Abstract
In this study, we developed a simple and fast typing procedure for 37 mucosotropic human papillomavirus (HPV) types using a nonradioactive reverse line blotting (RLB) procedure for general primer (GP5+/6+) PCR products. This system has the advantages not only that in a simple format, up to 42 PCR products can be simultaneously typed per membrane per day, but also that after stripping, the membranes can be easily rehybridized at least 15 times without a loss of signal. RLB appeared highly specific, and its sensitivity was identical to that of conventional typing performed with type-specific oligonucleotide probes in an enzyme immunoassay (EIA). The performance of RLB typing was evaluated with samples of HPV-positive cervical scrapings (n = 196) and biopsies of cervical premalignant lesions (n = 100). The distribution of HPV genotypes detected in these samples was in line with the distribution expected on the basis of literature data. In addition, RLB and EIA typing procedures were compared for the typing of high-risk HPV types in GP5+/6+ PCR products of 210 cervical scrapings from high-risk HPV-positive women who participated in a population-based screening program. The typing procedures had an excellent overall agreement rate of 96.5% (kappa value, 0.77). RLB was successful in detecting multiple HPV infections as well as single infections. In conclusion, the GP5+/6+ PCR-RLB procedure appeared to be a reliable and simple approach that may be of great value for large epidemiological studies, population-based cervical cancer screening programs, and vaccination trials that require high-throughput HPV typing.
High-risk human papillomavirus (HR-HPV) DNA has been shown to be present in 99.7% of cervical cancers worldwide (1, 29), and the persistence of HR-HPV infection appears necessary for the development of cervical premalignant lesions and invasive cervical cancer (14, 15, 20, 30). Therefore, HR-HPV testing may have implications for the clinical management of women with cervical lesions (25) and for primary screening for cervical cancer (17, 20). In addition, several human papillomavirus (HPV)-targeted therapies have been or are being developed, and the first trials with prophylactic HPV vaccines are being conducted.
To date, the detection of HPV genotypes has been done predominantly by L1 general- or consensus-primer PCR assays (3, 6, 12, 16, 24, 26) and by the commercially available liquid hybridization assay Hybrid Capture 2 (HC2) (25). General-primer PCR assays enable the detection of a broad spectrum of mucosotropic HPV types, since the primers anneal to a highly homologous region of the HPV types spanning a polymorphic inner region, allowing specific HPV typing. Of these general-primer PCR assays, the GP5+/6+ and the MY09/11 PCR systems (3, 16) are the most frequently used and clinically evaluated ones. Despite the existence of more than 70 HPV genotypes, HPV testing for clinical purposes has been greatly simplified and facilitated by testing for all HR-HPV types simultaneously in one assay, i.e., HR-HPV group detection. The latter can be accomplished by HR-HPV GP5+/6+ PCR-enzyme immunoassay (EIA) or HC2 (7, 8, 25). However, there is a strong need to identify individual HPV genotypes to investigate the epidemiology and clinical behavior of particular types. Moreover, HPV typing is of importance for characterizing study populations being used for HPV vaccination trials and for monitoring the efficiency of HPV-targeted therapies and vaccines.
HPV type determination after general-primer PCR assays has been done by nucleotide sequencing (19, 28) or oligonucleotide probe (oligoprobe) hybridization (Southern dot blotting [7, 16, 26] and EIA [8] formats) of the PCR products or by HPV type-specific PCR (27). However, all of these methods are very laborious (2). Recently, more convenient typing methods based on reverse hybridization have been described; in these methods, initially cloned HPV plasmids (4) and later HPV type-specific oligonucleotides (5, 13) were used. The most recently described assays, the MY09/11 reverse line blot assay (5) and the SPF line probe assay (13), can type 27 and 25 HPV genotypes, respectively, in a nonradioactive format using precipitating color substrates. However, these assays use individual hybridization strips for each PCR product, and these strips can be used only once. As a consequence, these assays are rather expensive and not ideal for high-throughput analyses.
Here we developed an easy and rapid reverse line blot (RLB) assay for large-scale identification of 37 HPV genotypes after GP5+/6+ PCR. The assay is based on the use of a miniblotter system to type GP5/6+ PCR products derived from up to 42 patient samples concurrently per hybridization membrane. Moreover, membranes can be easily stripped and rehybridized at least 15 times without a loss of signal.
