Abstract
Eleven monoclonal antibodies raised against recombinant Campylobacter jejuni hippurate hydrolase were tested for binding to lysates from 19 C. jejuni strains, 12 other Campylobacter strains, and 21 non-Campylobacter strains. Several monoclonal antibodies bound to C. jejuni but not to other Campylobacter species and may be useful in a species-specific immunoassay.
Campylobacter spp. are commonly implicated in gastroenteritis (1). Campylobacter jejuni is the species of Campylobacter most frequently associated with disease in humans, but other species are also capable of causing illness (6). Identification of Campylobacter isolates to the species level is important, both for distinguishing pathogenic species from those not associated with disease in humans and for studying the epidemiology of Campylobacter-induced illness. There is a need for a rapid, easy-to-use test, such as an immunoassay, for identification of suspect Campylobacter isolates to the species level. The ability of C. jejuni to hydrolyze N-benzoylglycine (hippurate) to benzoic acid and glycine is commonly used to distinguish it from other Campylobacter species (7). This ability is due to the presence of a hippurate hydrolase enzyme (EC 3.5.1.32) which appears to be unique to C. jejuni (3), suggesting that it would make a good target for an immunoassay used to identify C. jejuni. The purpose of this study was to generate monoclonal antibodies specific for the hippurate hydrolase of C. jejuni. The specificity of the monoclonal antibodies was determined by screening using enzyme-linked immunosorbent assays (ELISAs) and Western blots of whole-cell lysates from strains of C. jejuni, other Campylobacter species, other enteric bacteria, and other hippurate hydrolase-positive non-Campylobacter bacteria. The same strains were tested for hippurate hydrolase activity (4) and were screened by colony blotting and Southern blotting for binding to a DNA probe shown to be specific for the hippurate hydrolase enzyme of C. jejuni (3).
Glutathione S-transferase (GST)-tagged recombinant hippurate hydrolase was produced by amplifying the hipO gene of C. jejuni ATCC 43431 by PCR (with the primers 5′CTCGGATCCATGAATTTAATTCCAGAA3′ and 5′GAGGAATTCTTATTTTAAGTATTTTAAAG3′), using pHipO (3) as a template and introducing BamHI and EcoRI sites on either side of the gene. The amplified DNA was subcloned into BamHI and EcoRI sites of pGEX-2T (Amersham Pharmacia Biotech Inc., Baie d'Urfé, Quebec, Canada) and introduced into Escherichia coli JM101 by CaCl2 transformation (Bulk Redi-Pack GST Purification Modules; Amersham Pharmacia). The fusion protein was produced in E. coli and purified by glutathione-Sepharose affinity chromatography (Bulk Redi-Pack GST Purification Modules; Amersham Pharmacia). Fifty-microgram aliquots of GST-fusion protein in Freund's incomplete adjuvant were used to immunize adult BALB/c mice subcutaneously three to five times, at 2-week intervals. Monoclonal antibodies were prepared using polyethylene glycol fusion and hypoxanthine-aminopterin-thymidine supplement to select for fused cells (2, 5). Culture supernatants were screened for antibody binding to GST-hippurate hydrolase by using 3 μg of the antigen ml−1 on ELISAs and 0.75 μg of the antigen per lane on Western blots. Cells from positive wells were cloned twice by limiting dilution. Eleven monoclonal antibodies that recognized GST-hippurate hydrolase were isolated, and culture supernatants from these antibodies were harvested and frozen at −20°C. All of the antibodies were determined to be immunoglobulin G1 isotype using the Isostrip Mouse Monoclonal Antibody Isotyping Kit (Roche Diagnostics; Laval, Quebec, Canada).
Whole-cell lysates used to screen the monoclonal antibodies by ELISA and Western blotting were prepared from 19 strains of C. jejuni (ATCC 43431, ATCC 49349, ATCC 29428, and 16 nonreference strains), six nonreference strains of C. coli, six strains of C. lari (ATCC 35221, NCTC 11352, and four nonreference strains), five strains of C. hyointestinalis (ATCC 35212, ATCC 25217, and three nonreference strains), six strains of C. upsaliensis (ATCC 43954 and five nonreference strains), one strain of C. mucosalis (ATCC 43264), one strain of Arcobacter butzleri (ATCC 49616), two strains of E. coli (ATCC 25922 and ATCC 43894), two strains of Salmonella spp. (ATCC13076 and ATCC 8391), two strains of Shigella spp. (ATCC 12022 and ATCC 25931), five strains of Listeria monocytogenes (ATCC 19115 and four nonreference strains), five strains of Listeria innocua (ATCC 33091 and four nonreference strains), one nonreference strain of Corynebacterium matruchotii, two strains of Streptococcus agalactiae (ATCC 12386 and ATCC 13813), and one nonreference strain of Streptococcus uberis. Nonreference strains were isolated from food, environmental, or clinical samples and identified by established biochemical methods. Strains of C. jejuni, L. monocytogenes, L. innocua, C. matruchotii, S. agalactiae, and S. uberis, but not the other species, were found to possess hippurate hydrolase activity. To prepare whole bacterial lysates, the bacteria were grown on Trypticase soy agar plus 5% sheep blood agar at 37°C for 24 h. Campylobacter strains were grown in anaerobic jars that were evacuated and filled three times with a microaerophilic gas mixture. Bacteria were harvested, suspended in phosphate-buffered saline, lysed by sonication, and cleared by centrifugation at 12,000 × g for 5 min in a microcentrifuge. The lysates were used as antigens on ELISAs and Western blots at 100 μg ml−1 and 40 μg per lane, respectively.
