Abstract
Although substantial epidemiologic evidence links Streptococcus mutans to caries, the pathobiology of caries may involve more complex communities of bacterial species. Molecular methods for bacterial identification and enumeration now make it possible to more precisely study the microbiota associated with dental caries. The purpose of this study was to compare the bacteria found in early childhood caries (ECC) to those found in caries-free children by using molecular identification methods. Cloning and sequencing of bacterial 16S ribosomal DNAs from a healthy subject and a subject with ECC were used for identification of novel species or uncultivated phylotypes and species not previously associated with dental caries. Ten novel phylotypes were identified. A number of species or phylotypes that may play a role in health or disease were identified and warrant further investigation. In addition, quantitative measurements for 23 previously known bacterial species or species groups were obtained by a reverse capture checkerboard assay for 30 subjects with caries and 30 healthy controls. Significant differences were observed for nine species: S. sanguinis was associated with health and, in order of decreasing cell numbers, Actinomyces gerencseriae, Bifidobacterium, S. mutans, Veillonella, S. salivarius, S. constellatus, S. parasanguinis, and Lactobacillus fermentum were associated with caries. These data suggest that A. gerencseriae and other Actinomyces species may play an important role in caries initiation and that a novel Bifidobacterium may be a major pathogen in deep caries. Further investigation could lead to the identification of targets for biological interventions in the caries process and thereby contribute to improved prevention of and treatment for this significant public health problem.
Dental caries is the single most common chronic disease of childhood, with a rate five times greater than that seen for the next most prevalent disease, asthma (37). Early childhood caries (ECC) results in a considerable direct burden of pain and suffering as well as poorer general health (1, 16). Caries is disproportionately present in low-income children (37), although it is by no means limited to this group. The overall prevalence of ECC in the United States is estimated at 1 to 5%, although among high-risk populations the prevalence has been reported to be as high as 60% (29). Dental care was recently shown to be the most common unmet health care need among children in the United States (22). Treatment for children with caries can be expensive, often requiring extensive restorative treatment under general anesthesia. Despite efforts in restorative therapy, children who experience ECC continue to be at a higher risk for new lesions in both the primary and the permanent dentition (36). Interventions which disrupt the pathobiology of caries are needed to prevent and treat this aggressive infectious disease. In order to develop these strategies, however, it is important that all bacteria associated with dental caries and dental health be identified.
Considerable epidemiologic evidence links Streptococcus mutans to caries (40), and numerous laboratory investigations have demonstrated the ability of strains of this species to produce the lactic acid which causes dental caries (40). A closely related lactate-producing species, S. sobrinus, has also been linked to caries, although the prevalence is distinctly lower and this species is seldom found without S. mutans (40). Various Lactobacillus species have also been consistently associated with caries and are thought to be important secondary pathogens in dental caries (40). Actinomyces species have also been suspected to play a role in caries, with most evidence linking them to root surface caries (40). Other bacteria have been investigated as potential contributors to caries, and several investigators have suggested that the pathobiology of caries may involve more complex communities of bacterial species than previously thought [(3, 40, 42); M. K. Russell, M. F. J. Maiden, J. Lopman, S. K. Boches, J. L. Galvin, F. E. Dewhirst, and B. J. Paster., J. Dent Res. 79(IADR Abstr.):465, 2000].
Nearly all investigations into the microbial pathogenesis of caries have been conducted by cultivation of bacteria. Molecular methods for bacterial identification and enumeration now make it possible to more precisely study the microbiota associated with dental caries (12, 24). DNA sequence-based assays can be used to identify closely related species that are difficult to differentiate by traditional, culture-based approaches. In addition, 16S ribosomal DNA (rDNA) sequence-based clonal analysis allows for the detection and identification of species that are refractory to detection by traditional methods. These bacteria may escape detection either because they do not grow on the media used or because they are not distinguishable from similar species by observable, phenotypic characteristics.
Approximately 260 species of oral bacteria have been cultivated from humans, and actual diversity had been estimated at approximately 500 common species (21, 34). A major recent 16S rDNA sequencing effort with 2,522 oral clones by Paster et al. (26) and smaller projects by others (8, 14, 31, 35, 41) identified a total of over 500 human oral species or not-yet-cultivated phylotypes. These species and phylotypes were detected in the oral cavities of subjects with periodontitis and healthy controls (26, 31, 35), in dentoalveolar abscesses (41), and in a subject with mild gingivitis (14). Similar investigations have not been conducted with caries-associated biofilms (40), and it seems likely that important caries pathogens have yet to be identified.
The purpose of this study was to compare the bacteria found in children with severe caries to those found in caries-free children by using molecular identification methods. A PCR-based, reverse capture checkerboard hybridization method was used to detect and enumerate 23 known oral bacterial species or groups. 16S gene cloning and sequencing were used for identification of novel species or phylotypes and species not previously associated with dental caries.
MATERIALS AND METHODS
Subject selection.
Subjects were recruited from Columbus Children's Hospital Dental Clinic, Columbus, Ohio, and The Ohio State University College of Dentistry Dental Clinic, Columbus. Consent was obtained from the parents of all subjects for this institutionally approved study. The subjects ranged in age from 2 to 8 years. Thirty subjects with severe caries involving at least six teeth were identified for the experimental group. An age- and sex-matched healthy control group consisted of 30 subjects. Control subjects had no caries or existing restorations. One subject from the control group and one subject from the caries group were randomly selected for clonal analysis.
Sampling.
The healthy subjects were sampled at a minimum of four sites, including anterior and posterior teeth, and the samples were pooled. Pooled plaque from the caries subjects was collected separately from four types of sites: (i) surfaces of intact enamel, (ii) surfaces of white spot lesions, (iii) surfaces of cavitated lesions, and (iv) excavated carious dentin from cavitated lesions. For the enamel, white spot lesions, and cavitated lesions, the plaque was obtained by swiping the tooth surface with a dental explorer and wiping the plaque onto a coarse endodontic paper point. The carious dentin was excavated either with a spoon excavator or round bur in a slow-speed hand piece. Each sample was obtained by pooling material collected from at least three different teeth. Samples were placed in a sterile 1.5-ml microcentrifuge tube and transported to the laboratory, where they were frozen until further analysis.
