Abstract
Madurella mycetomatis is the main causative agent of human eumycetoma, a severe debilitating disease endemic in Sudan. It has been suggested that eumycetoma has a soil-borne or thorn prick-mediated origin. For this reason, efforts were undertaken to culture M. mycetomatis from soil samples (n = 43) and thorn collections (n = 35) derived from areas in which it is endemic. However, ribosomal sequencing data revealed that the black fungi obtained all belonged to other fungal species. In addition, we performed PCR-mediated detection followed by restriction fragment length polymorphism (RFLP) analysis for the identification of M. mycetomatis DNA from the environmental samples as well as biopsies from patients with mycetoma. In the case of the Sudanese soil samples, 17 out of 74 (23%) samples were positive for M. mycetomatis DNA. Among the thorn collections, 1 out of 22 (5%) was positive in the PCR. All PCR RFLP patterns clearly indicated the presence of M. mycetomatis. In contrast, 15 Dutch and English control soil samples were all negative. Clinically and environmentally obtained fungal PCR products share the same PCR RFLP patterns, suggesting identity, at least at the species level. These observations support the hypothesis that eumycetoma is primarily environmentally acquired and suggest that M. mycetomatis needs special conditions for growth, as direct isolation from the environment seems to be impossible.
Eumycetoma is a subcutaneous fungal infection in which the etiological agent occurs in the form of more or less compact mycelial grains. The immune response is a granulomatous foreign body reaction characterized by impressive local necrosis and the formation of extensive fistula (8, 9, 18). Areas where eumycetoma is endemic are arid and hot climate zones with limited rainfall. A genuine “mycetoma belt” is found in tropical and subtropical regions, with foci in southern India, Somalia, Senegal, Argentina, and Sudan (12, 17). Cases are particularly common in Sudan, where, for example, Abbott (1) reported 1,231 cases in a 2.5-year period. The main causative agent of eumycetoma in Sudan is Madurella mycetomatis (4, 15). The disorder is supposed to originate from traumatic inoculation of plant material or soil contaminated by these fungi. Abbott (1) noted a history of thorn pricks in 10 out of 62 cases of mycetoma, and in two of these, there were actually thorns found embedded in tissue. Basset et al. (3) reported two cases with thorns inside lesions from which only non-Madurella fungi could be isolated. It is unlikely that M. mycetomatis represents a primary pathogen for humans. Rather, the fungi involved probably are saprophytes, which are able to tolerate uncharacteristic conditions when inoculated by coincidence. However, direct evidence for this hypothesis is still lacking. Thirumalachar and Padhye (22) were barely able to recover M. mycetomatis from soil. This is rather unexpected for a saprophytic fungus.
The present paper reports efforts to find M. mycetomatis in the environment in Sudan. Isolating fungi by culturing soils and thorns, which are preconditioned for traumatic inoculation, was attempted. Isolated strains were compared to patient strains from Sudan that were identified as M. mycetomatis (2). Environmental fungal isolates with a colony appearance resembling that of Madurella were identified and compared by molecular methods. In addition, direct PCR-mediated testing of environmental material was performed.
MATERIALS AND METHODS
Strains and culture conditions.
Strains presented in the current study are listed in Table 1. Sudanese Madurella strains used for comparison were those of Ahmed et al. (2), isolated by direct culture of black grains obtained from deep biopsies of patient lesions at the Mycetoma Research Center, Khartoum, Sudan. They were maintained on malt extract agar (MEA) slants at 24°C and transferred to fresh tubes regularly to preserve viability. Reference strains, selected from genera being reported as agents of eumycetoma, were retrieved from the collection of the Centraalbureau voor Schimmelcultures (Utrecht, The Netherlands).
TABLE 1.
