Abstract
We have evaluated the VERSANT HCV RNA 3.0. Assay (HCV 3.0 bDNA assay) (Bayer Diagnostics, Berkeley, Calif.), which is an improved signal amplification procedure for the HCV 2.0 bDNA assay for the quantitation of hepatitis C virus (HCV) RNA in serum or plasma of HCV-infected individuals. The HCV 3.0 bDNA assay has a linear dynamic range of 2.5 × 103 to 4.0 × 107 HCV RNA copies per ml (c/ml). The performance of the HCV 3.0 bDNA assay was evaluated using three different test panels. An overall specificity of 96.8% relative to the detection limit of the HCV 3.0 bDNA assay was found. The intra- and interrun reproducibilities for both the dilution panel and the NAP (AcroMetrix, Benicia, Calif.) panel were consistent with coefficients of variation of less than 9%. Quantitation with the HCV 3.0 bDNA assay was linear over the entire range of both panels (ranges of 4.4 × 103 to 3.5 × 106 c/ml and 5 × 103 to 2 × 106 IU/ml, respectively), with correlation coefficients of 0.999, slopes close to one, and intercepts close to zero. The regression equation indicated that 1 IU corresponded to about 4.8 copies of HCV RNA. A correlation coefficient of 0.941 was found for HCV RNA values (in international units per milliliter) obtained from the HCV 3.0 bDNA assay and the HCV Monitor version 2.0 assay (HCV Monitor 2.0 assay) (Roche Diagnostic Systems, Branchburg, N.J.). Quantitative results obtained close to the lower limit of the HCV 3.0 bDNA assay might imply that its lower limit should be reconsidered and raised, if necessary. It appeared that quantitation values obtained from the HCV Monitor 2.0 assay of between 5 × 102 and 105 IU/ml were in general higher than those obtained from the HCV 3.0 bDNA assay, whereas values obtained from the HCV Monitor 2.0 assay were underestimated for samples with HCV RNA levels above 105 IU/ml.
Approximately 170 million individuals are chronically infected with hepatitis C virus (HCV) worldwide. The measurement of HCV RNA levels in plasma has become an important tool for monitoring individuals during antiviral therapy. Considerable advances have been made in the therapy of HCV during recent years. Pretreatment HCV RNA loads and genotypes have been found to be independent predictors of a sustained response after combination therapy with interferon and ribavirin (5, 11). Moreover, during induction therapies with interferon, measurement of HCV RNA early in therapy has been found to be predictive of the outcome of treatment (7, 14). With the introduction of new therapies and new antiviral drugs, such as pegylated interferon (8, 13), early detection, tailoring, and frequent quantitation of HCV RNA in plasma over a wide dynamic range will become extremely important.
Several versions of commercially available assays with different amplification and detection methods have been used for the quantitation of HCV RNA. Unfortunately, results of these assays are difficult to compare due to lack of standardization to a common or universal standard. These assays use different units of measurements (e.g., copies and genome equivalents) and differ in their linear ranges (1, 4, 10). The limitations of using different units can be overcome by the introduction of a World Health Organization (WHO) international standard, which ensures a consistent standard across all test methods (12). This international standard has been used as the primary reference for the development of an HCV RNA quantitation panel (NAP; AcroMetrix, Benicia, Calif.) in the range of 0 to 2 × 106 IU/ml for a variety of commercially available and in-house nucleic acid testing technologies (3). Roche Diagnostic Systems (Branchburg, N.J.) recently released the Cobas Amplicor HCV Monitor version 2.0 assay (HCV Monitor 2.0 assay), giving results in international units per milliliter. The introduction of an international standard will simplify the recommendation of the International Consensus Conference on HCV in 1999 to use a defined threshold of 2 × 106 c/ml copies/ml (as a guideline for determining the outcome of treatment with interferon and ribavirin in patients infected with HCV genotypes 1, 4, and 5 (2). A recent study showed that this clinically relevant threshold of 2 × 106 c/ml was equal to about 8 × 105 IU/ml (9). In the present study, we evaluated the specificity, reproducibility, and linearity of the recently developed and improved VERSANT HCV RNA 3.0 assay (HCV 3.0 bDNA assay) (Bayer Diagnostics, Berkeley, Calif.) for the quantitation of HCV RNA in plasma. In addition, a comparison was made between quantitation values (in international units per milliliter) obtained from the HCV Monitor 2.0 assay and the HCV 3.0 bDNA assay.