Isolation of bacterial DNA.
The DNA was isolated by using a bead beater apparatus. The samples were placed in 300 μl of Tris-EDTA buffer, beaten with 0.25 g of 0.1-mm glass beads for 60 s at 5,000 rpm in a Biospec Products bead beater, and chilled on ice immediately thereafter. The DNA was purified as previously described (15) and frozen for later analysis.
Amplification of 16S rDNA by PCR.
16S rDNA was amplified under standard conditions by using a universal forward primer (5′-GAG AGT TTG ATY CTG GCT CAG-3′) and a universal reverse primer (5′-GAA GGA GGT GWT CCA RCC GCA-3′) (26). PCR was performed with thin-walled tubes and a PE Applied Biosystems 9700 Thermocycler. One microliter of DNA template was added to a reaction mixture (50-μl final volume) containing 20 nmol of each primer, 40 nmol of deoxynucleoside triphosphates, and 1 U of Taq 2000 polymerase (Stratagene, La Jolla, Calif.) in buffer containing Taqstart antibody (Sigma Chemical Co.). In a hot-start protocol, samples were preheated at 95°C for 8 min followed by amplification under the following conditions: denaturation at 95°C for 45 s, annealing at 60°C for 45 s, and elongation for 1.5 min with an additional 5 s for each cycle. A total of 30 cycles were performed and then followed by a final elongation step at 72°C for 10 min. The results of PCR amplification were examined by electrophoresis in a 1% agarose gel. DNA was stained with ethidium bromide and visualized under short-wavelength UV light.
Checkerboard hybridization.
The reverse capture checkerboard assay was carried out as previously described (25). DNA probes were synthesized with multiple thymidines (T) at the 5′ end of the oligonucleotide (sequences are shown in Table 1). The poly(T) tails were cross-linked to the membrane support via UV irradiation, leaving the specific probe available for hybridization. The 16S rDNA sequences isolated from bacterial plaque were amplified by PCR with universal primers; the forward primer was labeled at the 5′ end with digoxigenin (sequences are shown in Table 1). The digoxigenin-labeled amplicon was hybridized to the capture probes bound to the membrane. The digoxigenin residues were complexed to antidigoxigenin antibody covalently bound to alkaline phosphatase. Standard chemifluorescence (Storm system) procedures were used for detection.
TABLE 1.
Primers and probes used for PCR and reverse capture checkerboard analysis
| Specificity | Starting base position (E. coli) | Sequence (5′-3′)a |
|---|---|---|
| Universal forward PCR primer (digoxigenin labeled) | 8 | Digoxigenin-AGAGTTTGATYMTGGC |
| Universal reverse PCR primer | 1492 | GYTACCTTGTTACGACTT |
| Actinomyces gerencseriae | 65 | TTTTTTTTTTTTTTTTTTTTACCCCAGAAGCCCGTT |
| Actinomyces israelii | 179 | TTTTTTTTTTTTTTTTTTTTGGCACAGCCAGAACAC |
| Actinomyces naeslundii serotype II | 87 | TTTTTTTTTTTTTTTTTTTTCACTCATCCAGAACCAG |
| Actinomyces naeslundii serotype III | 70 | TTTTTTTTTTTTTTTTTTTTAGCAGGCCCCTTCA |
| Actinomyces odontolyticus | 998 | TTTTTTTTTTTTTTTTTTTTCAGTGCCGCCGTGC |
| Bifidobacterium (All) | 228 | TTTTTTTTTTTTTTTTTTTTGGACGCGACCCCAT |
| Bifidobacterium dentium | 179 | TTTTTTTTTTTTTTTTTTTTCATCCAACCGGAGCAT |
| Lactobacillus fermentum | 70 | TTTTTTTTTTTTTTTTTTTTATCAATCAATTGGGCCAAC |
| Rothia dentocariosa | 995 | TTTTTTTTTTTTTTTTTTTTAAATGACGCAGTCCAGTATATG |
| Rothia dentocariosa or Stomatococcus mucilaginosus | 179 | TTTTTTTTTTTTTTTTTTTTGCGGAGATTGGTCGTAT |
| Streptococcus anginosus | 187 | TTTTTTTTTTTTTTTTTTTTCTTTCAAGCATCTAACATGTG |
| Streptococcus anginosus or S. gordonii | 87 | TTTTTTTTTTTTTTTTTTTTCAACTCACAGTYTATGGTGTAG |
| Streptococcus anginosus or S. intermedius | 445 | TTTTTTTTTTTTTTTTTTTATGG ATTCTCACACTTGTTCTTCCTb |
| Streptococcus constellatus | 445 | TTTTTTTTTTTTTTTTTATATTACTCTCACACACGATCTTCCTb |
| Streptococcus cristatus | 178 | TTTTTTTTTTTTTTTTTTTTCATGCAATAGTCAATGTTATG |
| Streptococcus intermedius or S. constellatus | 172 | TTTTTTTTTTTTTTTTTTTTCAGTAAATGTTCTTATGCGGTA |
| Streptococcus mitis biovar II | 175 | TTTTTTTTTTTTTTTTTTTTCAATAACTGCTATTATGCGG |
| Streptococcus mitis, S. oralis, or S. pneumoniae | 187 | TTTTTTTTTTTTTTTTTTTTCCTTTTAAGYAAATGTCATGC |
| Streptococcus mutans | 586 | TTTTTTTTTTTTTTTTTTTTTTTTACTCCAGACTTTCCTGACCG |
| Streptococcus parasanguinis | 170 | TTTTTTTTTTTTTTTTTTTTGTCGACTTTTATGCGGTATTA |
| Streptococcus salivarius | 175 | TTTTTTTTTTTTTTTTTTTTGTCATCCATTGTTATGCGG |
| Streptococcus salivarius, S. sanguinis, or group H6 | 89 | TTTTTTTTTTTTTTTTTTTATGCAACTCATCCAAGAAGAG |
| Streptococcus sobrinus | 175 | TTTTTTTTTTTTTTTTTTTTGTTAACTCCTCTTATGCGG |
| Streptococcus group H6 | 176 | TTTTTTTTTTTTTTTTTTTTATGCGATAATCCATTTTATGCG |
| Streptococcus (All) | 491 | TTTTTTTTTTTTTTTTTTTTTTAGCCGTCCCTTTCTGGT |
| Veillonella (All) | 217 | TTTTTTTTTTTTTTTTTTTTAATCCCCTCCTTCAGTGA |
| Universal 1 | 1089 | TTTTTTTTTTTTTTTTTTTTCTCGTTGCGGGACTTAAC |
| Universal 2 | 341 | TTTTTTTTTTTTTTTTTTTTCTGCTGCCTCCCGTAGG |
Sequences that target a specific species or phylogenetic group are shown in italics. Seventeen to 22 T's are covalently bound at the 5′ end.
