Abstract
The worldwide recrudescence of tuberculosis and the widespread appearance of antibiotic resistance have strengthened the need for rapid and specific diagnostic tools. The prevailing microbiological identification of Mycobacterium tuberculosis, the causative agent of tuberculosis, which implies the use of in vitro cultures and acid-fast staining microscopy, is time-consuming. Detection of M. tuberculosis directly in clinical samples through PCR amplification of mycobacterium-specific genes, designed to shorten diagnostic delay, demonstrated reliability and high sensitivity. However, the quality of the diagnosis depends on the specificity of the target sequence for M. tuberculosis complex strains. In the present study, we demonstrated the specificity of recA and pps1 inteins for this complex and thus the feasibility of using intein-coding sequences as a new target for PCR diagnosis. Indeed, the recA and pps1 genes of 36 clinical isolates of M. tuberculosis and 10 field strains of M. bovis were found to be interrupted by an intein sequence at the RecA-a and Pps1-b sites, respectively, while a large number of nontuberculous mycobacterial species failed to demonstrate these insertions. Besides, the MtuPps1, which was cloned and expressed in Escherichia coli, was shown to possess an endonuclease activity. The intein cleaves the 40-bp sequence spanning the intein insertion site Pps1-b in the inteinless pps1 gene. In addition to the PCR amplification of recA and pps1 intein genes as a tool for diagnosis, the specific endonuclease activity could represent a new molecular approach to identify M. tuberculosis.
Tuberculosis remains a major public health problem in developing countries. Despite the decline in incidence observed in the 1980s, resurgence has recently occurred. Furthermore, the recrudescence of infections with Mycobacterium tuberculosis and the emergence of multidrug-resistant strains represent a serious public health threat in developed countries, where the incidence of tuberculosis is related to immunodeficient patients, homeless people, and the advancing age of the population (World Health Organization web site at http://www.who.int/health-topics/tb.htm).
This worldwide resurgence of tuberculosis has strengthened the need for rapid and specific diagnostic tools to control its spread. Delay in diagnosis results, first, in late initiation of antitubercular therapy and, second, in prolonged transmission of the infection.
Until recently, the diagnosis of tuberculosis was based on clinical features and microbiological assays, radiological examinations, immunological tests, microscopy identification, or in vitro cultures (37). Acid-fast staining microscopy of specimens combined with isolation and culture of the bacilli remains the “gold standard” method to specifically identify mycobacteria. Because of the low growth rate of M. tuberculosis, this method is time-consuming, and the diagnosis can take up to 8 weeks.
More recently, molecular approaches have been designed to ensure early detection of M. tuberculosis in clinical samples. These methods include DNA amplification by strand displacement amplification (35) or by PCR of M. tuberculosis-specific genes. The use of adequate oligonucleotide pairs to amplify target sequences such as rRNA 16S, IS6110, mtp40, or the senX3-regX3 intergenic region has demonstrated the reliability and high sensitivity of PCR diagnosis (3). Moreover, technical adaptations of PCR protocols (13, 25, 40) have made it possible to detect mycobacteria directly from clinical samples, such as sputum, blood, urine, and cerebrospinal or pleural fluid. These nucleic acid-based methods have been found to be more sensitive than conventional methods and were able to detect even small numbers of M. tuberculosis cells (1, 12, 18). However, the choice of the most favorable target sequence is controversial, since results from different clinical studies evaluating the various amplification assays were divergent concerning their specificity and sensitivity (11, 17, 20, 31, 39; S. H. Gillespie, T. D. McHugh, and L. E. Newport, Letter, J. Clin. Microbiol. 35:799-801, 1997). To optimize specificity and sensitivity, the target sequence should be present in all strains of the M. tuberculosis complex, with a high degree of sequence conservation between strains to avoid false-negative reactions, and should be absent in all other mycobacterium species, thereby eliminating the chance of false-positive results.
In a previous work undertaken to clarify the role of RecA inteins in mycobacteria (32), we showed that the insertion site and the sequence of the recA intein from M. tuberculosis (8) are unique among mycobacteria. Similarly, the M. tuberculosis pps1 gene (6) possesses an insertion sequence with no homolog known to date in other mycobacterial species (Inbase, the New England Biolabs intein database, at http://www.neb.com/neb/inteins/). The species specificities of both of these invading sequences make them potential targets for PCR amplification. Moreover, since inteins are potential endonucleases (Inteins-Protein Introns web site at http://bioinfo.weizmann.ac.il/∼pietro/inteins/), the search for their specific functions could lead to a new molecular approach to diagnose tuberculosis.
In the present study, we show first that both intein coding sequences disrupting recA and pps1 genes from M. tuberculosis are specific to the M. tuberculosis complex and can thus effectively be used as DNA target sequences in a PCR-based diagnosis. In addition, we demonstrated that, unlike the RecA intein, the Pps1 intein MtuPps1 exhibits an endonuclease activity, which could provide a new tool to unequivocally detect and identify members of the M. tuberculosis complex.
MATERIALS AND METHODS
Mycobacterial strains, growth conditions, and genomic DNA isolation.
Thirty-three nontuberculous mycobacterial species (listed in Fig. 1) were used in this study. They correspond to the strains previously described (32), except that only one strain each of Mycobacterium avium (IP140310013), Mycobacterium aurum (ATCC 23366), Mycobacterium chitae (IP141160001), and Mycobacterium duvalii (IP141180002) was examined and that one strain of Mycobacterium xenopi (IP14035004) was added to the test. The strains were from the Reference Centre for Mycobacteria (Institut Pasteur, Paris) and were kindly provided by V. Vincent Lévy-Frébault. All of the strains were identified by conventional biochemical tests (38) and chemotaxonomic criteria, such as mycolic acid and species-specific glycolipid analyses (7). Strains were grown at their optimal temperature of growth (38) on Löwenstein-Jensen medium for a few days to several weeks, depending on the growth rate of the mycobacterial species. Genomic DNA was isolated from the 33 mycobacterial strains by the glass bead disruption method as described by Saves and collaborators (32). In addition, the cosmid B2235, which was used to amplify the recA gene from Mycobacterium leprae, was a gift from K. Eiglmeier (Institut Pasteur, Paris, France).