Survey of strains
| Strain | Species | Source | Similarity with reference M. mycetomatis rRNA seq.a |
|---|---|---|---|
| Reference strains | |||
| CBS 247.48 | M. mycetomatis | United States (Gammel) | 100% |
| CBS 201.38 | Madurella sp. 1 | Indonesia | Confident alignmentb |
| CBS 248.48 | Madurella sp. 1 | New Mexico | Confident alignment |
| CBS 217.55 | Madurella sp. 1 | Argentina | Confident alignment |
| CBS 216.29 | Madurella sp. 2 | Italy | Distantly related |
| Clinical isolates | |||
| DH11849 | M. mycetomatis | Sudan, patient | 0-bp difference |
| DH11850 | M. mycetomatis | Sudan, patient | 0-bp difference |
| DH11854 | M. mycetomatis | Sudan, patient | 0-bp difference |
| P2 | M. mycetomatis | Sudan, patient | 1-bp difference |
| DH11853 | M. mycetomatis | Sudan, patient | 2-bp difference |
| DH11862 | M. mycetomatis | Sudan, patient | 3-bp difference |
| Environmental isolates | |||
| DH11963 | Exserohilum mcginnisii | Sudan, soil | Nonrelevant |
| dH11959 | Bipolaris papendorfi | Sudan, soil | Nonrelevant |
| dH11984 | Bipolaris spicifera | Sudan, soil | Nonrelevant |
| 4 | Ulospora bilgrami | Sudan, thorn | Nonrelevant |
| 5 | Ulospora bilgrami | Sudan, thorn | Nonrelevant |
| 6 | Ulospora bilgrami | Sudan, thorn | Nonrelevant |
| 7 | Ulospora bilgrami | Sudan, thorn | Nonrelevant |
| 8 | Ulospora bilgrami | Sudan, thorn | Nonrelevant |
| 9 | Chaetomium brasiliense | Sudan, thorn | Nonrelevant |
| 10 | Chaetomium murorum | Sudan, soil | Nonrelevant |
| dH11971 | Phoma | Sudan, thorn | Nonrelevant |
| dH11983 | —c | Sudan, soil | Nonrelevant |
| dH11975 | — | Sudan, soil | Nonrelevant |
CBS 247.48 is considered to be the M. mycetomatis reference strain to which the ribosomal sequences for the other strains were matched for reasons of comparison.
“Confident alignment” indicates that difference was less than 10%.
—, isolates belong to currently undefined fungal species.
Cultivation of environmental pigmented fungi.
Strains were cultured from soil and thorn samples collected in the Gezira area along the Blue Nile southeast of Khartoum, Sudan (Fig. 1). From 43 samples (up to 20 cm in depth), 4 g of soil was suspended in 30 ml of sterile 0.9% saline by high-speed vortexing for 5 min. After settling, the supernatant was diluted 1:5 and 1:50 for inoculation of two or three MEA culture plates with 0.5 ml of the suspension.
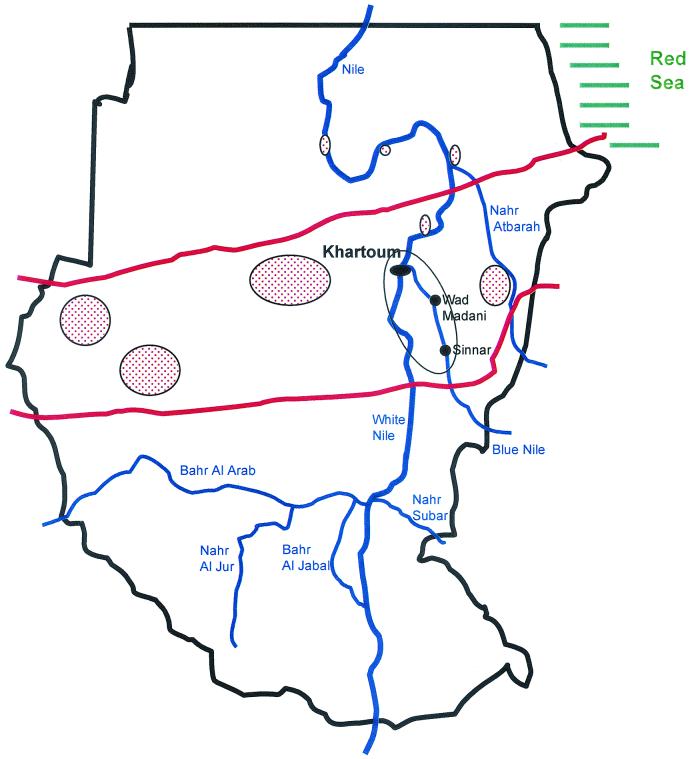
FIG. 1.