Additional sequence information from other strains of the target species has shown that the underlined base T should be C and the underlined base A should be T. However, specificity was observed with the potential mismatch.
Clonal analysis.
PCR-amplified 16S genes isolated from plaque were cloned by using a TOPO TA cloning kit (Invitrogen, San Diego, Calif.) in accordance with the manufacturer's instructions. Transformation was done by using competent Escherichia coli TOP10 cells provided by the manufacturer. The transformed cells were plated on Luria-Bertani agar plates supplemented with kanamycin and incubated overnight at 37°C. White colonies were placed in 40 μl of 10 mM Tris. The sizes of the inserts (approximately 1,500 bp) were determined by PCR with flanking vector primers followed by electrophoresis in a 1% agarose gel.
16S rDNA sequencing.
Purified DNA from PCR was sequenced by using an ABI Prism cycle sequencing kit (BigDye Terminator cycle sequencing kit with AmpliTaq DNA polymerase FS; Perkin-Elmer). The primers used for sequencing were previously described (7). Quarter dye chemistry was used with 80 μM primers and 1.5 μl of PCR product in a final volume of 20 μl. Cycle sequencing was performed by using a PE Applied Biosystems 9700 Thermocycler with 25 cycles of denaturation at 96°C for 10 s and annealing and extension at 60°C for 4 min. Sequencing reactions were run on an ABI model 377 DNA sequencer.
16S rDNA data analysis.
A total of 294 clones with the correctly sized insert (approximately 1,500 bases) were analyzed. Approximately 500 bases were obtained first to determine identity or approximate phylogenetic position. Full sequences of approximately 1,500 bases were obtained with five or six additional sequencing primers (26) for most of the novel species. For identification of closest relatives, sequences of the unrecognized inserts were compared to the 16S rRNA gene sequences of over 4,000 bacteria in our database and the 16,000 sequences in the Ribosomal Database Project (18, 19) and GenBank. Programs for data entry, editing, sequence alignment, secondary structure comparison, similarity matrix generation, and phylogenetic tree construction were written by F. E. Dewhirst (27). The similarity matrices were corrected for multiple base changes at single positions by the method of Jukes and Cantor (13). Similarity matrices were constructed from the aligned sequences by using only those sequence positions from which data were available for 90% of the strains. Phylogenetic trees were constructed by using the neighbor-joining method of Saitou and Nei (30). TREECON software was used for the construction and drawing of evolutionary trees (38).
Statistical analysis.
To determine if differences were present between healthy subjects and healthy sites in subjects with caries, levels of bacterial species found in the healthy group were compared to those at intact sites in the group with caries. Since distributions were skewed, the nonparametric Wilcoxon rank sum test was used for comparisons. To examine the relationship of bacterial levels to caries severity, paired, within-subject comparisons based on depth of caries penetration were made for subjects with caries. Specifically, levels found at intact sites in subjects with caries were compared to levels found at each of the disease sites in the same subjects (white spot, cavitated, and dentin penetration). The Wilcoxon signed rank test, a nonparametric test for paired data, was used. The Bonferroni correction was applied to adjust the alpha level for both the Wilcoxon rank sum test and the Wilcoxon signed rank test since multiple comparisons were made (with tests run for 23 species, the alpha level was lowered to 0.002).
RESULTS
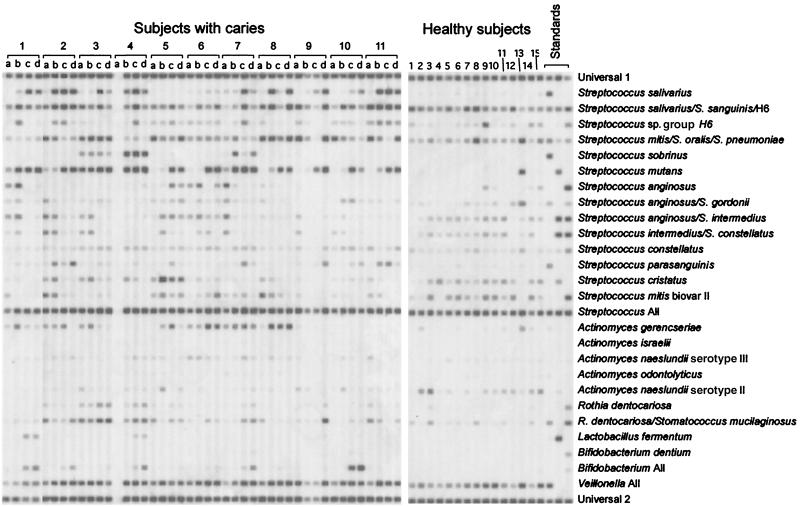
The association of 23 known bacterial species or species groups with caries was determined by a reverse capture checkerboard assay. For these studies, plaque was collected from 30 subjects with caries (four sites per subject) and 30 healthy controls. A hybridization blot demonstrating the bacterial population distribution in 11 subjects with caries and 15 healthy controls is shown in Fig. 1. Samples from one of these subjects with caries and one of these healthy subjects were used for clonal analysis as described below.
FIG. 1.