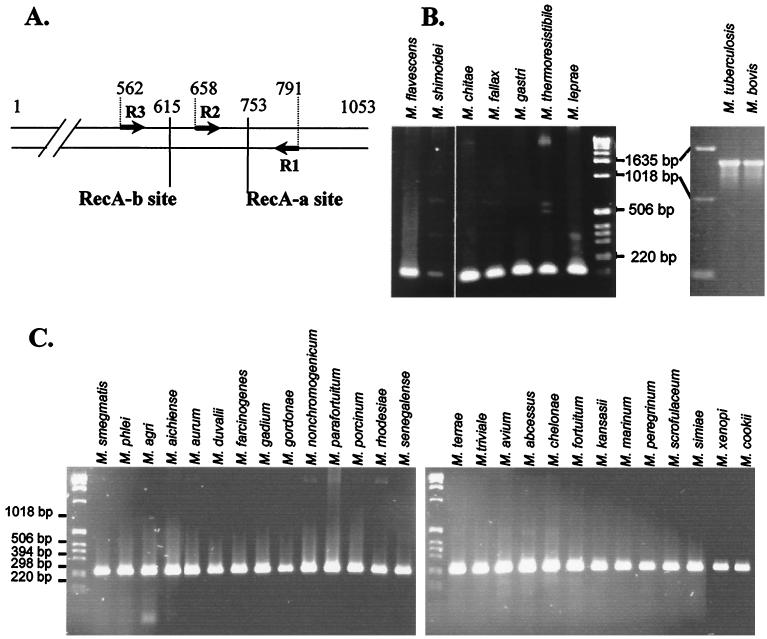
FIG. 1.
Specificity of the MtuRecA intein. (A) Location of intein insertion sites in mycobacterial recA genes. Arrows represent the oligonucleotides used to amplify gene fragments. The positions of the RecA-a and RecA-b sites and of oligonucleotides are indicated as the number of base pairs from the translation start site in the M. tuberculosis H37Rv gene. (B) PCR amplification with R1 and R2 oligonucleotides from genomic DNA of nine species of mycobacteria. (C) PCR amplification with R1 and R3 oligonucleotides from genomic DNA of 27 species of mycobacteria.
M. tuberculosis isolates were collected from patients attending health care clinics in two suburbs of Cape Town, South Africa. M. bovis isolates were a gift from A. Michel (Tuberculosis Laboratory, Onderstepoort Veterinary Institute, Onderstepoort, South Africa). Cultures were grown on Löwenstein-Jensen slants until confluent growth was achieved. The bacilli were then inactivated by being heated to 80°C for 1 h. Three milliliters of extraction buffer (50 mM Tris-HCl [pH 8], 25 mM EDTA, 5% sodium glutamate) was added to each culture, and the cells were gently scraped to release them from the Löwenstein-Jensen medium. This cell suspension was then transferred to a 50-ml tube containing 20 g of glass balls (each 0.5 mm in diameter). This process was repeated, and the cell suspensions were pooled. The cells were then dispersed by vigorous mixing. Lysozyme (25 mg) together with RNase A (25 μg) was added, and the cell suspension was incubated at 37°C for 2 h. Proteinase K buffer (0.65 ml of 500 mM Tris-HCl [pH 7.5], 50 mM CaCl2) and proteinase K (150 μg) were added, and the suspension was incubated at 45°C for 16 h. Proteins were removed by phenol-chloroform-isoamyl alcohol (25:24:1) extraction, and the aqueous phase was reextracted with an equal volume of chloroform-isoamyl alcohol (24:1). The DNA was precipitated with isopropanol, washed with 70% ethanol, and dried. Finally, DNA was resuspended in TE buffer (10 mM Tris-HCl [pH 8], 1 mM EDTA).
Genomic DNA from the MT103 strain of M. tuberculosis was obtained from Institut Pasteur (Paris, France).
PCR assays.
The primers R3 (5′ACCACGGCGATCTTCATCAACCAGCT 3′), R2 (5′ GCGTCGGTGCGCATGGACGTGCG 3′), and R1 (5′ AGGATGTCGAACTCGGCCAGCTTGAA 3′) correspond to the M. tuberculosis recA gene sequence or its complementary sequence at positions 562 to 587, 658 to 680, and 765 to 791, respectively, according to the M. tuberculosis H37Rv gene sequence. The primers P2 (5′ CATCCGCAACACCTACGACCGG 3′), P3 (5′ GAACATGGGCCZGTTCGAGCGGACG 3′), and P1 (5′ GTCGTTGTTCGACCAGTTCTGGATGGT 3′) correspond to the M. tuberculosis pps1 gene sequence or its complementary sequence at positions 357 to 378, 687 to 711, and 844 to 870, respectively, according to the M. tuberculosis H37Rv gene sequence.
PCR amplifications were performed with Taq DNA polymerase in a mixture containing 10 mM Tris-HCl [pH 8.3], 1.5 mM MgCl2, 50 mM KCl, and 0.2 mM deoxynucleoside triphosphates (dNTPs), with 5 μl of the genomic DNA preparation as a DNA template. Several PCR assays were performed under different reaction conditions with regard to oligonucleotide concentrations and hybridization temperature to ensure amplification specificity.
Cloning of MtuRecA and MtuPps1 coding sequences.
The entire coding sequences of MtuRecA and MtuPps1 inteins were amplified by PCR from genomic DNA of the MT103 strain of M. tuberculosis with oligonucleotide pairs MtuRecA-ATG (5′ ATGTGCCTCGCAGAGGGGACT 3′) and MtuRecA-3′ (5′ GTTGTGCACGACAACCCCTTC 3′) and MtuPps1-ATG (5′ ATGTGCCTGCCCGCCGGC 3′) and MtuPps1-3′ (5′ GTTGTGCACGGCGAACCCGT 3′), respectively. For this purpose, 1 μl (200 ng) of genomic DNA was incubated with Taq DNA polymerase in a mixture containing 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCl, 0.2 mM dNTPs, and 10 pmol of each oligonucleotide in a 50-μl mix, which was incubated as follows: 10 min at 92°C; 29 times for 1 min at 92°C, 1 min at 55°C, and 1.5 min at 72°C; and, finally, 5 min at 72°C.