MATERIALS AND METHODS
HPV test panel.
For the evaluation of the sensitivity and specificity of the RLB typing method, plasmid clones of the following HPV types were used as a test panel: HPV types 6, 11, 16, 18, 26, 31, 34, 35, 39, 40, 42, 43, 44, 45, 51, 52, 53, 54, 55, 56, 57, 58, 59, 61, 66, and 68 (for sources, see reference 8). To complement this test panel with types for which no HPV clones were available (HPV types 70, 71 [previously named CP8061], 72, 73, 81 [CP8304], 82/MM4, 82/IS39, 83 [MM7], 84 [MM8], and CP6108), we used cervical smears in which these types were identified in previous studies performed in our laboratory.
Clinical specimens.
For the evaluation of HPV typing of clinical specimens, both cervical scraping specimens and formalin-fixed, paraffin-embedded as well as frozen tissues were used; these specimens were derived from women participating in a cervical cancer screening program and from women attending a gynecologic clinic, respectively.
For detection of HPV in crude cell suspensions, a cervical brush was placed in 5 ml of phosphate-buffered saline containing 0.005% merthiolate (27). After vigorous vortexing, the suspensions were centrifuged for 10 min at 3,000 × g. The cell pellets were resuspended in 0.01 M Tris-HCl (pH 7.5) and frozen at −80°C. Aliquots (100 μl) were boiled for 10 min and cooled on ice. For PCR, 10 μl of the extract was used.
For detection of HPV in tissue specimens, 5-μm sections were cut using the so-called sandwich method (29). Briefly, outer sections were stained with hematoxylin-eosin for histological analyses, whereas inner sections (in total, approximately 1 cm2 of tissue) were placed in a tube and used for PCR. Lysis mix (10 mM Tris-HCl [pH 7.5], 0.45% Tween 20, 500 μg of proteinase K [Roche, Mannheim, Germany]) (250 μl) was added and incubated overnight at 37°C. The sample was boiled and centrifuged for 1 min in a standard tabletop centrifuge (8,000 × g). After cooling, 10 μl of the sample was used for PCR.
GP5+/6+ PCR.
Pretreated samples were subjected to GP5+/6+ PCR as previously described (3, 8) with some modifications in the PCR program, depending on the thermal cycler used (PTC225 [Biozyme, Landgraaf, The Netherlands] or PE9700 [Perkin-Elmer, Foster City, Calif.]) (Table 1). The modified ramping times between the temperatures used for denaturation, annealing, and elongation appeared essential for optimal PCR. PCRs were performed in duplicate, and PCR products were pooled to obtain sufficient material for subsequent typing by both EIA and RLB.
TABLE 1.
Conditions for GP5+/6+ PCR with PTC225 and PE9700
| Parameter | Conditions used with:
|
|
|---|---|---|
| PTC225 | PE9700a | |
| Start | 4 min at 94°C | 4 min at 94°C |
| Cycles (n = 40) | 20 s at 94°C (denaturation) | 20 s at 94°C (denaturation) |
| In 24 s to 90°C | In 33% of SA to 38°C | |
| In 66 s to 48°C | 30 s at 38°C (annealing) | |
| In 30 s to 38°C | In 44% of SO to 71°C | |
| 30 s at 38°C (annealing) | 80 s at 71°C (elongation) | |
| In 18 s to 42°C | In 62% of SO to 94°C | |
| In 42 s to 66°C | ||
| In 18 s to 71°C | ||
| 80 s at 71°C (elongation) | ||
| In 24 s to 69°C | ||
| In 90 s to 94°C | ||
| Final step | 4 min at 71°C | 4 min at 72°C |
| Hold | Forever at 4°C | Forever at 4°C |
SA, standard cool-down temperature (5.39°C/s); SO, standard warm-up temperature (4.43°C/s).
EIA of GP5+/6+ PCR products.