Reverse capture checkerboard hybridization blot for 11 subjects with caries and 15 healthy control subjects (without caries). For the caries samples, lanes a represent intact enamel, lanes b represent white spot lesions, lanes c represent cavitated lesions, and lanes d represent carious dentin. Species-specific probes were applied in horizontal rows, and samples or standards were then hybridized in vertical lanes. Probe specificities are listed on the right. The last three columns contained DNA isolated from laboratory strains at a concentration of 108 cells per ml. The first lane of standards contained S. salivarius, S. oralis, S. sobrinus, S. gordonii, S. parasanguinis, A. gerencseriae, A. odontolyticus, S. mucilaginosus, and Veillonella parvula. The second lane contained S. sanguinis, S. mitis, S. mutans, S. intermedius, A. israelii, A. naeslundii, and L. fermentum. The third lane contained S. pneumoniae, S. anginosus, S. constellatus, S. mitis biovar II, A. naeslundii serotype III, R. dentocariosa, and B. dentium.
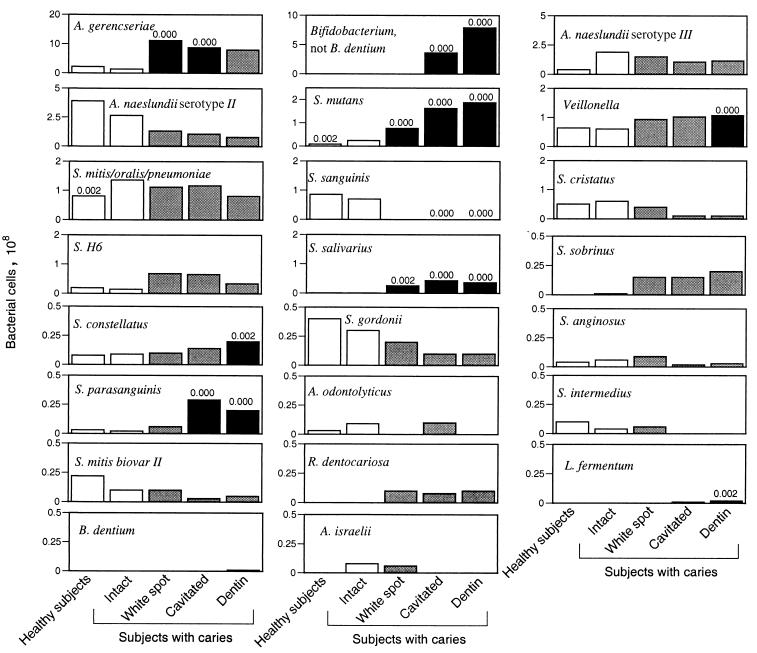
To determine if differences were present between healthy subjects and healthy sites in subjects with caries, between-group comparisons were made for bacterial levels in healthy subjects and intact enamel sites in diseased subjects. Significant differences were observed for two probes, as shown in Fig. 2. To examine the relationship of bacterial levels to caries progression, paired, within-subject comparisons based on depth of caries penetration were made for subjects with caries: bacterial levels found on intact enamel were compared to those found at each of the caries sites (white spot, cavitated lesions, and excavated carious dentin). Significant differences were observed for nine probes. Mean levels and P values are shown in Fig. 2.
FIG. 2.
Mean levels of 23 species or species groups as determined by a reverse capture checkerboard assay for 30 healthy subjects and 30 subjects with caries. Species are ordered by decreasing quantity detected, and scales have been adjusted for each species. Levels in healthy subjects and at intact enamel sites in subjects with caries are shown as white bars. They were compared by the Wilcoxon rank sum test; P values are shown for significant differences above the first white bar. Levels at caries sites were compared to those found at intact enamel sites in the same subject by the Wilcoxon signed rank test. Levels that were significantly different are shown as black bars with P values, and those that were not are shown as gray bars. For all tests, the Bonferroni correction for multiple comparisons was used, adjusting alpha levels to 0.002. S. H6, Streptococcus group H6.
The mean age for the healthy group was 4.3 years (standard deviation [SD], 1.6), and that for the group with caries was 4.8 years (SD, 1.6). This difference was not significant, as determined by a t test. The healthy group was 48% female, and the caries group was 43% female. This difference was not significant, as determined by chi-square analysis. The groups were not matched for race, and the control group contained significantly more African-American subjects (38%; P = 0.02) than did the caries group (10%). However, no significant differences by racial group were seen for levels of any species either in healthy subjects or in subjects with caries (as determined by the Wilcoxon rank sum test).
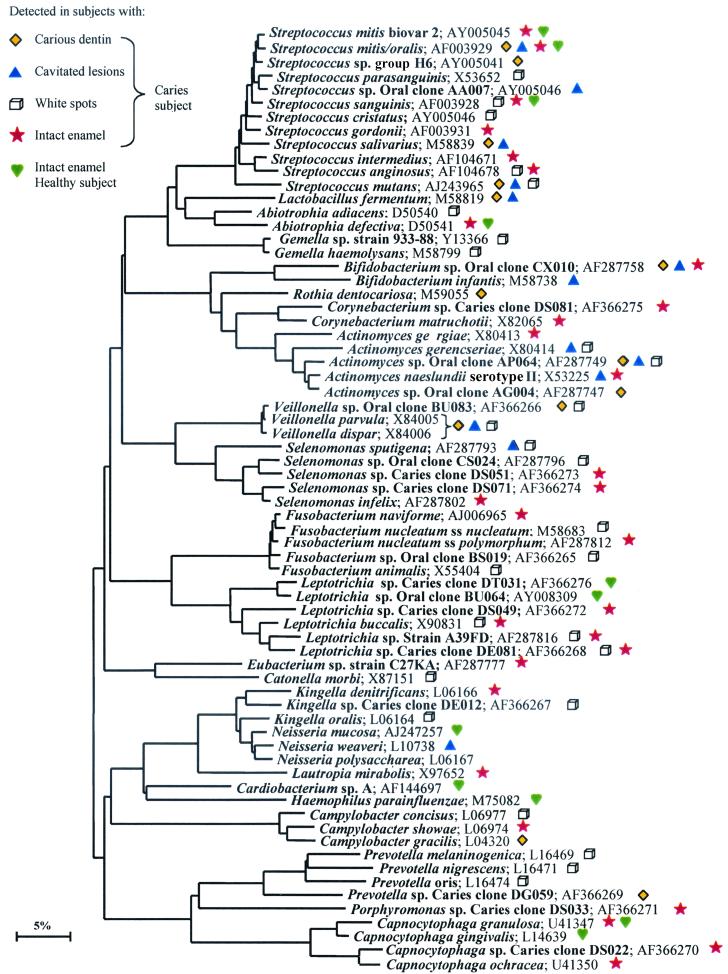
To determine if novel or previously unsuspected species are important in ECC, bacterial 16S gene PCR products from one healthy subject (healthy subject 1, Fig. 1) and all four site types from a single subject with caries (caries subject 1, Fig. 1) were cloned and sequenced. The healthy subject randomly selected for clonal analysis was a 6-year-old African-American male, and the subject with caries was a 4-year-old white female. A minimum of 50 clones were sequenced from one sample of each type, for a total of 294 sequences. A 16S gene distance tree (Fig. 3) shows the phylogenetic relationship of all species detected by clonal analysis. Table 2 shows all clones recovered more than once in order of decreasing frequency of detection and by type of site from which they were recovered.