These sequences were inserted in the pCR T7/CT-TOPO plasmid (Invitrogen). The resulting constructs allow the expression of both inteins fused to C-terminal V5 and six-His tags under the regulation of T7 promoter and T7 RNA polymerase in Escherichia coli.
Production and extraction of MtuPps1 and MtuRecA inteins.
E. coli BL21(De3)(pLysS) bacteria were transformed with the two expression vectors. Bacteria were grown at 30°C in Luria broth culture medium supplemented with 100 μg of ampicillin (Sigma) and 30 μg of chloramphenicol per ml until they reached the exponential phase. Thus, the induction of MtuRecA or MtuPps1 expression with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) (Sigma) was performed for 4 h at 37°C or for 6 h at 30°C, respectively. Cells were lysed in 20 mM sodium phosphate (pH 7.5) by six cycles of freezing and thawing. The lysate was centrifuged at 10,000 × g for 45 min to separate the soluble intein from insoluble proteins. The soluble fraction was dialyzed against a mixture containing 10 mM Tris-HCl (pH 7.5), 0.1 mM EDTA, 1 mM dithiothreitol, 200 μg of bovine serum albumin per ml, and 50 mM NaCl. A protein extract from nontransformed BL21(De3)(pLysS) was collected under similar culture and extraction conditions and used as a negative control in endonuclease assays.
Protein extracts were quantified by the Bradford's method (2). Samples containing 0.5 to 5 mg of proteins per ml were denatured at 95°C for 2 min in a sodium dodecyl sulfate (SDS)-urea-β-mercaptoethanol buffer and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) (homogenous, 10% polyacrylamide) according to the method of Laemmli (21). Separated proteins were electrically transferred to nitrocellulose membrane (BA45; Schleicher and Schuell) in a semidry apparatus (4). Membranes were incubated for 30 min in Tris-buffered saline (TBS; 10 mM Tris-HCl [pH 8], 137 mM NaCl) containing 1% Tween 20 and 10% nonfat dry milk for 1 h with anti-V5 antibodies (Invitrogen), diluted 1:5,000 in TBS containing 1% Tween 20 and 1% nonfat dry milk, and for 1 h with a peroxidase-labeled antimouse immunoglobulin G conjugate diluted 1:10,000 in the same buffer. A detection reaction with chemiluminescent substrate was performed with the ECL enhanced chemiluminescence detection kit according to the manufacturer's instructions (Amersham-Pharmacia Biotech).
Endonuclease assays.
To assay MtuRecA and MtuPps1 for endonuclease activity, the 40-bp DNA sequences spanning their homing site were inserted between XbaI and HindIII restriction sites of plasmid pUC19. Partially complementary oligonucleotide pairs MtuRecA-XbaI (5′ CTAGATCAAGGTCGTCAAGAACAAGTGTTCGCCCCCCTTCAAGCA 3′) and MtuRecA-HindIII (5′AGCTTGCTTGAAGGGGGGCGAACACTTGTTCTTGACGACCTTGAT3′) and MtuPps1-XbaI (5′ CTAGAACGTGCACTACGTAGAGGGTCGCACCGCACCGATCTACAAAA 3′) and MtuPps1-HindIII (5′ AGCTTTTGTAGATCGGTGCGGTGCAGCCCTCTACGTAGTGCACGTT 3′) were annealed by boiling a mix of 1 nmol of each oligonucleotide in a mixture of 10 mM Tris-HCl (pH 7.5) and 50 mM NaCl for 5 min and slow cooling to room temperature. The annealed oligonucleotides were then inserted in pUC19 overdigested by HindIII and XbaI (New England Biolabs). The resulting constructs, substrates 1 and 2, were sequenced. They were linearized by ScaI and extracted from a 1% agarose gel in TBE (90 mM Tris-borate, 2 mM EDTA) buffer, and the linear substrates were diluted in water to a concentration of 100 ng/μl for cleavage assays.
Endonuclease activity assays were performed in a final volume of 10 μl, in various reaction buffers, and at temperatures ranging from 15 to 50°C. Moreover, the quantity of proteins added to the reaction mix varied from 25 ng to 5 μg. The reaction mixtures were analyzed on a 1% agarose gel in TBE buffer.
Nucleotide sequence accession numbers.
The GenBank/EMBL accession numbers for the RecA and Pps1 inteins are X58485 and AL021184, respectively.
RESULTS
The MtuRecA intein sequence is specific for the M. tuberculosis complex.
The 1,320-bp sequence coding for RecA intein is inserted at the RecA-a site, immediately downstream of the 753rd nucleotide of the M. tuberculosis recA gene, while all of the seven other RecA intein sequences identified in M. chitae, Mycobacterium fallax, M. flavescens, M. gastri, M. leprae, Mycobacterium shimodei, and Mycobacterium thermoresistibile are inserted at the RecA-b site (32), 138 nucleotides upstream (Fig. 1A). PCR oligonucleotides were designed according to the conserved recA gene sequences between M. tuberculosis, M. leprae, and Mycobacterium smegmatis sequences. R1 and R2 primers were positioned on each side of the RecA-a site in order to amplify a 134-bp fragment spanning this intein insertion site (Fig. 1A). Hence, amplification reactions with primers R1 and R2 produced a 1,454-bp fragment in the presence of M. tuberculosis and M. bovis DNA, while the amplified fragment is only 134 bp long in the presence of DNA from different mycobacterial species outside of the complex (Fig. 1B).
In a parallel experiment, amplification with primers R1 and R3 generated a 230-bp fragment spanning both RecA-a and RecA-b sites, allowing the confirmation of the absence of intein in 27 other mycobacterial species (Fig. 1C).