The EIA was performed as previously described (8) with the exception that a different brand of streptavidin-coated microtiter plates (Roche) was used. In addition, two further changes were made in relation to the previous protocol (8) to minimize the possibility of cross-hybridization between related HPV types. First, the hybridization temperature was increased from 37 to 55°C. Second, the oligoprobe sequence of HPV type 45 was changed (Table 2), since the originally described 30-mer oligoprobe of HPV type 45 tended to cross-react with HPV type 43 DNA despite the presence of seven mismatches. The sequences of the EIA oligoprobes used in this study are shown in Table 2. For HR-HPV and low-risk HPV (LR-HPV) group-specific detection, oligoprobe cocktails for 14 HR-HPVs (i.e., HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68) and 23 LR-HPVs (i.e., HPV types 6, 11, 26, 34, 40, 42, 43, 44, 53, 54, 55, 57, 61, 70, 71 [CP8061], 72, 73, 81 [CP8304], 82/MM4, 82/IS39, 83 [MM7], 84 [MM8], and CP6108), respectively, were used.
TABLE 2.
HPV type-specific oligoprobes used for EIA and RLB typing of GP5+/6+ PCR products
| HPV type | Sequence of oligoprobe for:
|
|
|---|---|---|
| EIA | RLBa | |
| 6 | ATCCGTAACTACATCTTCCACATACACCAA | TCCGTAACTACATCTTCCA |
| 11 | ATCTGTGTCTAAATCTGCTACATACACTAA | TCTGTGTCTAAATCTGCTAC |
| 16 | GTCATTATGTGCTGCCATATCTACTTCAGA | CATTATGTGCTGCCATATC |
| 18 | TGCTTCTACACAGTCTCCTGTACCTGGGCA | TGCTTCTACACAGTCTCCT |
| 26 | AGTACATTATCTGCAGCATCTGCATCCACT | GTACATTATCTGCAGCATC |
| 31 | TGTTTGTGCTGCAATTGCAAACAGTGATAC | GCAATTGCAAACAGTGATAC |
| 33 | TTTATGCACACAAGTAACTAGTGACAGTAC | TGCACACAAGTAACTAGTGA |
| 34 | TACACAATCCACAAGTACAAATGCACCATA | TTTTCAGTTTGTGTAGGTACA |
| 35 | GTCTGTGTGTTCTGCTGTGTCTTCTAGTGA | CTGCTGTGTCTTCTAGTGA |
| 39 | TCTACCTCTATAGAGTCTTCCATACCTTCT | ATAGAGTCTTCCATACCTTC |
| 40 | GCTGCCACACAGTCCCCCACACCAACCCCA | AGTCCCCCACACCAACC |
| 42 | CTGCAACATCTGGTGATACATATACAGCTG | TGGTGATACATATACAGCTG |
| 43 | TCTACTGACCCTACTGTGCCCAGTACATAT | TCTACTGACCCTACTGTG |
| 44 | GCCACTACACAGTCCCCTCCGTCTACATAT | TACTAGTGAACAATATAAGCA |
| 45b | ACACAAAATCCTGTGCCAAGTACAT | TAATTTAACATTATGTGCCTC |
| 51 | AGCACTGCCACTGCTGCGGTTTCCCCAACA | TGCTGCGGTTTCCCCAA |
| 52 | TGCTGAGGTTAAAAAGGAAAGCACATATAA | GAATACCTTCGTCATGGC |
| 53 | GTCTATGTCTACATATAATTCAAAGCAAAT | TGTCTACATATAATTCAAAGC |
| 54 | TACAGCATCCACGCAGGATAGCTTTAATAA | CACGCAGGATAGCTTTAAT |
| 55 | CTGCTACAACTCAGTCTCCATCTACAACAT | TCAGTCTCCATCTACAACAT |
| 56 | GTACTGCTACAGAACAGTTAAGTAAATATG | CAGAACAGTTAAGTAAATATG |
| 57 | CCACTGTAACCACAGAAACTAATTATAAAG | CCACAGAAACTAATTATAAAG |
| 58 | ATTATGCACTGAAGTAACTAAGGAAGGTAC | TATGCACTGAAGTAACTAAG |
| 59 | TCTACTACTGCTTCTATTCCTAATGTATAC | TCTACTACTGCTTCTATTCC |
| 61 | TACTGCTACATCCCCCCCTGTATCTGAATA | CCCCCCCTGTATCTGAAT |
| 66 | TATTAATGCAGCTAAAAGCACATTAACTAA | AGCTAAAAGCACATTAACTAA |
| 68 | TCTACTACTACTGAATCAGCTGTACCAAT | CTGAATCAGCTGTACCAAT |
| 70 | CTGCCTGCACCGAAACGGCCATACCTGCTG | GAAACGGCCATACCTGCT |
| 72 | TGCCACAGCGTCCTCTGTATCAGAATATAC | AGCGTCCTCTGTATCAGAA |
| 73 | GTGTAGGTACACAGGCTAGTAGCTCTACTA | ACAGGCTAGTAGCTCTACT |
| 82/MM4 | CGACTCGTGTTACTCAATCTGTTGCACAAA | ACTGCTGTTACTCAATCTG |
| 83 (MM7) | CAGCTGCTGCTACACAGGCTAATGAATACA | TACACAGGCTAATGAATACA |
| 84 (MM8) | GTGCTGCTACCAACACCGAATCAGAATATA | TGCTACCAACACCGAATCA |
| 82/IS39 | GCACTGCTGCTACTCCATCAGTTGCACAGA | TGCTACTCCATCAGTTGC |