FIG. 3.
Phylogenetic tree showing the relationships of species and phylotypes detected by clonal analysis. Novel phylotypes are designated “Caries clone.” S. mitis and S. oralis as well as V. parvula and V. dispar could not be differentiated based on sequence comparisons and are grouped together. GenBank accession numbers are listed. The marker bar represents a 5% difference in nucleotide sequences.
TABLE 2.
Clones identified from a healthy subject and a subject with caries
| Species or phylotype | No. of times clones were recovered from the following subject and site:
|
|||||
|---|---|---|---|---|---|---|
| Healthy
|
Caries
|
Total | ||||
| Intact enamel | White spot lesions | Cavitated lesions | Dentin | |||
| Streptococcus mutans | 11 | 30 | 19 | 60 | ||
| Veillonella dispar or V. parvula | 4 | 21 | 7 | 32 | ||
| Streptococcus sanguinis | 25 | 1 | 1 | 27 | ||
| Bifidobacterium sp. clone CX010 | 2 | 4 | 11 | 17 | ||
| Corynebacterium matruchotii | 11 | 11 | ||||
| Abiotrophia defectiva | 7 | 1 | 8 | |||
| Leptotrichia buccalis | 7 | 1 | 8 | |||
| Actinomyces sp. clone AP064 | 3 | 2 | 2 | 7 | ||
| Fusobacterium animalis | 7 | 7 | ||||
| Streptococcus mitis or S. oralis | 1 | 2 | 1 | 2 | 6 | |
| Lactobacillus fermentum | 4 | 2 | 6 | |||
| Neisseria mucosa | 6 | 6 | ||||
| Streptococcus salivarius | 4 | 1 | 5 | |||
| Selenomonas sputigena | 1 | 4 | 5 | |||
| Streptococcus mitis biovar II | 2 | 2 | 4 | |||
| Actinomyces gerencseriae | 3 | 1 | 4 | |||
| Leptotrichia sp. clone DE081 | 3 | 1 | 4 | |||
| Leptotrichia sp. strain A39FD | 1 | 3 | 4 | |||
| Fusobacterium nucleatum subsp. polymorphum | 4 | 4 | ||||
| Streptococcus anginosus | 1 | 2 | 3 | |||
| Gemella haemolysans | 3 | 3 | ||||
| Leptotrichia sp. clone BU064 | 3 | 3 | ||||
| Capnocytophaga granulosa | 1 | 2 | 3 | |||
| Abiotrophia adiacens | 2 | 2 | ||||
| Gemella sp. strain 933-88 | 2 | 2 | ||||
| Additional species or phylotypesa | 4 | 17 | 13 | 4 | 6 | 43 |
| Total no. of species or phylotypes | 11 | 29 | 27 | 13 | 13 | 68 |
| No. of novel phylotypes | 1 | 7 | 2 | 0 | 1 | 10 |
| Total no. of clones | 50 | 58 | 61 | 74 | 51 | 294 |
Found only once; not listed here but included in Fig. 3.
DISCUSSION
For this study, bacterial samples were collected from a group of 30 children with severe dental caries. Samples were collected from healthy enamel and from lesions of various degrees of severity for each subject. To determine the association of bacterial species with caries severity, the samples obtained from healthy enamel were compared to those obtained from progressively deeper lesions. For these comparisons, a paired, within-subject statistical approach was used. Samples were also collected from a demographically similar control group with no previous history of decay, and between-group comparisons were made for samples collected from these healthy children and samples collected from healthy tooth enamel in children with decay.
Quantitative measurements for 23 bacterial species or species groups were obtained by a reverse capture checkerboard assay for all 60 subjects. As expected, nonnormal distributions were observed, so nonparametric tests were used for all comparisons. Since multiple tests were performed, a conservative approach was adopted, applying the Bonferroni correction to lower the alpha level to 0.002.
Bacterial levels in healthy subjects and at healthy sites in subjects with caries were compared by the Wilcoxon rank sum test for all species. Significantly higher levels were observed in subjects with caries than in healthy subjects only for S. mutans, as expected, and for the S. mitis-S. oralis-S. pneumoniae group, which was measured by using a probe that hybridized to all three species. S. mitis and S. oralis are indistinguishable by 16S gene sequences, and both have been associated with human dental caries. Although S. pneumoniae is frequently isolated from the oral cavity, it has not been associated with caries. Further development of molecular assays to distinguish among these species is needed to investigate their association with caries.
Within subjects with caries, bacterial levels found on intact enamel were compared to those found at each of the caries sites (white spot, cavitated lesions, and excavated carious dentin) by using a paired data approach, the Wilcoxon sign rank test. Significant associations were observed for nine species, as shown in Fig. 2. A large range in the levels of the different species was observed, and the species shown in Fig. 2 are arranged in descending order of bacterial levels, with scales adjusted for individual graphs. The most numerous were species of Actinomyces, Bifidobacterium, and Veillonella, S. mutans, the S. mitis-S. oralis-S. pneumoniae group, and S. sanguinis. The limit of detection for the assay was about 102 cells/PCR tube, or approximately 105 cells/ml.