MATERIALS AND METHODS
Quantitation of HCV RNA.
Quantitation of HCV RNA was performed according to each manufacturer's instructions.
HCV Monitor 2.0 assay.
The HCV Monitor 2.0 assay uses 200 μl of plasma or serum and has reported lower and upper detection limits of 5 × 102 and 5.0 × 105 IU/ml, respectively (4). Specimens yielding values above the upper limit were routinely diluted 100-fold and retested, and obtained values were multiplied by this dilution factor to obtain the actual HCV RNA concentration in international units per milliliter.
HCV 3.0 bDNA assay.
The HCV 3.0 bDNA assay uses 50μl of plasma or serum and has, according to the manufacturer, lower and upper detection limits of 2.5 × 103 and 4.0 × 107 c/ml, respectively. The bDNA technology is used in a sandwich nucleic acid hybridization procedure. Briefly, the HCV 3.0 bDNA assay provides an improved assay for singular measurements of plasma and serum samples. Greater sensitivity was achieved by improvements in probe design and number of probes, and assay background was reduced by the incorporation of synthetic nonnatural nucleosides. A second level of background reduction was achieved by a redesign of the label extenders. The whole process is performed in a semiautomated Bayer System 340 bDNA analyzer, which automatically performs all incubations, washing steps, readings, and data processing. The instrument can process two 96-microwell plates per run, i.e., 168 patient samples and 24 calibrators/day. The lot number of the kits used in this study was W004A1.
Test panels.
For testing the analytical performance of the HCV 3.0 bDNA assay, the following test materials were used: (i) 96 HCV-seronegative serum samples obtained from the Center for Blood Research, Sacramento, Calif.; (ii) a six-member dilution panel consisting of HCV-3a (ProMedDx, LLC, Norton, Mass.; provided by Bayer and quantified by the HCV 3.0 bDNA assay) diluted in HCV-seronegative plasma with quantitation values expressed in copies per milliliter (range, <2.5 × 103 to 2.7 × 106 c/ml); and (iii) a six-member HCV RNA quantitation panel (NAP) consisting of HCV-1b diluted in HCV-seronegative plasma with quantitation values expressed in international units per milliliter (range, 0 to 2.0 × 106 IU/ml). The NAP HCV RNA quantitation panel was calibrated against the WHO First International Standard (12) and was used to determine the factor for conversion of copies to international units for the HCV 3.0 bDNA assay.
Analytical performance of the HCV 3.0 bDNA assay.
Specificity, reproducibility, and linearity were assessed using the test panels. The same operator ran one full plate on four different days. A full plate run consisted of testing each panel member from either the dilution panel or the NAP panel in replicates of six as well as 48 seronegative samples in single wells. Both panel members were tested in replicates of six in two runs, whereas each seronegative sample was tested in two runs. Thus, after four runs there were 12 results for each panel member and 2 results for each seronegative sample. Results within the lower and upper quantitative limits of the HCV 3.0 bDNA assay were used for the evaluation of intra- and interrun reproducibilities and linearity. In addition, the linearity of the HCV 3.0 bDNA assay up to the upper threshold was assessed in single runs with twofold dilutions of five patient samples with HCV RNA levels above the upper limit.
Patient population and specimen collection.
Blood specimens were collected from HCV-infected individuals monitored at the Section of Hepatology of the Academic Medical Center. Blood specimens were drawn as part of usual follow-up and treatment monitoring, and approximately 25% of individuals were treatment naive. EDTA-treated plasma samples were processed and frozen at −70°C within 24 h of collection.
Quantitative relationship.
To compare the quantitative correlation of the assays, results for 151 patient samples were produced and stratified according to the dynamic ranges and the thresholds of both assays. Sixty-one of the specimens contained levels of HCV RNA that were within the dynamic ranges of both of the assays. Forty-one of the specimens required dilution to enable quantitation values to be within the dynamic range of the COBAS AMPLICOR HCV Monitor 2.0 assay.
Genotyping.
HCV genotypes of clinical samples of patients receiving treatment were determined by direct sequencing using a TruGene HCV genotyping assay and an OpenGene automated DNA sequencing system (Visible Genetics Inc., Toronto, Ontario, Canada).
Statistical analysis.