In order to check whether the R1-R2 primer pair can be used for tuberculosis screening, we performed an amplification reaction using genomic DNA preparations from 36 different M. tuberculosis strains isolated from patients with confirmed tuberculosis (36) and from 10 strains of M. bovis. The 1,454-bp specific fragment was generated with all of the M. tuberculosis or M. bovis strains (not shown), implying that all strains of M. tuberculosis and M. bovis tested possess the intein in the RecA-a site of the recA gene in their genomes.
The MtuPps1 intein gene is specific for the M. tuberculosis complex.
To date, three intein insertion sites have been described in the pps1 gene (33) (Inbase, the New England Biolabs intein database): Pps1-a, Pps1-b, and Pps1-c are the respective intein insertion sites of M. leprae, M. tuberculosis, and Mycobacterium gastri inteins (Fig. 2A). To control for the specificity of the M. tuberculosis intein amplification, we searched for the presence of intein sequences in 35 different mycobacterial species by PCR amplification of a 524-bp sequence spanning the three insertion sites. P1 and P2 primers were designed according to the conserved pps1 gene sequences in M. tuberculosis and M. leprae. As shown in Fig. 2B, intein was absent from 15 of our mycobacterial species, based on the amplification of a fragment of approximately 500 bp, while the presence of an intein in M. gastri, M. tuberculosis, and M. bovis was demonstrated by the amplification of larger fragments. In eight Mycobacterium species, namely M. chitae, M. fallax, M. farcinogenes, M. gordonae, M. parafortuitum, M. senegalense, M. thermoresistibile, and M. shimoidei, no amplified DNA was obtained, whatever the amplification conditions. Additionally, nonspecific amplified fragments were obtained from genomic DNA from the nine remaining species (M. gadium, M. chelonae, M. fortuitum, M. kansasii, M. marinum, M. peregrinum, M. scrofulaceum, M. xenopi, and M. cookii) under extremely permissive conditions of PCR, while no amplification was obtained under more stringent PCR conditions. That may be due to the lack of a pps1 gene or to the strong divergence of the sequence of the pps1 gene in these 17 mycobacterial species.
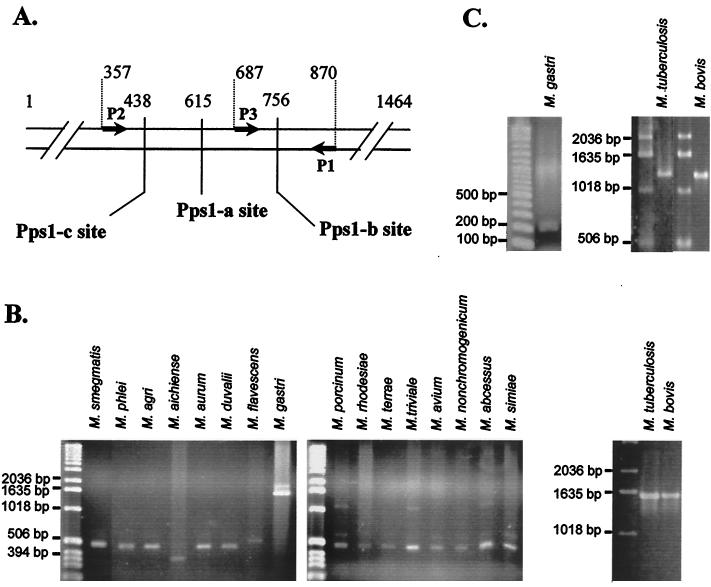
FIG. 2.
Specificity of MtuPps1 intein. (A) Location of intein insertion sites in mycobacterial pps1 genes. Arrows represent the oligonucleotides used to amplify gene fragments. The positions of Pps1-a, Pps1-b, and Pps1-c sites and of oligonucleotides are indicated as number of base pairs from the translation start site in the M. tuberculosis H37Rv gene. (B) PCR amplification with P1 and P2 oligonucleotides from genomic DNA of 18 species of mycobacteria. (C) PCR amplification with P1 and P3 oligonucleotides from genomic DNA of three species of mycobacteria.
A third primer, named P3 (Fig. 2A), was paired with primer P1 to specifically detect the intein from the M. tuberculosis complex. As expected, a 1,260-bp fragment was obtained from M. tuberculosis and M. bovis genomic DNA amplification, while a 183-bp fragment was amplified with all other mycobacterial strains and in particular with M. gastri (Fig. 2C).
Both oligonucleotide pairs (P1-P2 and P1-P3) were used to amplify genomic DNA from 36 M. tuberculosis and 10 M. bovis strains (not shown). Interestingly, the identification of approximately 1,600- and 1,300-bp fragments, respectively, in the 46 amplification reactions confirmed the presence of the pps1 intein in all of the M. tuberculosis and M. bovis strains tested.
Combined detection of recA and pps1 inteins by PCR.
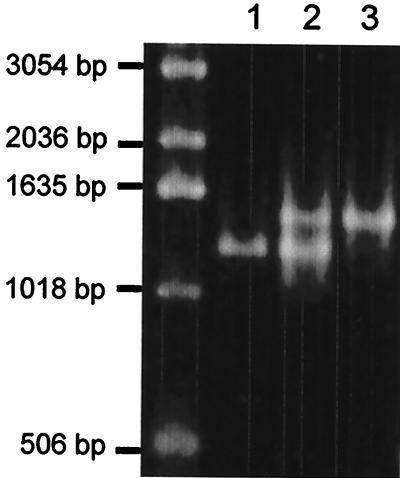
In an additional PCR assay, we tried to simultaneously amplify recA and pps1 genes in a one-tube reaction for a combined detection of both M. tuberculosis-specific inteins. pps1 and recA gene fragments were amplified from M. tuberculosis H37v genomic DNA by using P1 and P3 and R1 and R2 oligonucleotide pairs, respectively, either in separate tubes or in the same reaction tube. As expected, the two specific fragments of 1,260 and 1,454 bp, resulting from the amplification of pps1 and recA, respectively, were obtained when both oligonucleotide pairs were present in the reaction mix (Fig. 3).