A strong relationship to caries was observed for S. mutans at all lesion depths. This finding was expected, as the role of lactic acid production by S. mutans in caries pathogenesis is well documented and numerous clinical investigations have shown a strong association between caries and the presence of S. mutans (40).
Although S. sobrinus is closely related to S. mutans, often grouped with S. mutans as one of the mutans group streptococci, and is thought to be involved in dental caries (40), our data do not indicate a major role for S. sobrinus in ECC. S. sobrinus was detected at any site in only 9 of 30 subjects, was not significantly associated with caries, and was found at low levels compared to S. mutans and other caries-associated species.
In the earliest stage of caries, white spot formation, high mean levels of Actinomyces gerencseriae were observed (Fig. 2), suggesting a role for A. gerencseriae in caries initiation. The SD was large, and the bacteria were not present in all subjects (data not shown); however, in 18 out of 29 samples from white spot lesions, the bacteria were present at greater than 3 × 108 cells, making them the most numerous bacteria measured in these lesions. A. gerencseriae is a common inhabitant of the oral cavity of children once teeth have erupted (32), and closely related Actinomyces species have been shown to produce lactic acid (40). A. gerencseriae has been isolated from root caries (4) but has not been associated with caries in children. It is interesting that a closely related species, Actinomyces naeslundii serotype II (A. viscosus), has been associated with invasion into the tooth structure and early lesion formation (9). Actinomyces species have also been associated with early root caries lesions, although S. mutans has been found more commonly in advanced lesions (33). Further investigation is needed to explore the relationship of A. gerencseriae to ECC. Such investigation is important because this bacterium may provide a target for biological interventions to prevent caries initiation.
Another surprising finding was the strong relationship to deep caries that was observed for species of Bifidobacterium. Significant differences were observed for cavitated lesions and carious dentin but not for plaque from early, white spot lesions. This finding suggests an important role for Bifidobacterium in the progression of caries. Bifidobacterium species were the most numerous bacteria identified in both cavitated and deep dentinal caries, outnumbering S. mutans and far outnumbering Lactobacillus fermentum. Bifidobacterium species were present in plaque isolated from cavitated lesions in 16 of 30 subjects and in the dentin of 21 of 30 subjects with ECC. Levels ranged from 5 × 107 to 4 × 109 bacterial cells (data not shown). Bifidobacterium species have been isolated from caries lesions, including dentin (6, 11), and are known to produce lactate (6), but they have not been thought to play a major role in caries. Our data suggest that bifidobacteria may be major pathogens in deep caries in young children. Further investigation is needed, as these bacteria may provide additional targets for biological interventions to prevent caries progression. The “Bifidobacterium All” probe used in the checkerboard analysis is specific to the Bifidobacterium genus, except for B. dentium. A B. dentium-specific probe was also used, but only very low levels of this species were detected, indicating that the species of interest is not B. dentium. Indeed, a novel species of Bifidobacterium was identified by using clonal analysis (see below).
Lactobacillus species have been implicated as secondary pathogens in deep carious lesions (39). Although L. fermentum was found to be statistically significantly associated with deep lesions by the checkerboard analysis, only low and possibly clinically insignificant levels were observed. The subject randomly selected for clonal analysis had the highest levels of L. fermentum of any subject (2 × 107), and even in this subject, Bifidobacterium far outnumbered L. fermentum. These data suggest that the major secondary pathogens in ECC are not Lactobacillus species but rather is a novel species of Bifidobacterium.
Veillonella was found at relatively high levels in all subjects and was significantly more numerous in deep dentinal lesions than at any other site. Veillonella metabolizes lactic acid produced by other species to form propionic and acetic acids, both weaker acids than lactic acid and so less capable of dissolving tooth enamel. Species of Veillonella have been detected in high proportions in progressing incipient lesions (20), and in vitro studies have shown that the combination of Veillonella and S. mutans allows more acid production and greater demineralization than does S. mutans alone (23). Veillonella has also been shown to stimulate glycolysis in S. salivarius by acting as a sink for the lactic acid produced (10). These data suggest that the utilization of lactic acid by Veillonella may be an important factor in the caries process, providing protection for acid-producing bacteria.
Three other species found at low levels showed a statistically significant relationship to caries. S. salivarius was significantly associated with all depths of caries, and S. parasanguinis and S. constellatus were associated with deeper lesions. All of these species have been linked to caries, although none are thought to play a major role.
High levels and large between-subject variations were observed for A. naeslundii serotype II and A. naeslundii serotype III, but no significant relationship to caries was seen. Bacteria identified as A. naeslundii have been associated with root caries in an elderly population (2), but based on our data, A. naeslundii species do not appear to play a role in childhood caries.
S. sanguinis was the only species tested that was significantly associated with health, and this association is consistent with previous reports (39). This bacterium is acquired in most children shortly after tooth eruption, and its presence at high levels delays the acquisition of S. mutans (5). It is also known to modulate pH by producing ammonia (28).
In order to identify the predominant bacterial species, both cultivable and not yet cultivated, that were present in the supragingival plaque of a healthy subject and at different sites in one subject with caries, partial sequences of about 500 bp were obtained for 294 16S rDNA clones. Overall, 68 species or phylotypes were detected. Fifty were known species or cultivated strains within known genera, and 18 were phylotypes or not-yet-cultivated species. Of those 18 oral clones, 10 were novel phylotypes first reported in this study. The term phylotype was used for clusters of clone sequences that differed from known species by approximately 30 bases or more and were at least 99% similar to members of their cluster. The diversity of known species and novel phylotypes with respect to health status is shown in Table 2.
Half of all clones recovered from the healthy subject were S. sanguinis. This finding is consistent with the results of the checkerboard analysis and provides further evidence for the role of S. sanguinis in healthy plaque. A few unexpected species were observed at high levels: Abiotrophia defectiva and Neisseria mucosa were recovered in large numbers from the healthy subject. The most complex (29 species or phylotypes) and poorly characterized population was found on intact enamel sites in the caries subject, with 7 of the 10 novel phylotypes seen in this study being found at these sites. Again, unexpected species were seen: Corynebacterium matruchotii and Leptotrichia buccalis were found in large numbers. Further investigation of health-associated biofilm constituents may provide a better understanding of the pathobiology of caries and could lead to strategies for the establishment of a stable, healthy flora to prevent caries.