Variability between replicate tests and runs was described as the mean HCV RNA levels obtained with the HCV 3.0 bDNA assay, the standard deviation (SD), and the coefficient of variation (CV) by using the statistical functions of SPSS version 9.01 software. Linear regression analysis was done using scatter plots for log-transformed HCV RNA levels. The correlation coefficient, slope, and intercept were obtained by least-squares linear regression analysis.
RESULTS
Reproducibility of the HCV 3.0 bDNA assay.
Reproducibility was evaluated with two different panels without retesting any sample. The intra- and interrun reproducibilities were assessed by testing each panel member in replicates of six in two different runs. The CVs for quantitation ranged from 4.9 to 9% for the dilution panel and from 5.9 to 8.7% for the NAP panel.
Linearity of the HCV 3.0 bDNA assay.
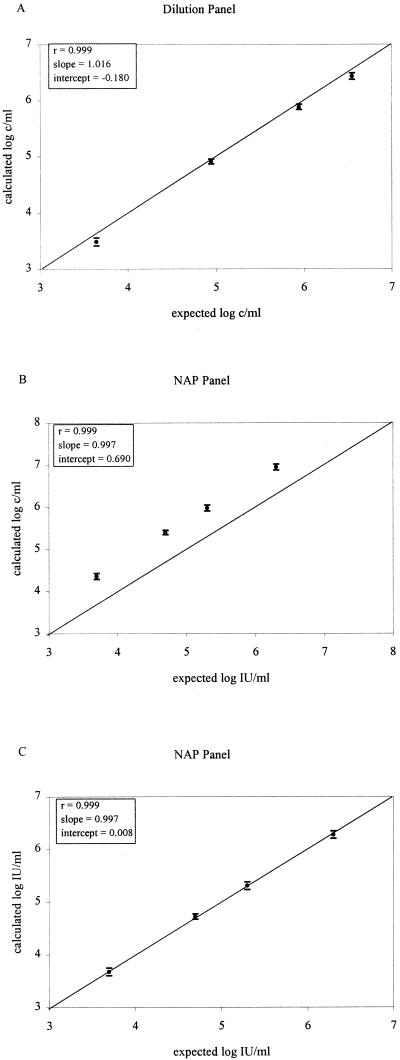
Linearity was determined with 12 replicates of each member of the dilution panel or NAP panel. The comparison was done using the mean from the log-transformed values of the calculated HCV RNA concentrations and two times the SD (95% confidence interval) and log-transformed values of the expected HCV RNA input concentrations. Regression analysis showed a good correlation for the dilution panel, but there was a downward quantitation bias relative to the expected values expressed in copies per milliliter (Fig. 1A; r, 0.999; slope, 1.016; intercept, −0.180). The results for the NAP panel are shown in Table 1. Panel members with HCV RNA levels below the lower limit of 2.5 × 103 c/ml were excluded from regression analysis. Following logarithmic transformation of these results, a good correlation was found with the values expected for the panel members with a slope close to one but with an intercept of 0.690, representing the difference between copies and international units with the HCV 3.0 bDNA assay (Fig. 1B). Based upon the resulting linear regression analysis, the following regression equation was found: log c/ml = (0.997 log IU/ml) + 0.690. This equation indicated that 1 IU is equal to about 4.8 copies. After conversion to IU, an excellent correlation was found with the expected values expressed in international units per milliliter for the NAP panel (Fig. 1C; r, 0.999; slope, 0.997; intercept, 0.008). The conversion factor of 4.8 was used in all subsequent experiments by an adaptation of the software (calculating results in both copies per milliliter and international units per milliliter).
FIG. 1.
Linearity determined using the mean from the log-transformed values for 12 replicates of the two panels and two times the SD (95% confidence interval). (A) Calculated values expressed as copies per milliliter for the dilution panel (genotype 3a) in comparison with expected values, in the range of 3.0 × 103 to 2.7 × 106 c/ml. (B) Calculated values expressed as copies per milliliter for the NAP panel (genotype 1b) in comparison with expected values, in the range of 4.7 × 103 to 2.0 × 106 IU/ml. (C) Comparison of calculated values for the NAP panel (genotype 1b) after conversion to international units [after using the regression equation log c/ml = (0.9970 log IU/ml) + 0.690] with expected values, in the range of 4.7 × 103 to 2.0 × 106 IU/ml. The ideal regression line (y = x) is depicted.
TABLE 1.