| CP6108 | GTGCTGCTTCCCAGTCTGCCACAGAATACA | CTTCCCAGTCTGCCACA |
| 71 (CP8061) | GTGCTACCAAAACTGTTGAGTCTACATATA | TGCTACCAAAACTGTTGAG |
| 81 (CP8304) | GCACAGCTACATCTGCTGCTGCAGAATACA | GCTACATCTGCTGCTGC |
Except for HPV types 34, 44, 45, and 52, oligoprobes for RLB were selected from the oligoprobe sequence used in EIA and underlined in the EIA oligoprobe sequence.
Modified from a previously published sequence (8).
RLB of GP5+/6+ PCR products.
RLB analyses were performed using a previously described system (10, 11, 23). The system is based on the use of a miniblotter for spotting in parallel up to 42 different oligoprobes containing a 5′-amino group on a carboxyl-coated nylon membrane. Subsequently, up to 42 PCR products can be pipetted into the parallel channels of the miniblotter in such a way that the channels are perpendicular to the rows of oligoprobes deposited previously. In this format, hybridization is followed by incubation of the membrane with antibiotin conjugate and enhanced chemiluminescence (ECL) detection. Most of the HPV-specific 5′-amino-linked oligonucleotides (Isogen, Maarssen, The Netherlands) selected for RLB were adapted from the oligonucleotides selected for use in EIA detection (8) (Table 2) in such a way that melting temperature was (approximately) 55°C and the length was approximately 20 bp. Only for HPV types 34, 44, 45, and 52 were oligoprobe sequences selected outside the EIA probe region to fulfill the above-mentioned criteria without a loss of specificity.
The sequences of the RLB oligoprobes are shown in Table 2. The oligoprobes were covalently bound to negatively charged membranes (Biodyne C; Pall BioSupport, East Hills, N.Y.). Briefly, membranes were activated by using 16% (wt/vol) EDAC [1-ethyl-3-(3-dimethylaminopropyl)carbodiimide; Sigma, St. Louis, Mo.] and placed in a miniblotter system (MN 45; Immunetics, Cambridge, Mass.). The oligonucleotides were applied in parallel lines; after 1 min of incubation, the channels were aspirated and the membranes were inactivated using 100 mM NaOH, followed by washing in 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7); Life Technologies, Rockville, Md.) supplemented with 0.1% sodium dodecyl sulfate (SDS) (BDH, Dorset, England). For hybridization, 10μl of biotinylated GP5+/6+ PCR product was diluted in 150 μl of 2× SSPE-0.1% SDS, heat denatured (96°C), and rapidly cooled on ice. The PCR products were pipetted into the parallel channels perpendicular to the rows of oligoprobes. Hybridization was performed at 42°C for 1 h, followed by two washings in 2× SSPE-0.5% SDS at 51°C. Subsequently, membranes were incubated with streptavidin-peroxidase conjugate for 45 to 60 min at 42°C (1:4,000 dilution in 2× SSPE-0.5% SDS; Roche), washed twice in 2× SSPE-0.5% SDS for 10 min at 42°C, and rinsed twice in 2× SSPE for 5 min at room temperature. Detection was done by using ECL detection liquid (Amersham, Buckinghamshire, England) followed by exposure to film (Hyperfilm; Amersham) for 1 to 10 min. Films were subsequently developed. For repeated use, the membranes were stripped by washing two times in 1% SDS at 80°C for 30 min each time followed by incubation in 20 mM EDTA (pH 8) at room temperature for 15 min. Membranes were sealed in plastic and stored at 4°C until further use.