FIG. 3.
PCR amplification with the P1-P3 oligonucleotide pair (lane 1), the R1-R2 oligonucleotide pair (lane 3), or both oligonucleotide pairs (lane 2) from genomic DNA of M. tuberculosis H37Rv.
MtuPps1 intein is a specific endonuclease.
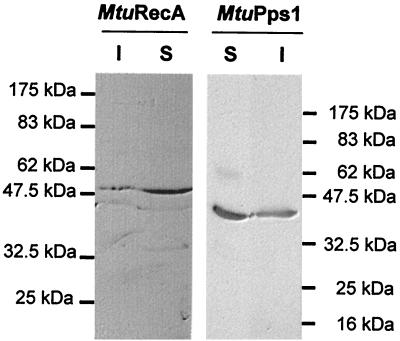
Both MtuRecA and MtuPps1 coding sequences were cloned in the pCR T7/CT-TOPO cloning vector in fusion with the DNA sequence encoding the C-terminal V5 and six-His epitopes. These constructs allowed the expression of the inteins in E. coli strain BL21(De3)(pLysS). Induction of exponential cultures with 1 mM IPTG resulted in the production of soluble inteins as determined by Western blot analysis with anti-V5 antibodies (Fig. 4).
FIG. 4.
Expression of MtuPps1 and MtuRecA inteins in E. coli. Tagged inteins were detected by Western blotting of the SDS-PAGE (10% polyacrylamide) gels used to separate proteins from soluble (S) and insoluble (I) fractions of MtuPps1 and MtuRecA extracts and immunologic detection by using anti-V5 antibody.
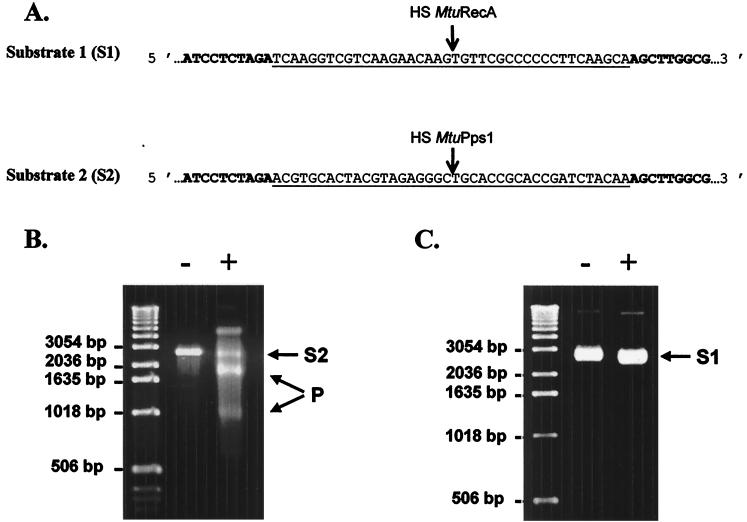
Few inteins are known to possess an endonuclease activity that recognizes and cuts a DNA sequence spanning the junction site created when the intein coding region is deleted from the host gene (homing site). We thus constructed substrate 1 (S1) for MtuRecA and substrate 2 (S2) for MtuPps1 by cloning synthetic 40-bp homing sites in pUC19 plasmid (Fig. 5A). The ScaI-linearized form of these DNA substrates was purified and used to assay the endonuclease activity of each intein, under various experimental conditions with regard to buffer pH and composition, to mono- and bivalent ions used as cofactors, and to temperature. Linear substrate S2 (2,730 bp) was cleaved in two specific products (940 and 1,790 bp) in the presence of MtuPps1 extract in a 10 mM Tris-HCl (pH 8) buffer containing 10 mM MgCl2 and 25 mM KCl at 37°C (Fig. 5B), while substrate S1 could not be cleaved in the presence of MtuRecA, independent of the reaction conditions (Fig. 5C). Hence, MtuPps1 exhibits a specific endonuclease activity, which was named PI-MtuI according to the current nomenclature.
FIG. 5.
Endonuclease assays. (A) DNA substrates for MtuPps1 and MtuRecA. The sequences of MtuPps1 and MtuRecA homing sites (HS) inserted in pUC19 (the sequence of which appears in boldface) are underlined. (B) Cleavage assay for MtuPps1. One hundred nanograms of linearized substrate 2 was incubated with either 2.5 μg of the crude extract of MtuPps1 (+) or 2.5 μg of a crude extract of nontransformed E. coli BL21(De3)(pLysS) (−), for 1 h at 37°C, in 10 mM Tris-HCl (pH 8) buffer containing 10 mM MgCl2 and 25 mM KCl. Substrate (S2; 2,730 bp) and products (P; 940 and 1,790 bp) were separated on a 1% agarose gel in TBE buffer. (C) Cleavage assay for MtuRecA. One hundred nanograms of linearized substrate 1 was incubated with either 2.5 μg of the crude extract of MtuRecA (+) or 2.5 μg of a crude extract of nontransformed E. coli BL21(De3)(pLysS) (−), for 1 h at 37°C, in 10 mM Tris-HCl (pH 8) buffer containing 10 mM MgCl2 and 25 mM KCl. The reaction mixture was analyzed on a 1% agarose gel in TBE buffer.
DISCUSSION
One of the limitations in the control of tuberculosis is the lack of highly sensitive and specific assays to rapidly identify M. tuberculosis, the causative agent of tuberculosis. While the conventional methods (i.e., microscopy and cultural identification) still represent the most reliable way to diagnose the infection, these time-consuming methods do not always allow for the early detection and subsequent initiation of therapy. Indeed, a few weeks delay can account for increased morbidity and prolonged transmission of the disease.
In order to reduce the delay in diagnosis, several methods have been developed. Semiautomated liquid culture systems, such as the BACTEC radiometric method or MB/Bact and MGIT systems, are able to detect mycobacterial slow growers within 10 to 14 days. While several of these growth detection methods have demonstrated good performance comparable to standard methods (23, 26), the necessity for a heavy and expensive apparatus limits their use in poor countries. Moreover, additional tests may be necessary to finally identify the mycobacterial species. Immunologic or enzymatic assays and phage systems (5, 30) were also proposed, but are generally nonuniformly applicable or lack specificity and/or sensitivity.