Evaluation of the HCV 3.0 bDNA assay with the NAP panel
| Panel member (n = 12) | Expected IU/ml | Calculated c/ml (mean ± 2 SD) | Ratio (copies/IU) |
|---|---|---|---|
| 000 | 0 | <2,500 | |
| 002 | 500 | <2,500 | |
| 003 | 5,000 | 22,804 ± 3,776 | 4.56 ± 0.76 |
| 004 | 50,000 | 250,882 ± 29,556 | 5.02 ± 0.59 |
| 005 | 200,000 | 968,075 ± 159,266 | 4.84 ± 0.80 |
| 007 | 2,000,000 | 9,025,995 ± 1,444,736 | 4.51 ± 0.72 |
Specificity of the HCV 3.0 bDNA assay.
Ninety-six unique seronegative serum samples were tested in single wells in four independent runs (48 specimens per run, i.e., 96 results in duplicate) to assess the specificity relative to the lower limit of 520 IU/ml of the HCV 3.0 bDNA assay. Results for 90 samples were below 520 IU/ml, whereas 6 samples had discordant results just above the lower limit of 520 IU/ml (range, 596 to 991 IU/ml) in duplicate runs, revealing a specificity of 93.8%. When results from the first and second runs were discrepant, a singular retest of the particular sample was done. After retesting of the six discordant samples, five samples had HCV RNA levels below 520 IU/ml, whereas 1 had a result of 573 IU/ml; the original results for this sample were <520 and 596 IU/ml, revealing an overall specificity of at least 96.8%.
Correlation between the HCV 3.0 bDNA assay and the HCV Monitor 2.0 assay.
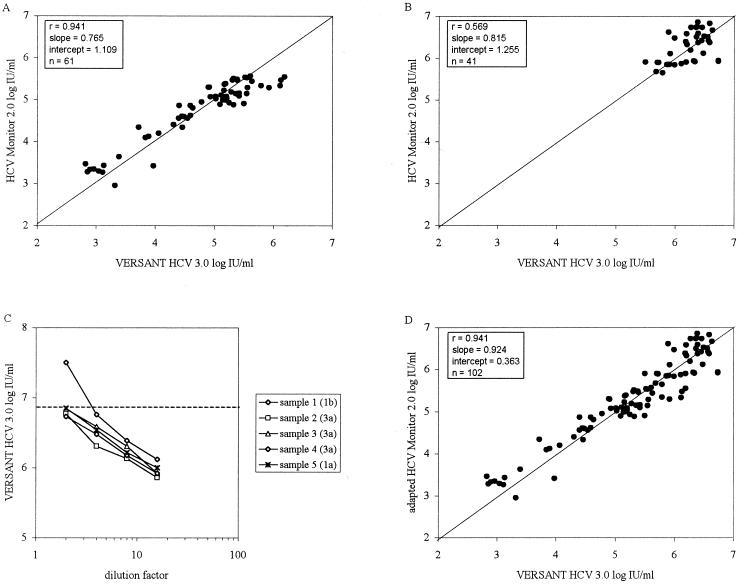
To determine the correlation between the HCV 3.0 bDNA assay and the HCV Monitor 2.0 assay, 151 patient samples were quantified. Of these patient samples, 102 had results available for analyses, i.e., values within the dynamic ranges of both assays (n = 61) and values that required dilution of the plasma specimens in the HCV Monitor 2.0 assay (n = 41). The correlation within the dynamic range of the HCV Monitor 2.0 assay was assessed with 61 paired patient samples. As shown in Fig. 2A, results obtained with both assays correlated well, but results obtained with the HCV Monitor 2.0 assay above approximately 105 IU/ml were nonlinear, resulting in a slope of 0.765 and an intercept of 1.019. Samples with HCV RNA levels above the upper limit of the HCV Monitor 2.0 assay (n = 41) were diluted 100-fold in seronegative plasma and were retested to extend the upper limit of the HCV Monitor 2.0 assay for comparison. The relationship between the two assays for values above 5.0 × 105 IU/ml showed a correlation of 0.569, a slope of 0.815, and an intercept of 1.255 (Fig. 2B). To determine if results above 5 × 105 IU/ml in the two assays were influenced by the potential nonlinearity of the HCV 3.0 bDNA assay, serial twofold dilutions of plasma samples from five patients with high levels of HCV RNA (i.e., above the upper limit of the HCV 3.0 bDNA assay) were quantified. A linear association was found for dilution series for all five plasma samples, independent of the genotype (Fig. 2C). Combining all data from the 102 patients (using results obtained by the HCV Monitor 2.0 assay for undiluted and diluted specimens) showed a high overall correlation of 0.924, a slope close to one, and an intercept of 0.363 (Fig. 2D).