Detailed map of the region in Sudan where the thorns were collected and the soil was sampled. Main sampling sites were in the encircled region comprising the region southeastern to Khartoum (Wad Madani and Sinnar). Other regions where mycetoma is highly endemic are highlighted by the elliptic red-dot regions. The red bars indicate the boundaries of the Sudanese mycetoma belt.
Thirty-five thorn samples were collected and divided into two groups: fresh ones from green bushes (15 samples) and old ones lying on the ground or from old dry bushes (20 samples). Thorns primarily derived from the abundant species Acacia mallifera. Two or more thorns, depending on their size, were crushed in 2 ml of 0.9% sodium chloride solution. Two dilutions were made to inoculate two plates each (fresh thorns, 1:1 and 1:10; old thorns, 1:10 and 1:100). To decrease the number of contaminants in the case of several samples, two thorns were briefly rubbed with a cotton swab with 70% ethanol before dilution (fresh thorns, 1:1; old thorns, 1:10) and used to inoculate two plates. Aliquots of 0.5 ml were used per culture plate with Sabouraud's dextrose agar (SDA) containing 100 mg of chloramphenicol/liter. Plates were incubated at 37°C and checked for appropriate colonies at 3-day intervals. Strains with a colony appearance similar to that of M. mycetomatis and exuding a rusty brown, diffusible pigment into the agar were selected (16, 22).
DNA isolation of fungi isolated from soil.
For DNA extraction, about 1 cm2 of mycelium was transferred to a 2:1 mixture of silica gel and Celite 545 with 300 μl of cetyltrimethylammonium bromide (CTAB) buffer (200 mM Tris-HCl, [pH 7.5], 200 mM Na-EDTA, 8.2% NaCl, 2% CTAB). The material was ground with a micro-pestle (Eppendorf). After adding an additional 200 μl of CTAB buffer and vigorous shaking, the sample was incubated for 10 min at 65°C in a water bath. An equal volume of chloroform was added, vortexed briefly, and centrifuged for 5 min at 14,000 rpm (model 5915C centrifuge; Eppendorf, Hamburg, Germany). After transferring the aqueous supernatant to a new Eppendorf tube, 2 volumes (≈800 μl) of 96% ethanol were added and mixed gently. DNA was precipitated at −20°C for at least 30 min. The pellet, obtained by centrifugation, was washed twice with 500 μl of 70% ethanol. DNA was dried overnight at room temperature and suspended in 97.5 μl of TE buffer (10 mM Tris, 10 mM Na-EDTA, pH 8.0) with 2.5 μl of RNase solution (200 U of pancreatic RNase A/ml in 0.01 M sodium acetate). Samples were incubated for 5 to 30 min at 37°C and stored at −20°C.
Ribosomal PCR and sequencing for identification of soil organisms.
DNA samples isolated from unknown strains isolated from soil or thorns was analyzed by a general primer-mediated ribsomal PCR test. The rDNA internal transcribed spacer (ITS) domain was amplified with primers V9G and LS266 (11). Other primer combinations (V9G/ITS4, LS266/ITS5) were used in case of negative results. The near-complete SSU rDNA gene was amplified with primers NS1 and NS24. Of several programs used, optimal results were obtained with the following: 94°C for 5 min followed by 30 cycles of 94°C for 1 min, 48°C for 1 min, and 72°C for 2 min, followed by 72°C for 3 min. One unit of Super-Taq polymerase (Sphaero Q, Leiden, The Netherlands) was used for a 50-μl reaction mixture with 2 μl of DNA. In some cases, 6% dimethylsulfoxide (Merck, Darmstadt, Germany) was added. Amplicons were purified using the GFX PCR DNA and Gel Band Purification Kit according to the manufacturer's instructions (Amersham Pharmacia, Roosendaal, The Netherlands). Concentrations of amplicons were estimated by comparison with a marker (SmartLadder; Eurogentec, Seraing, Belgium) on a 1% agarose gel.