One of the most important technical advances in diagnosis of tuberculosis is the development of molecular methods. The detection, through PCR amplification, of a DNA sequence specifically present in the genomes of members of the M. tuberculosis complex has the potential to overcome the sensitivity and specificity limitations of other diagnostic methods. However, while protocols used to amplify DNA from mycobacteria present in a wide range of samples have been greatly optimized, the choice of the target sequence remains controversial. Various PCR assays have been suggested; these include amplification of sequences encoding mycobacterial antigens, insertion sequences, rRNA, and intergenic regions.
Among the proposed targets, the 16S rRNA gene is the sequence amplified by commercial kits: i.e., the Roche AMPLICOR MTB (Roche Diagnostic Systems, Somerville, N.J.) and the Gen-Probe AMTB (Gen-Probe, Inc., San Diego, Calif.). Although the performance of each of these tests is excellent, their specificity cannot be optimal, since the target gene is present in all mycobacterial species. Hence, the discrimination between mycobacteria necessitates hybridization with specific DNA probes (16, 41). Indeed, the use of these tests was initially restricted to acid-fast bacillus-positive specimens (1).
The most popular target sequence is the insertion sequence IS6110, also known as IS986 or IS987 (27). Even if some studies demonstrated excellent diagnostic results (15, 17), the specificity of IS6110-based assays is controversial (11; Gillespie et al., Letter). The multicopy IS6110 sequence, identified in the majority of M. tuberculosis strains, accounts for the high sensitivity of the amplification procedure; however, strains with few or no copies of this insertion sequence have also been isolated (42). Moreover, since this sequence is a member of the IS3 family, the most widespread group of insertion sequences in bacteria, homologous sequences have been found in nontuberculous mycobacteria (20, 28). Hence, even if the use of various primer sets to amplify different segments of the sequence may enhance the specificity of the test, the existence of strains lacking this sequence seriously compromises the use of IS6110-based PCR for tuberculosis diagnosis.
Little is known about the diagnosis based on the amplification of the MPB64- and Hsp65-encoding genes, but these procedures have shown poor sensitivity and require an additional oligonucleotide hybridization step for the detection of the amplified fragments. Therefore, the specificity remains rather uncertain (3, 22). The mtp40 gene detection has been thoroughly studied. A comparative study showed that mtp40 is more reliable than the IS6110-based amplification test (19). However, Weil et al. (39) demonstrated that this gene is not present in all strains of M. tuberculosis. Moreover, since this gene is absent in the other species of the M. tuberculosis complex (9), the diagnosis could fail to recognize many cases of tuberculosis. Hence, it should instead be used to discriminate between M. tuberculosis and other species of the M. tuberculosis complex (10).
More recently, an interesting alternative was found in the amplification of the senX3-regX3 intergenic region (24, 34), since this sequence is distributed in all strains of the M. tuberculosis complex. However, one should note that, first, this sequence is also found in M. leprae genome, and, second, the length and the sequence of this intergenic region are variable, making interpretation of PCR results quite ambiguous.
In the present study, we demonstrated the feasibility of using intein coding sequences as a target for PCR diagnosis, since the M. tuberculosis recA and pps1 intein sequences are specific for members of the M. tuberculosis complex (Fig. 1 and 2). Indeed, only the recA genes of M. tuberculosis and M. bovis were found to be interrupted by an intein sequence at the RecA-a site, while a total of 34 different mycobacterial species have failed to demonstrate this insertion. Likewise, the MtuPps1 intein sequence embedded at the Pps1-b site of the M. tuberculosis pps1 gene was absent from 16 other mycobacterial species and was not detected in 17 other species. Moreover, all of the M. tuberculosis and M. bovis strains tested in this study harbor both intein sequences, suggesting a widespread distribution of these invading sequences among the members of the M. tuberculosis complex.
The primer pairs were designed in order to amplify a fragment of the host gene, spanning the intein insertion site, rather than the intein sequence itself. Hence, a DNA fragment has to be amplified even in absence of an intein coding sequence, so that it is possible to control the absence of inhibitory substances in the PCRs. The recA gene was efficiently amplified in all of the mycobacterial species and strains tested, and a 1,320-bp-longer fragment was obtained only in presence of M. tuberculosis or M. bovis DNA (Fig. 1). These results indicated that the recA gene is present in all 36 of the species, a result probably related to the essentiality of RecA recombinase in mycobacteria, as suggested by Frischkorn and collaborators and Papavinasasundaram and collaborators (14, 29). Moreover, the amplification yields were rather high, suggesting that the sensitivity of the PCR diagnosis would be quite good, even if the target sequences are present in single copy in the M. tuberculosis genome.
In the case of pps1, no amplification was obtained for some mycobacterial species. Since recA amplification was performed from the same DNA preparations, these negative reactions were not attributed to the presence of any inhibitory substances. The absence of amplified fragment could be explained by the absence of the pps1 gene in these mycobacteria. However, this hypothesis is unlikely, since inteins are generally present in essential host proteins, and a previous study suggested an important role of Pps1 in mycobacteria (33). Hence, a probable explanation may reside in sequence divergences of pps1 genes between various mycobacterial species, hindering the hybridization of primers. Nevertheless, the detection of a 1,300-bp-long fragment in the presence of M. tuberculosis and M. bovis DNA, compared to the 200-bp fragment obtained in presence of other mycobacterial species (Fig. 2), allowed the specific identification of M. tuberculosis and M. bovis.
As formerly suggested by Brisson-Noël and collaborators (3), two amplification assays should be done in parallel to avoid misinterpretation of PCR results, without the need for an additional internal amplification control. Here, we propose the simultaneous detection of MtuRecA- and MtuPps1-specific sequences. The combination of amplifications of both recA and pps1 genes, in a multiprimer PCR (Fig. 3), can ensure the rapid and specific detection of M. tuberculosis.