HPV E7 type-specific PCR.
The HPV E7 type-specific PCR was used for analysis of the few cases that showed discrepant typing results in the RLB and EIA. PCR primers and conditions were described previously (29). Detection of PCR products was performed by EIA under the same conditions as those mentioned above. However, some of the E7 oligoprobe sequences were different from those described earlier (29). Oligonucleotide sequences of the E7 primers and probes (Eurogentec, Liege, Belgium) for 14 HR-HPV types are shown in Table 3.
TABLE 3.
Oligonucleotide sequences used in the HPV E7 type-specific PCR
| Primer | Sequence (5′-3′) | Probe | Sequence (5′-3′) |
|---|---|---|---|
| HPV16E7.667Bio | GATGAAATAGATGGTCCAGC | 16E7DIGPRO | CGGACAGAGCCCATTACAATATTGTAACCT |
| HPV16E7.774Bio | GCTTTGTACGCACAACCGAAGC | ||
| HPV18E7.696 | AAGAAAACGATGAAATAGATGGA | 18E7DIGPRO | CCCGACGAGCCGAACCACAACGTCACACAA |
| HPV18E7.799Bio | GGCTTCACACTTACAACACA | ||
| HPV31E7.811 | GGGCTCATTTGGAATCGTGTG | 31E7DIGPRO | TACCTGCTGGATCAGCCATTGTAGTTACAG |
| HPV31E7.890Bio | AACCATTGCATCCCGTCCCC | ||
| HPV33E7.671Bio | TGAGGATGAAGGCTTGGACC | 33E7DIGPRO | TGTGACAACAGGTTACAATGTAGTAATCAG |
| HPV33E7.761 | TGACACATAAACGAACTGTG | ||
| HPV35E7.674Bio | CTATTGACGGTCCAGCT | 35E7DIGPRO | CAACAGGACGTTACAATATTATAATTGGAG |
| HPV35E7.752 | TACACACAGACGTAGTGTCG | ||
| HPV39E7.601Bio | CCAAAGCCCACCTTGCAGGA | 39E7DIGPRO | TCCTAATTGCTCGTGACATACAAGGTCAAC |
| HPV39E7.703 | ATGGTCGGGTTCATCTATTTC | ||
| HPV45E7.741Bio | CCCACGAGCCGAACCACAG | 45E7DIGPRO | AGCTCAATTCTGCCGTCACACTTACAACAT |
| HPV45E7.822 | TCTAAGGTCCTCTGCCGAGC | ||
| HPV51E7.718 | TACGTGTTACAGAATTGAAG | 51E7DIGPRO | TCAAGTGTAGTACAACTGGCAGTGGAAAGC |
| HPV51E7.841Bio | AACCAGGCTTAGTTCGCCCATT | ||
| HPV52E7.691Bio | GCAGAACAAGCCACAAGCAA | 52E7DIGPRO | ATAGCCGTAGTGTGCTATCACAACTGTGAC |
| HPV52E7.776 | TAGAGTACGAAGGTCCGTCG | ||
| HPV56E7.784 | GGTGCAGTTGGACATTCAGAG | 56E7DIGPRO | CAAAGAGGACCTGCGTGTTGTACAACAGCT |
| HPV56E7.886Bio | GTTACTTGATGCGCAGAGTG | ||
| HPV58E7.98 | CGAGGATGAAATAGGCTTGG | 58E7DIGPRO | ACAGCTAATTACTACATTGTAACTTGTTGT |
| HPV58E7.761Bio | ACACAAACGAACCGTGGTGC | ||
| HPV59E7.646 | CTCCGAGAATGAAAAAGATGAA | 59E7DIGPRO | ATCATCCTTTGCTACTAGCTAGACGAGCTG |
| HPV59E7.749Bio | GCTGAAGTTGATTATTACA | ||
| HPV66E7.641 | AATGCAATGAGCAATTGGACAG | 66E7DIGPRO | AGGATGAAATAGACCATTTGCTGGAGCGGC |
| HPV66E7.742Bio | CTTATGTTGTTCAGCTTGTC | ||
| HPV68E7.4604 | AACAACAGCGTCACACAATTCA | 68E7DIGPRO | AGTGTAACAACCTACTGCAACTAGTAGTAG |
| HPV68E7.4704Bio | AGTTGTACGTTCCGCAGGTT |
RESULTS
Development of RLB. (i) RLB oligoprobes.