The sequencing reactions were carried out with 15 to 50 ng of DNA for a 10-μl reaction mixture, including 4 pmol of primer and 4 μl of BigDye RR Mix (Applied Biosystems) using primers ITS4 and ITS1 (ITS domain) or Oli1, Oli5, Oli9-11, Oli13, and Oli14 (reference 21 and Table 2) and NS24 (SSU gene) with 25 cycles, as follows: 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min. Sequence reaction products were analyzed on an automated capillary sequencer (ABI, Gouda, The Netherlands). The sequences were adjusted using SeqMan of Lasergene software (DNASTAR Inc., Madison, Wis.) and analyzed using BioNumerics (Applied Maths, Kortrijk, Belgium). Sequences were compared to a set of dedicated sequences available at CBS (ITS fragment ITS 1+2, including 5.8S and the nearly complete SSU). Sequences of putative agents of eumycetoma were included (e.g., Acremonium spp., Chaetomium, Neotestudina spp., Pseudallescheria spp., Bipolaris, Leptosphaeria, and Madurella spp.). The sequences alignment was considered confident when less than a 10% difference was observed. In case no match was found, sequences were run against the EMBL databank using BLAST.
TABLE 2.
Universal and Madurella-specific primers used for PCR and sequencinga
| Primer | Position (gene/nucleotides) | Reference | Primer sequence (5′ → 3′)b |
|---|---|---|---|
| ITS4 | LSU/41-60 | 23 | ←TCCTCCGCTTATTGATATGC |
| ITS5 | SSU/1745-1767 | 23 | GGAAGTAAAAGTCGTAACAAGG→ |
| V9D | SSU/1609-1627 | 6 | TTAAGTCCCTGCCCTTTGTA→ |
| LS266 | LSU/287-266 | 19 | ←GCATTCCCAAACAACTCGACTC |
| NS1 | SSU/20-38 | 23 | GTAGTCATATGCTTGTCT→ |
| NS24 | SSU/1769-1750 | 10 | ←AAACCTTGTTACGACTTTTA |
| Oli5 | SSU/83-102 | 14 | GAAACTGCGAATGGCTCATT→ |
| Oli9 | SSU/573-591 | 14 | CGCGGTAATTCCAGCTCCA→ |
| Oli10 | SSU/951-935 | 14 | ←TGGYRAATGCTTTCGC |
| Oli11 | SSU/962-980 | 14 | TTRATCAAGAACGAAAGT→ |
| Oli13 | SSU/1418-1438 | 14 | ←ATAACAGGTCTGTGATGCCC |
| Oli14 | SSU/1418-1438 | 14 | ATAACAGGTCTGTGATGCCC→ |
| 26.1Ac | ITS2/111-130 | 2 | ←AATGAGTTGGGCTTTAACGG |
| 28.3Ac | ITS1/474-494 | 2 | TCCCGGTAGTGTAGTGTCCCT→ |
Positions correspond to the Saccharomyces cerevisiae consensus sequence.
R = purine A or G; Y = pyrimidine C or T.
Primer sequences and positions are based on the M. mycetomatis ITS sequence (GenBank accession no. AF162133).
Processing of soil and thorn samples for direct DNA testing.
Two grams of soil (74 Sudanese samples, 14 Dutch controls, and 1 United Kingdom control) or four or five thorns (n = 22) were suspended in 5 or 2 ml of samples, sterile physiological saline containing 2% sodium dodecyl sulfate (SDS) (Merck, Darmstadt, Germany) and 5 mg of proteinase K (Merck). The samples were incubated at room temperature on a horizontal shaker for 10 min and then incubated at 37°C (1 h). One milliliter of the clear upper layer of the suspensions was extracted with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1). The DNA was precipitated from the aqueous layer with cold absolute ethanol (Merck). The pellet was washed once with 70% ethanol, air dried, and dissolved in 100 μl of sterile distilled water. This crude extract was further purified with the QIAmp Tissue kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations. The extracted DNA was then immediately tested by PCR.
PCR RFLP analysis of DNA extracted from thorns and soil samples.