Moreover, the detection of the specific endonuclease activity of MtuPps1 intein could represent a new molecular approach to identify M. tuberculosis. One can imagine a PCR amplification followed by in vitro translation of the intein and a specific cleavage assay by using linear substrate S2 (Fig. 5). Effectively, the detection of the cleavage activity is even more specific than the sole PCR amplification of the intein sequence and, concerning specificity, should provide excellent results. However, the sensitivity of such a test is still to be determined. In vitro transduction assays have to be performed and optimized with this particular mycobacterial sequence in order to validate this new diagnostic approach.
In conclusion, the results described here highlight that the specificity of M. tuberculosis and M. bovis inteins found in RecA and Pps1 host proteins should be considered as an important new molecular tool for diagnosis of tuberculosis.
Acknowledgments
We thank A. Michel (Tuberculosis Laboratory, Onderstepoort Veterinary Institute, Onderstepoort, South Africa), V. Vincent Lévy-Frébault (Institut Pasteur, Paris, France), and K. Eiglmeier (Institut Pasteur) for providing the mycobacterial strains or genomic DNA; M. A. Lanéelle (IPBS/CNRS, Toulouse, France) for the cultivation of the mycobacterial strains; D. Zerbib (IPBS/CNRS) for helpful discussions; and N. Trinchero for technical assistance.
REFERENCES
- 1.Bergmann, J. S., and G. L. Woods. 1996. Clinical evaluation of the Roche AMPLICOR PCR Mycobacterium tuberculosis test for detection of M. tuberculosis in respiratory specimens. J. Clin. Microbiol. 34:1083-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Brisson-Noel, A., C. Aznar, C. Chureau, S. Nguyen, C. Pierre, M. Bartoli, R. Bonete, G. Pialoux, B. Gicquel, and G. Garrigue. 1991. Diagnosis of tuberculosis by DNA amplification in clinical practice evaluation. Lancet 338:364-366. [DOI] [PubMed] [Google Scholar]
- 4.Burnette, W. N. 1981. “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 112:195-203. [DOI] [PubMed] [Google Scholar]
- 5.Chang, C. L., E. Y. Lee, H. C. Son, and S. K. Park. 2000. Evaluating the usefulness of the ICT tuberculosis test kit for the diagnosis of tuberculosis. J. Clin. Pathol. 53:715-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell et al. 1998.Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 7.Daffé, M., and P. Draper. 1998. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39:131-203. [DOI] [PubMed] [Google Scholar]
- 8.Davis, E. O., S. G. Sedgwick, and M. J. Colston. 1991. Novel structure of the recA locus of Mycobacterium tuberculosis implies processing of the gene product. J. Bacteriol. 173:5653-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Del Portillo, P., L. A. Murillo, and M. E. Patarroyo. 1991. Amplification of a species-specific DNA fragment of Mycobacterium tuberculosis and its possible use in diagnosis. J. Clin. Microbiol. 29:2163-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Portillo, P., M. C. Thomas, E. Martinez, C. Marañón, B. Valladares, M. E. Patarroyo, and M. Carlos López. 1996. Multiprimer PCR system for differential identification of mycobacteria in clinical samples. J. Clin. Microbiol. 34:324-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doucet-Populaire, F., V. Lalande, E. Carpentier, A. Bourgoin, M. Dailloux, C. Bollet, A. Vachee, D. Moinard, J. Texier-Maugein, B Carbonelle, and J. Grasset. 1996. A blind study of the polymerase chain reaction for the detection of Mycobacterium tuberculosis DNA. Tuber. Lung Dis. 77:358-362. [DOI] [PubMed] [Google Scholar]
- 12.Folgueira, L., R. Delgado, E. Palenque, J. M. Aguado, and A. R. Noriega. 1996. Rapid diagnosis of Mycobacterium tuberculosis bacteremia by PCR. J. Clin. Microbiol. 34:512-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Folgueira, L., R. Delgado, E. Palenque, and A. R. Noriega. 1993. Detection of Mycobacterium tuberculosis DNA in clinical samples by using a simple lysis method and polymerase chain reaction. J. Clin. Microbiol. 31:1019-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frischkorn, K., P. Sander, M. Scholz, K. Teschner, T. Prammananan, and E. C. Bottger. 1998. Investigation of mycobacterial recA function: protein introns in the RecA of pathogenic mycobacteria do not affect competency for homologous recombination. Mol. Microbiol. 29:1203-1214. [DOI] [PubMed] [Google Scholar]
- 15.Githui, W. A., S. M. Wilson, and F. A. Drobniewski. 1999. Specificity of IS 6110-based DNA fingerprinting and diagnostic techniques for Mycobacterium tuberculosis complex. J. Clin. Microbiol. 37:1224-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glennon, M., T. Smith, M. Cormican, D. Noone, T. Barry, M. Maher, M. Dawson, J. J. Gilmartin, and F. Gannon. 1994. The ribosomal intergenic spacer region: a target for the PCR based diagnosis of tuberculosis. Tuberc. Lung Dis. 75:353-360. [DOI] [PubMed] [Google Scholar]
- 17.Hellyer, T. J., L. E. DesJardin, M. K. Assaf, J. H. Bates, M. D. Cave, and K. D. Eisenach. 1996. Specificity of IS6110-based amplification assays for Mycobacterium tuberculosis complex. J. Clin. Microbiol. 34:2843-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hemal, A. K., N. P. Gupta, T. P. Rajeev, R. Kumar, L. Dar, and P. Seth. 2000. Polymerase chain reaction in clinically suspected genitourinary tuberculosis: comparison with intravenous urography, bladder biopsy, and urine acid fast bacilli culture. Urology 56:570-574. [DOI] [PubMed] [Google Scholar]