A database search (BLAST from National Center for Biotechnology Information [NCBI] and NTI [Informax, Oxford, United Kingdom]) of all oligoprobe sequences selected for RLB revealed complete matches with corresponding HPV types, at least two mismatches with other HPV genotypes, and at least four mismatches with non-HPV sequences present in the NCBI nucleotide sequence database (GenBank). Also, the selected probes showed perfect matches to all known HPV variants (18, 31, 32), as determined by sequence alignments.
In order to determine the optimal concentration of the oligoprobes to be used for RLB, oligoprobe concentrations ranging from 25 to 400 pmol/μl were applied to RLB membranes. Subsequently, membranes were hybridized with GP5+/6+ PCR products derived from clinical samples which, on the basis of EIA typing, were found to contain HPV type 11, 16, 18, and/or 33 DNA. For all the HPV types, the most optimal signals were obtained with 100 to 200 pmol of oligoprobe/μl (Fig. 1, upper panel). Consequently, 200 pmol/μl was chosen for generating the RLB membranes for subsequent experiments. No signal was obtained with HPV-negative samples, providing evidence that the signals were HPV specific (Fig. 1, upper panel).
FIG. 1. .
Optimization of oligoprobe concentration for RLB. (Upper panel) Different concentrations of oligoprobes were spotted onto the membrane and subsequently hybridized with GP5+/6+ PCR products derived from clinical specimens containing HPV type 16, HPV types 16 and 33, HPV types 11 and 18, HPV type 16, no HPV (−), HPV type 33, and HPV type 16 from (top to bottom). Optimal signals were derived when 100 to 200 pmol of oligoprobe/μl was used. (Lower panel) Dilutions of cloned HPV type 16 (pHPV16) DNA were subjected to GP5+/6+ PCR and RLB, showing that 100 ag of HPV type 16 DNA can easily be detected.
(ii) Sensitivity and specificity.
First, the sensitivity of RLB detection was compared with that of conventional EIA detection. For that purpose, GP5+/6+ PCR products were generated from 100 pg of cloned DNAs of HPV types 6, 11, 16, 18, 31, and 58 and subjected to both EIA using specific probes for the respective types and RLB. Hybridization with serial dilutions of the GP5+/6+ PCR products showed that the sensitivity of RLB detection was identical to that of EIA detection. Subsequently, serial dilutions ranging from 100 ag to 100 pg of cloned representatives of phylogenetically distant HPV groups, i.e., HPV types 6, 11, 16, 18, 33, 39, and 51, were tested by GP5+/6+ PCR and the two different readout systems. RLB and EIA reached the same detection level, and the sensitivities for these HPV types were identical to those previously described for EIA (8), as shown for HPV type 16 (detection level, 100 ag) in Fig. 1 (lower panel). It was further evident that dilutions showing borderline EIA signals gave rise to clearly visible signals with RLB after a standard 5-min ECL exposure.