The monoclonal antibodies could be arranged into three groups, based on their species specificities. Group I contained three monoclonal antibodies that bound to lysates from C. jejuni strains and from two out of five strains of the hippurate hydrolase-positive L. monocytogenes strains tested. Group II contained five monoclonal antibodies that bound not only to lysates from C. jejuni strains and from the two L. monocytogenes strains but also bound to lysates from other Campylobacter species and the closely related A. butzleri. Group III contained three antibodies. All three antibodies showed a poor ability to distinguish C. jejuni from other species on ELISAs. On Western blotting, however, one of the antibodies from group III recognized lysates only from C. jejuni strains and the two L. monocytogenes strains, and the remaining two antibodies, 345-1-15 and 167-3-1, recognized lysates only from C. jejuni and not from the two L. monocytogenes strains. The antibodies recognized a single protein of 42.4 ± 0.8 kDa (average ± standard deviation) on Western blots of C. jejuni lysates. This was similar to the predicted molecular mass of the hippurate hydrolase enzyme (3). In the two L. monocytogenes strains, a single protein of 46.7 ± 0.1 kDa was recognized by the antibodies. The proteins recognized in the other Campylobacter strains and the A. butzleri strain were of variable sizes. None of the antibodies recognized whole-cell lysates from C. jejuni strain ATCC 43431 in which the hipO gene was interrupted by insertion of a kanamycin resistance gene. A summary of the binding of the group I monoclonal antibody 364-10-9 to Western blots of the strains tested in this study is shown in Fig. 1.
FIG. 1.
Monoclonal antibody 364-10-9 binding of whole-cell lysates of C. jejuni, other Campylobacter species, and other bacterial species. Lane 1, prestained low-molecular-weight standards; lane 2, C. jejuni ATCC 43431; lane 3, C. jejuni ATCC 43431 hip− mutant; lane 4, C. coli; lane 5, C. lari ATCC 35221; lane 6, C. hyointestinalis; lane 7, E. coli ATCC 25922; lane 8, L. monocytogenes ATCC 19119.
Colony blots and Southern blots were prepared as described elsewhere (DIG User's Guide for Filter Hybridization; Boehringer Mannheim GmbH), hybridized overnight at 42°C in standard buffer plus 50% formamide containing 37.5 ng of probe ml−1 and using UV light to cross-link DNA to nylon membranes. Southern blots were prepared from 5 μg of HindIII-digested genomic DNA per lane as described previously (8) and quantitated with the PicoGreen quantification reagent for double-stranded DNA (Molecular Probes, Eugene, Oreg.). The hipO-specific DNA probe was prepared by digesting a pUC19 plasmid containing a 0.8-kb HindIII fragment of hipO (3) with HindIII, gel purifying the fragment, and labeling it with digoxigenin using a Digoxigenin DNA Labeling and Detection Kit (Boehringer Mannheim, Indianapolis, Ind.). Blots were developed using a DIG Luminescent Detection Kit for Nucleic Acids (Boehringer Mannheim) with CSPD as substrate. The colony blots showed strong binding of the probe only to C. jejuni strains. A single band of 1.71 to 2.43 kb was observed on Southern blots of HindIII-digested genomic DNA isolated from C. jejuni strains.
Several of the monoclonal antibodies showed promise for use in an immunoassay. The group I monoclonal antibody 364-10-9 generated a strong signal on both ELISAs and Western blots of C. jejuni whole-cell lysates but did not display detectable binding to lysates from other Campylobacter species, suggesting that this antibody had the potential to be used in an assay for identification of C. jejuni among presumptive Campylobacter spp. isolates. While this antibody also recognized two of the hippurate hydrolase-positive L. monocytogenes strains tested, this binding would not interfere with an assay used for identification of C. jejuni from pure cultures of potential Campylobacter spp. isolates, since Listeria spp. are unlikely to be coisolated on selective media with Campylobacter spp. The binding of 364-10-9 to the L. monocytogenes strains did not correlate with hippurate hydrolase activity, as all of the Listeria spp. isolates included in the study were hippurate hydrolase-positive and only two strains were recognized by the monoclonal antibodies. The protein recognized by 364-10-9 in these strains could be a different enzyme isotype or an unrelated protein with structural similarity to hippurate hydrolase. The colony blotting and Southern blotting confirmed that the hipO gene appears to be restricted to C. jejuni and is absent from other hippurate hydrolase-positive bacteria, such as the two L. monocytogenes strains.