The major disease-associated species detected by the reverse capture checkerboard assay were also detected by clonal analysis, confirming the high levels and caries association of S. mutans, Veillonella, Bifidobacterium, and S. salivarius detected by checkerboard analysis. A. gerencseriae was also observed by clonal analysis, although it did not appear to be as numerous as indicated by checkerboard analysis. L. fermentum was observed in deeper lesions by clonal analysis, but the subject who was selected for clonal analysis had extreme levels of lactobacilli relative to the other subjects, as discussed above. In addition, known, cultivable species that were not previously thought to play a role in dental caries (and therefore were not included in checkerboard analysis) were detected in substantial numbers by clonal analysis. For example, Fusobacterium animalis was the second most numerous species detected in white spot lesions. Some species of Fusobacterium produce lactate, so F. animalis may play a role in caries initiation. Several novel phylotypes were detected at caries sites, including an uncharacterized Actinomyces clone and a novel Bifidobacterium species. Based on the checkerboard analysis, this previously unknown species of Bifidobacterium may be one of the major pathogens in ECC. Further investigation of these newly implicated phylotypes or species will be interesting.
Although there are many advantages to molecular approaches over traditional culturing techniques, there are some limitations. Bias can be introduced by bacterial lysis procedures. Because of the high fraction of gram-positive species in supragingival dental plaque, a bead beater was chosen for this study to minimize the bias toward more easily lysed bacteria. Reverse capture checkerboard analysis was selected as the approach for this study because it is an efficient technique for the analysis of multiple bacterial species, even though it is less precise than more labor-intensive quantitative PCR methods. Although molecular methods have been developed that allow total quantitation of oral bacteria and calculation of relative compositions of particular species (17), these approaches were not suitably efficient for the large volume of samples and number of species examined in this study.
In conclusion, both checkerboard analysis and clonal analysis confirmed the relationship of S. mutans to dental caries and S. sanguinis to oral health. However, a number of additional species or phylotypes that may also play a role in health or disease were identified and warrant further investigation. Ten novel phylotypes were identified, demonstrating that the supragingival flora in children has not yet been completely described. It appears that A. gerencseriae or other Actinomyces species may play an important role in caries initiation, and a novel Bifidobacterium species may be a major pathogen in deep caries. Further studies are needed to investigate the possibility that previously unsuspected species are important in caries pathogenesis. Larger sample sizes for clonal analysis of unsuspected and novel species and studies with highly accurate methods for bacterial quantitation are needed. These studies could lead to the identification of targets for biological interventions in the caries process and thereby contribute to improved prevention of and treatment for this significant public health problem.
Acknowledgments
This work was supported by Public Health Service grants DE11443 and DE10374 to B.J.P. and F.E.D. and DE10467 to E.J.L. and A.L.G. from the National Institute of Dental and Craniofacial Research.
REFERENCES
- 1.Acs, G., R. Shulman, M. W. Ng, and S. Chussid. 1999. The effect of dental rehabilitation on the body weight of children with early childhood caries. Pediatr. Dent. 21:109-113. [PubMed]
- 2.Bowden, G. H. 1990. Microbiology of root surface caries in humans. J. Dent. Res. 69:1205-1210. [DOI] [PubMed] [Google Scholar]
- 3.Bradshaw, D. J., and P. D. Marsh. 1998. Analysis of pH-driven disruption of oral microbial communities in vitro. Caries Res. 32:456-462. [DOI] [PubMed] [Google Scholar]
- 4.Brailsford, S. R., R. B. Tregaskis, H. S. Leftwich, and D. Beighton. 1999. The predominant Actinomyces spp. isolated from infected dentin of active root caries lesions. J. Dent. Res. 78:1525-1534. [DOI] [PubMed] [Google Scholar]
- 5.Caufield, P. W., A. P. Dasanayake, Y. Li, Y. Pan, J. Hsu, and J. M. Hardin. 2000. Natural history of Streptococcus sanguinis in the oral cavity of infants: evidence for a discrete window of infectivity. Infect. Immun. 68:4018-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crociani, F., B. Biavati, A. Alessandrini, C. Chiarini, and V. Scardovi. 1996. Bifidobacterium inopinatum sp. nov. and Bifidobacterium denticolens sp. nov., two new species isolated from human dental caries. Int. J. Syst. Bacteriol. 46:564-571. [DOI] [PubMed] [Google Scholar]
- 7.Dewhirst, F. E., C. C. Chien, B. J. Paster, R. L. Ericson, R. P. Orcutt, D. B. Schauer, and J. G. Fox. 1999. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl. Environ. Microbiol. 65:3287-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dymock, D., A. J. Weightman, C. Scully, and W. G. Wade. 1996. Molecular analysis of microflora associated with dentoalveolar abscesses. J. Clin. Microbiol. 34:537-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Firestone, A. R., F. F. Feagin, T. J. Heaven, J. Sheetz, and F. Denys. 1993. In vitro demineralization by strains of Actinomyces viscosus and Streptococcus sobrinus of sound and demineralized root surfaces. J. Dent. Res. 72:1180-1183. [DOI] [PubMed] [Google Scholar]
- 10.Hamilton, I. R., and S. K. C. Ng. 1983. Stimulation of glycolysis through lactate consumption in a resting cell mixture of Streptococcus salivarius and Veillonella parvula. FEMS Microbiol. Lett. 20:61-65. [Google Scholar]