FIG. 2.
Comparison of the HCV 3.0 bDNA assay with the HCV Monitor 2.0 assay within the dynamic ranges of the assays. The ideal regression line (y = x) is depicted. (A) Correlation of the HCV 3.0 bDNA assay with values within the dynamic range of the HCV Monitor 2.0 assay. (B) Correlation of the HCV 3.0 bDNA assay with values obtained for 100-fold-diluted plasma samples with HCV RNA loads above the upper limit of the HCV Monitor 2.0 assay. (C) Serial twofold dilutions of plasma samples from five patients with values above the upper limit of the HCV 3.0 bDNA assay. The genotypes are indicated in parentheses. The broken line indicates the upper limit of detection of the HCV 3.0 bDNA assay. (D). Overall correlation between the HCV 3.0 bDNA assay and values within the dynamic range as well as values obtained for 100-fold-diluted plasma samples with HCV RNA loads above the upper limit of the HCV Monitor 2.0 assay.
Influence of HCV genotypes on both quantitative assays.
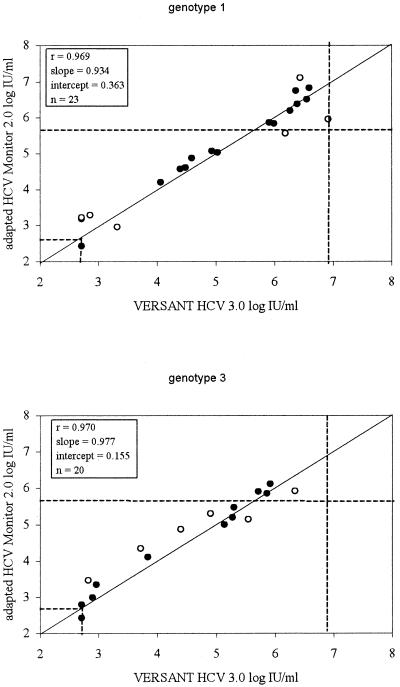
Genotypes were determined for samples from patients receiving antiviral treatment for HCV. Of the 102 samples, 50 samples from different patients were genotyped. We found genotype 1 in 23 samples (46%), genotype 3 in 20 samples (40%), genotype 4 in 4 samples (8%), and genotype 2 in 3 samples (6%). In 37 of 50 samples (74%), equivalent quantitation (i.e., less than a 0.3-log-unit difference in quantitation values between methods) was found with both quantitative assays, independent of the genotypes found. A good correlation between the assays for genotypes 1 and 3 was found, with slopes approaching one and intercepts close to zero (Fig. 3). Regression analysis for patients infected with genotypes 2 and 4 was not performed due to the small sample size. Differences of more than 0.3 log unit in quantitation values for the two methods were found in samples from 13 patients: 6 infected with genotype 1, 6 infected with genotype 3, and 1 infected with genotype 2.
FIG. 3.
Correlation of the HCV 3.0 bDNA assay with values obtained for 100-fold-diluted plasma samples with HCV RNA loads above the upper limit of the HCV Monitor 2.0 assay in relation to genotypes 1 and 3. The ideal regression line (y = x) is depicted, and the broken lines indicate the lower limits and upper limits of detection for both assays. Discordant results (i.e., more than a 0.3-log-unit difference between values for both assays) are indicated by open circles.
Lower limit of the HCV 3.0 bDNA assay.
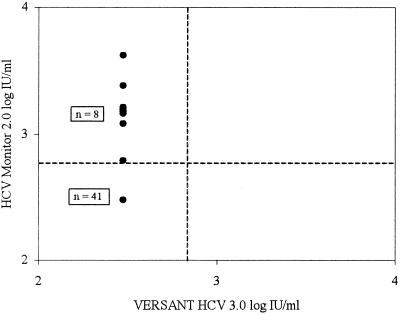
For the 49 samples with HCV RNA levels below the lower limit of either one of the assays, discordant results were analyzed. Of these 49 samples, 41 were found not to have detectable HCV RNA by both assays. Eight samples had HCV RNA levels below the lower limit of the HCV 3.0 bDNA assay while having HCV RNA levels just above the lower limit of the HCV Monitor 2.0 assay, ranging between 6.2 × 102 and 4.2 × 103 IU/ml (Fig. 4).