This analysis aimed at the broad-spectrum amplification of fungal DNA, followed by an M. mycetomatis-specific nested PCR. In the first PCR, ITS4 and ITS5 primers were used: 5′-TCCTCCGCTTATTGATATGC-3′ and 5′-GGAAGTAAAAGTCGTAACAAGG-3′, respectively (23). For the nested PCR, M. mycetomatis specific primers 26.1A (5′-AATGAGTTGGGCTTTAACGG-3) and (AATGAGTTGGGCTTTAACGG-3) and 28.3A (5′-TCCCGGTAGT′GTAGTGT-CCCT′-3) were employed (2). The PCRs were performed in 50-μl reaction volumes containing 0.2 U of SuperTaq polymerase (HT Biotech, Cambridge, United Kingdom), PCR buffer, deoxynucleotide triphosphate mix, 50 pmol of each primer, and 10 μl of extracted DNA (approximately 100 ng of DNA) in the first PCR round and 5 μl of the amplified material in the nested PCR. Cycling was performed in a model 60 thermocycler (Biomed, Theres, Germany) for 40 cycles (94°C for 1 min, 58°C for 1 min, 72°C for 3 min). PCR products were examined by electrophoresis in 1% agarose gels containing ethidium bromide (0.3μg/ml). The electrophoresis was carried out in 0.5× Tris-borate-EDTA (TBE) buffer at a constant current of 100 mA for 1 h.
Restriction fragment length polymorphism (RFLP) in the M. mycetomatis ITS regions was studied by analysis of the PCR products generated by primers 26.1A and 28.3A by cleavage with CfoI, MspI (Boehringer Mannheim GmbH, Mannheim, Germany), and HaeIII (New England Biolabs, Boston, Mass.) restriction enzymes. The restriction enzymes were used as recommended by the manufacturers. Fifteen microliters of PCR product was incubated overnight at 37°C with the restriction enzyme mixtures, which contained enzyme's buffer and 5 U of each enzyme per reaction. RFLP was determined by electrophoresis in 3% Nusieve GTG agarose gels (FMC Bioproducts, Rockville, Md.). The electrophoresis was performed in 0.5× TBE buffer at a constant current of 100 mA for 3 h.
RESULTS
Cultivation and identification of soil- and thorn-borne fungi.
Overall, 43 soil samples and 35 batches of thorns were analyzed for the presence of black-pigmented fungi resembling M. mycetomatis. Ultimately, 52 colonies from 24 samples were subcultured in a pure form on SDA. Strains were studied microscopically and by molecular analysis. None of the strains turned out to represent M. mycetomatis based on either morphological criteria or ribosomal sequencing. Some of the strains were excluded prior to sequencing based on ribosomal gene RFLP analyses (result not shown). Sequence results are listed in Table 1, presenting data for 13 environmental, 6 clinical, and 5 reference isolates. M. mycetomatis CBS 247.48 was found to be practically identical (0- to 3-bp difference) to six strains of cases of mycetoma from Sudan. CBS 247.48 might be Gammel's strain from Cleveland, Ohio, dating from 1927, which is the type strain of Madurella americana. Three further clinical strains putatively representing M. mycetomatis (CBS 201.38 from Indonesia, CBS 248.48 from New Mexico, and CBS 217.55 from Argentina) were found at some distance but could be aligned with confidence. M. mycetomatis CBS 216.29 from Italy was found to be different from both clusters. About 105 out of 190 positions of ITS1 and about 140 out of 160 positions of ITS2 of M. mycetomatis could be aligned with environmental Chaetomium species from Sudan, which were identified down to the species level by morphology and by comparison with sequences from the public domain. Three strains from Sudanese soil were found to be members of Pleosporales with distoseptate conidia, viz. Exserohilummcginnisii, Bipolaris papendorfii, and Bipolaris spicifera. Their ITS sequences showed no homology with Leptosphaeria. Five strains from Sudanese thorns and showing superficial resemblance to one of the known agents of mycetoma, Neotestudina rosatii, were morphologically identified as Ulospora bilgramii (13). Five reference strains of N. rosatii were clearly different but could be aligned with confidence. The type strain of N. rosatii CBS 427.62 originated from a case of mycetoma, whereas the remaining four strains were environmental (not shown in Table 1).
PCR-mediated detection of M. mycetomatis from various sources.