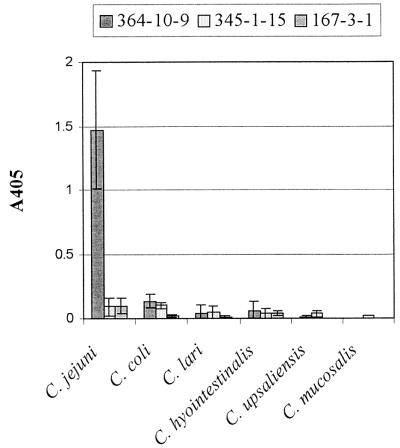
The two group III monoclonal antibodies that recognized the C. jejuni strains but did not recognize the two L. monocytogenes strains on Western blotting, 345-1-15 and 167-3-1, might also be used in an assay for identification of C. jejuni isolates. The usefulness of these two monoclonal antibodies, however, is limited by the failure of these antibodies to differentiate C. jejuni from other Campylobacter species by ELISA (Fig. 2).
FIG. 2.
Binding of monoclonal antibodies 364-10-9, 345-1-15, and 167-3-1 to whole-cell lysates of Campylobacter species by ELISA. Columns represent the average ± standard deviation of the average values obtained from all strains of each species tested. Each average value was originally determined from four replicate tests after subtraction of sample blank values. The number of strains tested for C. jejuni, C. coli, C. lari, C. hyointestinalis, C. upsaliensis, and C. mucosalis was 19, 6, 6, 5, 6, and 1, respectively.
Acknowledgments
We gratefully acknowledge the financial support of the Ontario Ministry of Agriculture, Food, and Rural Affairs and Vita-tech Canada Inc.
We thank Eric Hani for his contribution of the pHipO plasmid and the pUC19 plasmid containing the 0.8-kb HindIII fragment of the hipO gene, David Woodward and Lawrence Price of the National Laboratory for Enteric Pathogens, Health Canada, for their contribution of nonreference C. jejuni, C. coli, C. lari, C. hyointestinalis, C. upsaliensis, and C. mucosalis strains, and Anna Lammerding of the Health of Animals Laboratory, Health Canada, for her contribution of nonreference L. monocytogenes and L. innocua strains.
REFERENCES
- 1.Altekruse, S. F., N. J. Stern, P. I. Fields, and D. L. Swerdlow. 1999. Campylobacter jejuni—an emerging foodborne pathogen. Emerg. Infect. Dis. 5:28-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galfre, G., and C. Milstein. 1975. Preparation of monoclonal antibodies: strategies and procedures. Methods Enzymol. 73:3-46. [DOI] [PubMed] [Google Scholar]
- 3.Hani, E. K., and V. L. Chan. 1995. Expression and characterization of Campylobacter jejuni benzoylglycine amidohydrolase (hippurate hydrolase) gene in Escherichia coli. J. Bacteriol. 177:2396-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunt, J. M., C. Abeyta, and T. Tran. 1998. Campylobacter, p. 7.01-7.24. In U.S. Food and Drug Administration bacteriological analytical manual, 8th ed. AOAC International, Gaithersburg, Md.
- 5.Lam, J. S., L. A. MacDonald, M. Y. C. Lam, L. G. M. Duchesne, and G. G. Southam. 1987. Production and characterization of monoclonal antibodies against serotype strains of Pseudomonas aeruginosa. Infect. Immun. 55:1051-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mishu, B., C. M. Patton, and R. V. Tauxe. 1992. Clinical and epidemiological features of non-jejuni, non-coli Campylobacter species, p. 31-41. In I. Nachamkin, M. J. Blaser, and L. S. Tompkins (ed.), Campylobacter jejuni: current status and future trends. American Society for Microbiology, Washington, D.C.
- 7.Nachamkin, I. 1995. Campylobacter and Arcobacter, p. 483-491. In P. M. Murray (ed.), Manual of clinical microbiology, 6th ed. ASM Press, Washington, D.C.
- 8.Wilson, K. 1992. Preparation of genomic DNA from bacteria, p. 2-10-2-12. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Short protocols in molecular biology, 2nd ed. John Wiley & Sons, New York, N.Y.