FIG. 4.
Correlation of the HCV 3.0 bDNA assay with the HCV Monitor 2.0 assay in relation to the lower limit of the HCV 3.0 bDNA assay. The number of samples is indicated, and the broken lines indicate the lower limits of detection for both assays.
DISCUSSION
In the present study, we evaluated the HCV 3.0 bDNA assay for the quantitation of HCV RNA in plasma. The performance characteristics were established using well-standardized panels. Excellent reproducibility (intra- and interassay run variability of less than 9% [CV]) and specificity (≥96.8%) relative to the lower detection limit of 520 IU/ml were found. Overall, the HCV 3.0 bDNA assay displayed good linearity within the dynamic range of the quantitation panels because the slopes were close to one and the intercepts approached zero (with a 95% confidence interval).
Differences in standardization for the quantitation of HCV RNA make comparison between assays difficult. Standardization for the measurement of HCV RNA levels is therefore a mandatory step, especially when a clinically relevant threshold is used as a guideline to define the duration of treatment (2). The limitations of standardization can be overcome in part by the introduction of a WHO international standard to ensure a consistent standard across all test methods (12). The introduction of the international unit as the standard measurement for HCV RNA levels makes comparison between different test formats possible and easier to interpret (3). Therefore, standardization will be important for more reliable quantitation of HCV RNA levels in clinical studies and in the decisions made during treatments.
Our results obtained with the NAP HCV RNA quantitation panel showed that quantitation with the HCV 3.0 bDNA assay was linear over a wide range, and a factor of about 4.8 was found for conversion from copies to international units. Using this conversion factor, a direct comparison between the HCV Monitor 2.0 assay and the HCV 3.0 bDNA assay was possible.
From this comparison, it appeared that values above approximately 105 IU/ml were underestimated by the HCV Monitor 2.0 assay and linearity was lost in comparison to the HCV 3.0 bDNA assay. This underestimation in viral quantitation by the HCV Monitor 2.0 assay may have consequences for reliable identification of individuals with viral levels above the important guideline threshold of 8 × 105 IU/ml.
This problem can be overcome by repeating the HCV Monitor 2.0 assay with diluted samples close to and above the upper threshold of the HCV Monitor 2.0 assay, giving HCV RNA levels well within the dynamic range. Considering the reported upper limit of 5 × 105 IU/ml for the HCV Monitor 2.0 assay, we routinely diluted 100-fold all samples with results above this upper limit. The linear correlation between both assays when the diluted samples were reanalyzed in the HCV Monitor 2.0 assay was poor, although the slope was closer to one and the majority of data points were well within the 0.3-log-unit difference between both assays. However, in a small number of cases, values obtained from the HCV Monitor 2.0 assay with diluted specimens seemed to be underestimated relative to values obtained from the HCV 3.0 bDNA assay. Thus, 100-fold dilution may not always be sufficient for high-titer specimens to obtain values within the linear range of the HCV Monitor 2.0 assay. A recent study on the quantitation range of the HCV Monitor 2.0 assay showed a similar “saturation” effect influencing the quantitation of clinical specimens independent of the genotypes (6).
In our study, a few samples, after 100-fold dilution, had values that were clearly higher in the HCV Monitor 2.0 assay than in the HCV 3.0 bDNA assay. To examine whether a nonlinear relationship around the upper limit also influenced the HCV 3.0 bDNA assay, we analyzed (besides two well-defined panels) twofold serial dilutions of five clinical samples with HCV RNA levels above the upper limit of 8.3 × 106 IU/ml. This experiment showed that the HCV 3.0 bDNA assay was linear in the upper range of quantitation for these clinical specimens independent of the genotypes present in the specimens. A possible explanation for values being higher in the HCV Monitor 2.0 assay with diluted samples is that at viral loads of between 103 and 105 IU/ml, values from the HCV Monitor 2.0 assay are in general higher than those from the HCV 3.0 bDNA assay. After dilution of specimens, many of the quantitation values from the HCV Monitor 2.0 assay will fall in this lower range; therefore, after multiplication by the dilution factor, the values will be higher than those from the HCV 3.0 bDNA assay.