The first round of PCR, involving the ITS4-ITS5 primer pair, done directly on nucleic acid extracts from soil and thorn samples produced clearly visible products in all instances. Frequently mixtures of various PCR products were produced, indicating the presence of DNA from various fungal species. These results demonstrated that the PCR tests were not inhibited by contaminants from soil of thorns. The combination of primers V9G and LS266 generated the expected DNA amplicons of 1,100 bp as well.
Out of 22 thorn batches tested in this study, only one reacted with the Madurella-specific primers in the nested PCR and produced the characteristic 424-bp fragment matching the positive control (Fig. 2). The M. mycetomatis DNA was detected in significantly higher rates in the soil samples, since 17 positives (23%) were observed among the 74 soil samples from endemic foci. All the fourteen Dutch and the single British control soil samples remained negative in all PCR experiments. All the PCR products thus obtained (n = 18) displayed the same RFLP pattern, which exactly matched those of the clinical M. mycetomatis strains (Fig. 3). Patterns generated with all different restriction enzymes were identical and differed from those of the other species mentioned in Table 1.
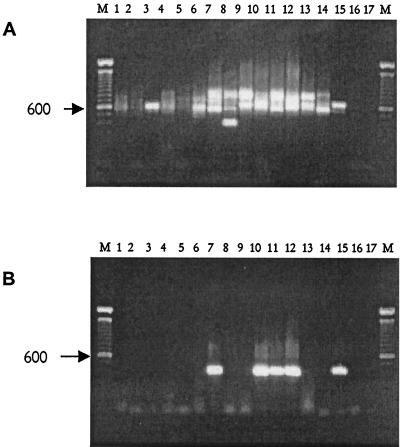
FIG. 2.
Molecular detection of M. mycetomatis in soils and thorn samples. (A) Results of the broad-spectrum ITS PCR employing primers ITS4 and ITS5. Note that amplicons can be observed in all lanes. (B) Result of the nested PCR using M. mycetomatis-specific primers. Lanes 1 to 6, amplicons derived from Dutch control samples: lanes 7 to 14, amplified material from Sudanese samples; lane 15, results obtained with purified M. mycetomatis DNA; lanes 16 and 17, negative controls; lanes M, 100-bp molecule sizing ladder, and the 600-bp fragment is indicated (arrow).
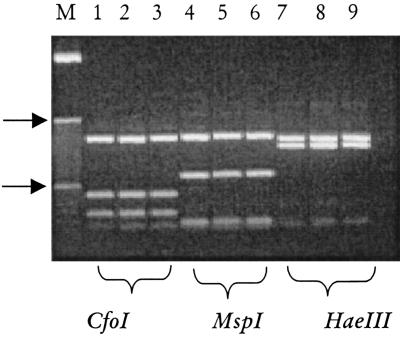
FIG. 3.
Molecular identification of M. mycetomatis from the environment and clinical biopsies. Shown are digests for the specific 26.1A/28.3A PCR products obtained for two clinical isolates (lanes 2 and 3, 5 and 6, and 8 and 9) and a single environmental isolate (lanes 1, 4, and 7). Note that per restriction digest, the patterns are identical. On the left (lane M), the 10-bp molecule sizing ladder is shown, and the 100- and 1,000-bp markers are indicated (arrow).
DISCUSSION
The classical etiology of human eumycetoma involves a relatively limited set of agents as established pathogens typically causing mycetoma (20). This view is corroborated by the overrepresentation of a small number of species involved in the reported cases of eumycetoma, such as Acremonium spp., Pseudallescheria boydii, and Madurella spp. An environmental source has been suggested for the latter microorganism, but direct proof thereof is largely lacking. Presumably, when more ecological data become available, it could become clear that M. mycetomatis actually has another important ecological niche, rendering subcutaneous growth in humans insignificant from a fungal perspective (7). In addition, there are large problems with the correct identification of M. mycetomatis. It shows poor morphological differentiation, is sterile, and easily causes confusion among clinicians and microbiologists (5). Hence, reliable identification of M. mycetomatis is still problematic. In order to solve this problem, Ahmed et al. (2) undertook a molecular study into Madurella-like agents of mycetoma in Sudan. These authors found morphological dissimilarity among strains originating from cases of mycetoma but noted that all clinical isolates proved to be identical. A single ribosomal spacer genotype was predominant, and this species was identified as M. mycetomatis. These findings now enable the targeted search, by molecular techniques, for the environmental source of this important cause of eumycetoma.
