. [DOI] [PubMed] [Google Scholar]
- 56.Subbarao, S., K. Limpakarnjanarat, T. D. Mastro, J. Bhumisawasdi, P. Warachit, C. Jayavasu, N. L. Young, C. C. Luo, N. Shaffer, M. L. Kalish, and G. Schochetman. 1998. HIV type 1 in Thailand, 1994-1995: persistence of two subtypes with low genetic diversity. AIDS Res. Hum. Retrovir. 14:319-327. [DOI] [PubMed] [Google Scholar]
- 57.Subbarao, S., C. C. Luo, K. Limpakarnjanarat, J. Bhumisa-wasdi, N. L. Young, T. D. Mastro, G. Schochetman, and M. L. Kalish. 1996. Evaluation of oligonucleotide probes for the determination of the two major HIV-1 env subtypes in Thailand. AIDS 10:350-351. [PubMed] [Google Scholar]
- 58.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Triques, K., A. Bourgeois, S. Saragosti, N. Vidal, E. Mpoudi-Ngole, N. Nzilambi, C. Apetrei, M. Ekwalanga, E. Delaporte, and M. Peeters. 1999. High diversity of HIV-1 subtype F strains in Central Africa. Virology 259:99-109. [DOI] [PubMed] [Google Scholar]
- 60.van Harmelen, J., E. van der Ryst, R. Wood, S. F. Lyons, and C. Williamson. 1999. Restriction fragment length polymorphism analysis for rapid gag subtype determination of human immunodeficiency virus type 1 in South Africa. J. Virol. Methods 78:51-59. [DOI] [PubMed] [Google Scholar]
- 61.Viazov, S., A. Zibert, K. Ramakrishnan, A. Widell, A. Cavicchini, E. Schreier, and M. Roggendorf. 1994. Typing of hepatitis C virus isolates by DNA enzyme immunoassay. J. Virol. Methods 48:81-91. [DOI] [PubMed] [Google Scholar]
- 62.Vidal, N., M. Peeters, C. Mulanga-Kabeya, N. Nzilambi, D. Robertson, W. Ilunga, H. Sema, K. Tshimanga, B. Bongo, and E. Delaporte. 2000. Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggests that the HIV-1 pandemic originated in Central Africa. J. Virol. 74:10498-10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wasi, C., B. Herring, S. Raktham, S. Vanichseni, T. D. Mastro, N. L. Young, H. Rubsamen-Waigmann, H. von Briesen, M. L. Kalish, C. C. Luo, et al. 1995. Determination of HIV-1 subtypes in injecting drug users in Bangkok, Thailand, using peptide-binding enzyme immunoassay and heteroduplex mobility assay: evidence of increasing infection with HIV-1 subtype E. AIDS 9:843-849. [DOI] [PubMed] [Google Scholar]