Overall, nearly equivalent quantitation (less than a 0.3-log-unit difference) for HCV genotypes 1 and 3 was found with both assays. Patients infected with genotypes 2 and 4 showed similar results, but the number of samples with these genotypes was too low for analysis. Although the number of samples with known genotypes was limited, our results may imply that the differences obtained between the quantitation assays were independent of the genotypes and were related more to the upper limit of the HCV Monitor 2.0 assay and the lower limits of the HCV Monitor 2.0 assay and the HCV 3.0 bDNA assay.
The specificity relative to the lower limit of 520 IU/ml of the HCV 3.0 bDNA assay was assessed using 96 unique seronegative serum samples, which were tested in single wells in four independent runs (48 specimens per run), providing 96 duplicate results. Results for 90 samples were below 520 IU/ml, whereas 6 samples had discordant results between duplicate runs, just above the lower limit of 520 IU/ml (range, 596 to 991 IU/ml), for a specificity of 93.8%. After the six discordant samples were retested, five samples had HCV RNA levels below 520 IU/ml, whereas 1 had a result of 573 IU/ml (the original results for this sample were <520 and 596 IU/ml), for an overall specificity of at least 96.8%. To examine whether this high specificity could be used for comparison with the HCV Monitor 2.0 assay, 49 samples with HCV RNA levels below the lower limit of either one of the assays and with discordant results were analyzed. Among the 49 samples, 41 samples had results below the lower limit of both assays, and 8 samples had HCV RNA levels below the lower limit of the HCV 3.0 bDNA assay; however, the latter had HCV RNA levels in the HCV Monitor 2.0 assay ranging between 6.2 × 102 and 4.2 × 103 IU/ml. Assuming 100% specificity for the HCV Monitor 2.0 assay (4), these results may imply that the lower limit of the HCV 3.0 bDNA assay should be reconsidered and raised, if necessary. In general, quantitation values obtained from the HCV Monitor 2.0 assay (between 5 × 102 and 1 × 105 IU/ml) were higher than those obtained from the HCV 3.0 bDNA assay. The reason for the overall remaining subtle differences between the assays is not known but may be related to the intrinsic differences between target amplification-based and signal amplification-based assays.
In summary, this study describes the performance of the recently introduced HCV 3.0 bDNA assay for the quantitation of HCV RNA in plasma and serum, expressed in copies per milliliter. Regression analysis revealed a conversion factor of about 4.8 between international units per milliliter and HCV RNA copies per milliliter. The assay uses a 50-μl sample volume and has a high throughput of up to 168 patient results per day. The HCV 3.0 bDNA assay is highly reproducible and specific and is able to quantify HCV RNA over a broad range of between 5.2 × 102 and 8.3 × 106 IU/ml. Quantitative results obtained close to the lower limit of the HCV 3.0 bDNA assay may imply that its lower limit should be reconsidered and raised, if necessary. The comparison of the HCV 3.0 bDNA assay with the HCV Monitor 2.0 assay showed a good correlation within the linear ranges of both assays. The HCV Monitor 2.0 assay showed a nonlinear association for HCV RNA loads above approximately 105 IU/ml, leading to an underestimation of HCV RNA loads. The HCV 3.0 bDNA assay will be clinically relevant in the decision of duration and tailoring of therapy for patients chronically infected with HCV.
ADDENDUM IN PROOF
While the manuscript was being reviewed, Bayer Diagnostics decided to change the reportable range of the HCV RNA 3.0 assay (bDNA) and the relationship between HCV RNA copies/ml and International Units (IU). The following advisory was issued: “Based upon on-going analysis of data generated in many assays and across multiple product lots, Bayer has decided to change the assay’s reportable range to 3,200 to 40,000,000 copies/ml to enhance the overall performance of the assay at the lower limit of quantitation. Similarly, the relationship between the Assay’s HCV RNA copies/ml and IU/ml was reevaluated after Bayer completed additional multi-lot experiments. Initial data resulted in a conversion factor of 4.8 HCV RNA copies/IU. Additional analyses incorporating the newly obtained data result in a revised conversion factor of 5.2 HCV RNA copies per International Unit” (Customer Bulletin of August 2001, Bayer Diagnostics, Berkeley, Calif.).