Although it is unlikely that M. mycetomatis in nature is present in the same form it is in patients, it is suggested that the isolation of this species directly from the environment is very difficult using the current protocols. Despite failed attempts to culture the organism, we now present data showing that it is possible to detect M. mycetomatis in the environment of areas in Sudan where it is endemic by molecular methods. In addition, thorny A. mallifera bushes, which are very common in the region sampled and are responsible for a high number of thorn pricks, were shown to occasionally harbor the same species of fungus. This supports the observation of Abbott (1) that the frequency of mycetoma is associated, albeit weakly, with the presence of thorny trees. The low recovery rate suggests, however, that M. mycetomatis is not an ordinary saprobe on the thorns.
Human subcutaneous inoculation apparently is an efficient selective means for recovery of the fungus. A comparable situation is known in black yeast-like fungi, where Cladophialophora bantiana (11) and Fonsecaea pedrosoi (De Hoog et al., unpublished data) are homologous examples. These fungi are thus far known only from mammals and can be obtained from the environment after mouse passage only. Interestingly, F. pedrosoi is one of the agents of chromoblastomycosis, which is also a mycosis associated with (minor) trauma.
Other fungal species (anamorph genera Bipolaris, Drechslera, and Exserohilum) were frequently isolated from soil in Sudan. Among these was E. mcginnisii, which thus far is only known from a single strain from a human patient and hence has been regarded as a true pathogen. However, its occurrence in the environment suggests that this is also one of the saprobes which only occasionally is able to infect a human host.
In conclusion, we have demonstrated the presence of M. mycetomatis DNA on or in Acacia thorns and in soil samples. The fungal prevalence in the soil especially suggests that this may be the prime reservoir from which infections originate. This is further corroborated by the fact that clinical isolates and environmental strains seem to share the same ribosomal genotype, confirming that at least the same fungal species is concerned. The challenge for current research is to elucidate the processes that underlie the culture-refractory nature of environmental M. mycetomatis isolates. Whether or not the fungus readily adapts to humans provides another interesting and important research topic.
Acknowledgments
Part of this research was financed through a Young Investigators award provided to A.V. by the European Society of Clinical Microbiology and Infectious Diseases. D.A. thanks Boehringer-Ingelheim for financial support.
A. Aptroot and G. Verkleij are acknowledged for their help with identification of fungi. R. C. Summerbell and A. A. Padhye are thanked for strains and for comments on the manuscript. K. Luijsterburg and A. H. G. Gerrits van den Ende are thanked for technical assistence.
REFERENCES
- 1.Abbott, P. 1956. Mycetoma in the Sudan. Trans. R. Soc. Trop. Med. Hyg. 50:11-30. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed, A. O. A., M. M. Mukhtar, M. Kool-Sijmons, A. H. Fahal, G. S. De Hoog, A. H. G. Gerrits van den Ende, E. E. Zijlstra, H. Verbrugh, A. M. El Sir Abugroun, A. M. Elhassan, and A. Van Belkum. 1999. Development of a species-specific PCR-restriction fragment length polymorphism analysis procedure for identification of Madurella mycetomatis. J. Clin. Microbiol. 37:3175-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basset, A., R. Camain, R. Baylet, and D. Lambert. 1965. Role des épines de mimosacées dans l'inoculation des mycétomes (à propos de deux observations). Bull. Soc. Pathol. Exot. 58:22-24. [PubMed] [Google Scholar]
- 4.Brumpt, E. 1906. Les mycétomes. Asselin & Houzeau, Paris, France.