- 11.Hoshino, E. 1985. Predominant obligate anaerobes in human carious dentin. J. Dent. Res. 64:1195-1198. [DOI] [PubMed] [Google Scholar]
- 12.Hugenholtz, P., and N. R. Pace. 1996. Identifying microbial diversity in the natural environment: a molecular phylogenetic approach. Trends Biotechnol. 14:190-197. [DOI] [PubMed] [Google Scholar]
- 13.Jukes, T. H. 1969. Evolution of protein molecules, p. 406-425. In H. N. Munro (ed.), Mammalian protein metabolism, vol. 3. Academic Press, Inc., New York, N.Y. [Google Scholar]
- 14.Kroes, I., P. W. Lepp, and D. A. Relman. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 96:14547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leys, E. J., A. L. Griffen, S. J. Strong, and P. A. Fuerst. 1994. Detection and strain identification of Actinobacillus actinomycetemcomitans by nested PCR. J. Clin. Microbiol. 32:1288-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Low, W., S. Tan, and S. Schwartz. 1999. The effect of severe caries on the quality of life in young children. Pediatr. Dent. 21:325-326. [PubMed] [Google Scholar]
- 17.Lyons, S. R., A. L. Griffen, and E. J. Leys. 2000. Quantitative real-time PCR for Porphyromonas gingivalis and total bacteria. J. Clin. Microbiol. 38:2362-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, J. M. Stredwick, G. M. Garrity, B. Li, G. J. Olsen, S. Pramanik, T. M. Schmidt, and J. M. Tiedje. 2000. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 28:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maidak, B. L., G. J. Olsen, N. Larsen, R. Overbeek, M. J. McCaughey, and C. R. Woese. 1997. The RDP (Ribosomal Database Project). Nucleic Acids Res. 25:109-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milnes, A. R., and G. H. Bowden. 1985. The microflora associated with developing lesions of nursing caries. Caries Res. 19:289-297. [DOI] [PubMed] [Google Scholar]
- 21.Moore, W. E. C., and L. V. H. Moore. 1994. The bacteria of periodontal diseases. Periodontology 2000 5:66-77. [DOI] [PubMed] [Google Scholar]
- 22.Newacheck, P. W., D. C. Hughes, Y. Y. Hung, S. Wong, and J. J. Stoddard. 2000. The unmet health needs of America's children. Pediatrics 105:989-997. [PubMed] [Google Scholar]
- 23.Noorda, W. D., D. J. Purdell-Lewis, A. M. van Montfort, and A. H. Weerkamp. 1988. Monobacterial and mixed bacterial plaques of Streptococcus mutans and Veillonella alcalescens in an artificial mouth: development, metabolism, and effect on human dental enamel. Caries Res. 22:342-347. [DOI] [PubMed] [Google Scholar]
- 24.Pace, N. R., D. A. Stahl, D. J. Lane, and G. J. Olsen. 1986. The analysis of natural microbial populations by ribosomal RNA sequences. Adv. Microb. Ecol. 9:1-55. [Google Scholar]
- 25.Paster, B. J., I. M. Bartoszyk, and F. E. Dewhirst. 1998. Identification of oral streptococci using PCR-based, reverse-capture checkerboard hybridization. Methods Cell Sci. 20:223-231. [Google Scholar]
- 26.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paster, B. J., and F. E. Dewhirst. 1988. Phylogeny of campylobacters, wolinellas, Bacteroides gracilis and Bacteroides ureolyticus by 16S rRNA sequencing. Int. J. Syst. Bacteriol. 38:56-62. [Google Scholar]
- 28.Quivey, R. G., Jr., W. L. Kuhnert, and K. Hahn. 2000. Adaptation of oral streptococci to low pH. Adv. Microb. Physiol. 42:239-274. [DOI] [PubMed] [Google Scholar]
- 29.Ripa, L. W. 1988. Nursing caries: a comprehensive review. Pediatr. Dent. 10:268-282. [PubMed] [Google Scholar]
- 30.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 31.Sakamoto, M., M. Umeda, I. Ishikawa, and Y. Benno. 2000. Comparison of the oral bacterial flora in saliva from a healthy subject and two periodontitis patients by sequence analysis of 16S rDNA libraries. Microbiol. Immunol. 44:643-652. [DOI] [PubMed] [Google Scholar]
- 32.Sarkonen, N., E. Kononen, P. Summanen, A. Kanervo, A. Takala, and H. Jousimies-Somer. 2000. Oral colonization with Actinomyces species in infants by two years of age. J. Dent. Res. 79:864-867. [DOI] [PubMed] [Google Scholar]
- 33.Schupbach, P., V. Osterwalder, and B. Guggenheim. 1996. Human root caries: microbiota of a limited number of root caries lesions. Caries Res. 30:52-64. [DOI] [PubMed] [Google Scholar]
- 34.Socransky, S. S., and A. D. Haffajee. 1994. Evidence of bacterial etiology: a historical perspective. Periodontology 2000 5:7-25. [DOI] [PubMed] [Google Scholar]
- 35.Spratt, D. A., A. J. Weightman, and W. G. Wade. 1999. Diversity of oral asaccharolytic Eubacterium species in periodontitis—identification of novel phylotypes representing uncultivated taxa. Oral Microbiol. Immunol. 14:56-59. [DOI] [PubMed] [Google Scholar]
- 36.Tinanoff, N., and D. M. O'Sullivan. 1997. Early childhood caries: overview and recent findings. Pediatr. Dent. 19:12-16. [PubMed] [Google Scholar]
- 37.U.S. Department of Health and Human Services. 2000. Oral health in America: a report of the Surgeon General—executive summary. National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, Md.
- 38.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 39.van Houte, J. 1980. Bacterial specificity in the etiology of dental caries. Int. Dent. J. 30:305-326. [PubMed] [Google Scholar]
- 40.van Houte, J. 1994. Role of micro-organisms in caries etiology. J. Dent. Res. 73:672-681. [DOI] [PubMed] [Google Scholar]
- 41.Wade, W. G., D. A. Spratt, D. Dymock, and A. J. Weightman. 1997. Molecular detection of novel anaerobic species in dentoalveolar abscesses. Clin. Infect. Dis. 25(Suppl. 2):S235-S236. [DOI] [PubMed] [Google Scholar]
- 42.Zambon, J. J., and S. A. Kasprzak. 1995. The microbiology and histopathology of human root caries. Am. J. Dent. 8:323-328. [PubMed] [Google Scholar]