Acknowledgments
We are very grateful to Pauline Wertheim-Van Dillen (Academic Medical Center) for critical reading of the manuscript and David Hendricks, Lorine Tanimoto, Reinhard Oesterle, and Hans Sluimer (Bayer Diagnostics) for providing reagents and technical assistance.
REFERENCES
- 1.Detmer, J., R. Lagier, J. Flynn, C. Zayati, J. Kolberg, M. Collins, M. Urdea, and R. Sanchez-Pescador. 1996. Accurate quantification of hepatitis C virus (HCV) RNA from all HCV genotypes by using branched-DNA technology. J. Clin. Microbiol. 34:901-907. [DOI] [PMC free article] [PubMed]
- 2.EASL International Consensus Conference on Hepatitis C. 1999. Consensus statement. J. Hepatol. 31(Suppl. 1):3-8. [PubMed] [Google Scholar]
- 3.Jorgensen, P. A., and P. D. Neuwald. 2001. Standardized hepatitis C virus RNA panels for nucleic acid testing assays. J. Clin. Virol. 20:35-40. [DOI] [PubMed] [Google Scholar]
- 4.Lee, S. C., A. Antony, N. Lee, J. Leibow, J. Q. Yang, S. Soviero, K. Gutekunst, and M. Rosenstraus. 2000. Improved version 2.0 qualitative and quantitative AMPLICOR reverse transcription-PCR tests for hepatitis C virus RNA: calibration to international units, enhanced genotype reactivity, and performance characteristics. J. Clin. Microbiol. 38:4171-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, J. K. Albrecht, et al. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 6.Mellor, J., A. Hawkins, and P. Simmonds. 1999. Genotype dependence of hepatitis C virus load measurement in commercially available quantitative assays. J. Clin. Microbiol. 37:2525-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neumann, A. U., N. P. Lam, H. Dahari, D. R. Gretch, T. E. Wiley, T. J. Layden, and A. S. Perelson. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282:103-107. [DOI] [PubMed] [Google Scholar]
- 8.Nieforth, K. A., R. Nadeau, I. H. Patel, and D. Mould. 1996. Use of an indirect pharmacodynamic stimulation model of MX protein induction to compare in vivo activity of interferon alfa-2a and a polyethylene glycol-modified derivative in healthy subjects. Clin. Pharmacol. Ther. 59:636-646. [DOI] [PubMed] [Google Scholar]
- 9.Pawlotsky, J. M., M. Bouvier-Alias, C. Hezode, F. Darthuy, J. Remire, and D. Dhumeaux. 2000. Standardization of hepatitis C virus RNA quantification. Hepatology 32:654-659. [DOI] [PubMed] [Google Scholar]
- 10.Pawlotsky, J. M., M. Martinot-Peignoux, J. D. Poveda, A. Bastie, V. Le Breton, F. Darthuy, J. Remire, S. Erlinger, D. Dhumeaux, and P. Marcellin. 1999. Quantification of hepatitis C virus RNA in serum by branched DNA-based signal amplification assays. J. Virol. Methods 79:227-235. [DOI] [PubMed] [Google Scholar]
- 11.Poynard, T., P. Marcellin, S. S. Lee, C. Niederau, G. S. Minuk, G. Ideo, V. Bain, J. Heathcote, S. Zeuzem, C. Trepo, J. Albrecht, et al. 1998. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. Lancet 352:1426-1432. [DOI] [PubMed] [Google Scholar]
- 12.Saldanha, J., N. Lelie, A. Heath, et al. 1999. Establishment of the first international standard for nucleic acid amplification technology (NAT) assays for HCV RNA. Vox Sang. 76:149-158. [DOI] [PubMed] [Google Scholar]
- 13.Zeuzem, S., S. V. Feinman, J. Rasenack, E. J. Heathcote, M. Y. Lai, E. Gane, J. O'Grady, J. Reichen, M. Diago, A. Lin, J. Hoffman, and M. J. Brunda. 2000. Peginterferon alfa-2a in patients with chronic hepatitis C. N. Engl. J. Med. 343:1666-1672. [DOI] [PubMed] [Google Scholar]
- 14.Zeuzem, S., J. H. Lee, A. Franke, B. Ruster, O. Prummer, G. Herrmann, and W. K. Roth. 1998. Quantification of the initial decline of serum hepatitis C virus RNA and response to interferon alfa. Hepatology 27:1149-1156. [DOI] [PubMed] [Google Scholar]