- 5.De Hoog, G. S., A. Buiting, C. S. Tan, A. B. Stroebel, C. Ketterings, E. J. De Boer, B. Naafs, R. Brimicombe, M. K. E. Nohlmans-Paulssen, G. T. J. Fabius, A. H. Klokke, and L. G. Visser. 1993. Diagnostic problems with imported cases of mycetoma in The Netherlands. Mycoses 36:81-87. [DOI] [PubMed] [Google Scholar]
- 6.De Hoog, G. S., and G. van den Ende. 1998. Molecular diagnostic of clinical strain of filamentous Basidomycetes. Mycoses 41(5-6):183-189. [DOI] [PubMed] [Google Scholar]
- 7.De Hoog, G. S., F. D. Marvin-Sikkema, G. A. Lahpor, J. C. Gottschall, R. A. Prins, and E. Gueho. 1994. Ecology and physiology of the emerging opportunistic fungi Pseudallescheria boydii and Scedosporium prolificans. Mycoses 37:71-78. [DOI] [PubMed] [Google Scholar]
- 8.El Hassan, A. M., A. H. Fahal, I. A. El Hag, and E. A. G. Khalil. 1994. The pathology of mycetoma. Sudan Med. J. 32(Suppl. 1):23-45. [Google Scholar]
- 9.Fahal, A. H., and S. H. Suliman. 1994. Clinical presentation of mycetoma. Sudan Med. J. 32:46-66. [Google Scholar]
- 10.Gargas, A., and J. W. Taylor. 1995. Phylogeny of Discomycetes and early radition of apothecial Ascomycotina inferred from SSU rDNA sequence data. Exp. Mycol. 19:7-15. [DOI] [PubMed] [Google Scholar]
- 11.Gerrits van den Ende, A. H. G., and G. S. De Hoog. 1999. Variability and molecular diagnostics of the neurotropic species Cladophialophora bantiana. Stud. Mycol. 43:152-162. [Google Scholar]
- 12.Gumaa, S. A. 1994. The aetiology and epidemiology of mycetoma. Sudan Med. J. 32(Suppl. 1):14-22. [Google Scholar]
- 13.Hawksworth, D. L. 1979. Ascospore sculpturing and generic concepts in the Testudinaceae (syn. Zopfiaceae). Can. J. Bot. 57:91-99. [Google Scholar]
- 14.Hendriks, L., Y. van de Peer, M. van Herck, J. M. Neefs, and R. De Wachter. 1990. The 18S ribosomal RNA sequence of the sea anemone Anemonia sulcata and its evolutionary position among other eukaryotes. FEBS Lett. 269:445-449. [DOI] [PubMed] [Google Scholar]
- 15.Laveran, M. A. 1902. Au sujet d'un cas de mycetoma à grains noirs. Bull. Acad. Med. Paris 47:773-776. [Google Scholar]
- 16.Mackinnon, J. E. 1954. A contribution to the causal organisms of maduromycosis. Trans. R. Soc. Trop. Med. Hyg. 48:470-480. [DOI] [PubMed] [Google Scholar]
- 17.Mahgoub, E. S., and I. G. Murray. 1973. Mycetoma. William Heinemann, London, United Kingdom.
- 18.Mariat, F., P. Destombes, and G. Segretain. 1977. The mycetomas: clinical features, pathology, etiology, and epidemiology. Contrib. Microbiol. Immunol. 4:1-39. [PubMed] [Google Scholar]
- 19.Masclaux, F., E. Gueho, G. S. de Hood, and R. Christen. 1995. Phylogenetic relationship of human-pathogenic Cladosporium (Xylohypha) species inferred from partial LS rRNA sequences. J. Med. Vet. Mycol. 33:327-338. [DOI] [PubMed] [Google Scholar]
- 20.Rippon, J. W. 1988. Medical mycology. The pathogenic fungi and the pathogenic actinomycetes, 3rd ed. Saunders, Philadelphia, Pa.
- 21.Sterflinger, K., R. De Baere, G. S. De Hoog, R. De Wachter, W. E. Krumbein, and G. Haase. 1997. Coniosporium perforans and C. apollinis, two new rock-inhabiting fungi isolated from marble in the Sanctuary of Delos (Cyclades, Greece). Antonie Leeuwenhoek 72:349-363. [DOI] [PubMed] [Google Scholar]
- 22.Thirumalachar, M. J., and A. A. Padhye. 1968. Isolation of M. mycetomi from soil in India. Hindustan Antibiot. Bull. 10:314-318. [PubMed] [Google Scholar]
- 23.White, T. J., T. Burns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, Calif.