- 19.Herrera, E. A., and M. Segovia. 1996. Evaluation of mtp40 genomic fragment amplification for specific detection of Mycobacterium tuberculosis in clinical specimens. J. Clin. Microbiol. 34:1108-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kent, L., T. D. McHugh, O. Billington, J. W. Dale, and S. H. Gillespie. 1995. Demonstration of homology between IS6110 of Mycobacterium tuberculosis and DNAs of other Mycobacterium spp. J. Clin. Microbiol. 33:2290-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lee, B. W., J. A. Tan, S. C. Wong, C. B. Tan, H. K. Yap, P. S. Low, J. N. Chia, and J. S. Tay. 1994. DNA amplification by the polymerase chain reaction for the rapid diagnosis of tuberculous meningitis. Comparison of protocols involving three mycobacterial DNA sequences, IS6110, 65 kDa antigen, and MPB64. J. Neurol. Sci. 123:173-179. [DOI] [PubMed] [Google Scholar]
- 23.Liu, Y. C., T. S. Huang, and W. K. Huang. 1999. Comparison of a nonradiometric liquid-medium method (MB REDOX) with the BACTEC system for growth and identification of mycobacteria in clinical specimens. J. Clin. Microbiol. 37:4048-4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magdalena, J., A. Vachee, P. Supply, and C. Locht. 1998. Identification of a new DNA region specific for members of Mycobacterium tuberculosis complex. J. Clin. Microbiol. 36:937-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manjunath, N., P. Shankar, L. Rajan, A. Bhargava, S. Saluja, and Shriniwas. 1991. Evaluation of a polymerase chain reaction for the diagnosis of tuberculosis. Tubercle 72:21-27. [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Sanchez, L., J. Ruiz-Serrano, E. Bouza, L. Torres, M. Diaz, L. Alcala, and M. Rodriguez-Creixems. 2000. Utility of the BACTEC Myco/F lytic medium for the detection of mycobacteria in blood. Diagn. Microbiol. Infect. Dis. 38:223-226. [DOI] [PubMed] [Google Scholar]
- 27.McAdam, R. A., P. W. Hermans, D. van Soolingen, Z. F. Zainuddin, D. Catty, J. D. van Embden, and J. W. Dale. 1990. Characterization of a Mycobacterium tuberculosis insertion sequence belonging to the IS3 family. Mol. Microbiol. 4:1607-1613. [DOI] [PubMed] [Google Scholar]
- 28.McHugh, T. D., L. E. Newport, and S. H. Gillespie. 1997. IS6110 homologs are present in multiple copies in mycobacteria other than tuberculosis-causing mycobacteria. J. Clin. Microbiol. 35:1769-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papavinasasundaram, K. G., M. J. Colston, and E. O. Davis. 1998. Construction and complementation of a recA deletion mutant of Mycobacterium smegmatis reveals that the intein in Mycobacterium tuberculosis RecA does not affect RecA function. Mol. Microbiol. 30:525-534. [DOI] [PubMed] [Google Scholar]
- 30.Perkins, M. D. 2000. New diagnostic tools for tuberculosis. Int. J. Tuber. Lung Dis. 4:S182-S188. [PubMed] [Google Scholar]
- 31.Ritis, K., D. Tzoanopoulos, M. Speletas, E. Papadopoulos, K. Arvanitidis, S. Kartali, and P. Sideras. 2000. Amplification of IS6110 sequence for detection of Mycobacterium tuberculosis complex in HIV-negative patients with fever of unknown origin (FUO) and evidence of extrapulmonary disease. J. Intern. Med. 248:415-424. [DOI] [PubMed] [Google Scholar]
- 32.Saves, I., M. A. Laneelle, M. Daffé, and J. M. Masson. 2000. Inteins invading mycobacterial RecA proteins. FEBS Lett. 480:221-225. [DOI] [PubMed] [Google Scholar]
- 33.Saves, I., F. Westrelin, M. Daffé, and J. M. Masson. 2001. Identification of the first eubacterial endonuclease coded by an intein allele in the pps1 gene of mycobacteria. Nucleic Acids Res. 29:4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Supply, P., J. Magdalena, S. Himpens, and C. Locht. 1997. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol. Microbiol. 26:991-1003. [DOI] [PubMed] [Google Scholar]
- 35.Walker, G. T., J. G. Nadeau, P. A. Spears, J. L. Schram, C. M. Nycz, and D. D. Shank. 1994. Multiplex strand displacement amplification (SDA) and detection of DNA sequences from Mycobacterium tuberculosis and other mycobacteria. Nucleic Acids Res. 22:2670-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warren, R. M., S. L. Sampson, M. Richardson, G. D. Van Der Spuy, C. J. Lombard, T. C. Victor, and P. D. van Helden. 2000. Mapping of IS6110 flanking regions in clinical isolates of Mycobacterium tuberculosis demonstrates genome plasticity. Mol. Microbiol. 37:1405-1416. [DOI] [PubMed] [Google Scholar]
- 37.Watt, B. 2000. Issues facing TB control (5.1). (a). Diagnostic issues: laboratory diagnosis of tuberculosis present techniques. Scott. Med. J. 45: 38-39, 43. [DOI] [PubMed] [Google Scholar]
- 38.Wayne, L. G., and G. P. Kubica. 1986. Family Mycobacteriaceae Chester 1897, 63AL, p. 1436-1457. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. William & Wilkins, Baltimore, Md. [Google Scholar]
- 39.Weil, A., B. B. Plikaytis, W. R. Butler, C. L. Woodley, and T. M. Shinnick. 1996. The mtp40 gene is not present in all strains of Mycobacterium tuberculosis. J. Clin. Microbiol. 34:2309-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson, S. M., R. McNerney, P. M. Nye, P. D. Godfrey-Faussett, N. G. Stoker, and A. Voller. 1993. Progress toward a simplified polymerase chain reaction and its application to diagnosis of tuberculosis. J. Clin. Microbiol. 31:776-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woods, G. L. 2001. Molecular techniques in mycobacterial detection. Arch. Pathol. Lab. Med. 125:122-126. [DOI] [PubMed] [Google Scholar]
- 42.Yuen, L. K. W., B. C. Ross, K. M. Jackson, and B. Dwyer. 1993. Characterization of Mycobacterium tuberculosis strains from Vietnamese patients by Southern blot hybridization. J. Clin. Microbiol. 31:1615-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]