Subsequently, the specificity of RLB was evaluated with GP5+/6+ PCR products for a panel of HPV types (14 HR-HPV types, i.e., HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68, and 23 LR-HPV types, i.e., HPV types 6, 11, 26, 34, 40, 42, 43, 44, 53, 54, 55, 57, 61, 70, 71 [CP8061], 72, 73, 81 [CP8304], 82/MM4, 82/IS39, 83 [MM7], 84 [MM8], and CP6108) derived from either cloned HPVs or well-characterized clinical specimens. RLB analysis with membranes on which oligoprobes of these 37 HPV types were applied clearly showed specific hybridization without any cross-reaction. Even when large amounts of PCR products for phylogenetically closely related HPV types (i.e., HPV types 6 and 11, HPV types 39 and 45, and HPV types 33 and 58) were used in combination with longer ECL exposure times, no cross-reactivity was observed. In contrast, with EIA, weak, mutual cross-hybridization was occasionally found between HPV types 33 and 58 when hybridization was performed at 37°C, the condition previously described (8). This cross-hybridization disappeared without a loss of sensitivity when EIA hybridization was performed at 55°C. Therefore, an EIA hybridization temperature of 55°C was applied for this study.
In order to determine to what extent membranes can be reused, we repeatedly stripped and rehybridized the filters by applying the PCR products for the respective HPV types in different channels per experiment. It appeared that the membranes could be stripped and rehybridized at least 15 times without a loss of signal.
Evaluation of RLB. (i) Comparison of RLB typing with HR-HPV or LR-HPV group-specific EIA.
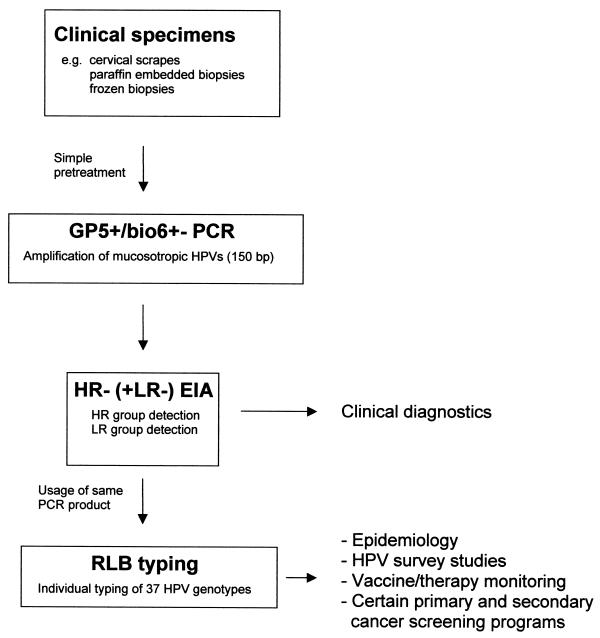
The performance of RLB for the typing of 37 HPV types in clinical specimens was first evaluated with HPV-positive cervical scraping (n = 196) and tissue biopsy (n = 100) specimens which were pretested by GP5+/6+ PCR followed by HR-HPV and LR-HPV group-specific EIA identification. A schematic outline of this procedure is given in Fig. 2. Results are shown in Table 4. Representative RLB results are shown in Fig. 3. RLB was performed for 40 samples concurrently per membrane. Clear RLB signals were obtained for all samples, and the identified types belonged to the HR-HPV and/or LR-HPV groups for which EIA scored positive. In addition to single HPV infections, multiple infections of up to four HPV types were found, and HR-HPV-LR-HPV multiple infections determined by EIA were successfully typed as HR-HPV-LR-HPV mixed infections by RLB (Table 4). In HPV-containing cytomorphologically normal scraping specimens (n = 163), HPV type 16 (35%), HPV type 31 (13%), and HPV type 18 (9%) were most frequently found among the HR-HPV types. Among the LR-HPV types, HPV types 42, 6, 70, and 73 (12.3, 2.5, 2.5, and 2.5%, respectively) were predominantly found, whereas HPV types 34, 44, 57, 61, MM8, IS39, and CP8061 were not detected. Twenty-two percent of the HPV-positive normal scraping specimens showed mixed infections. Interestingly, multiple infections appeared to be age dependent, being most frequent (50%) in women 15 to 24 years old (data not shown). Of these mixed infections, 75% were double infections, 22% were triple, and 3% were quadruple. In cytomorphologically abnormal scraping specimens (n = 33; predominantly mild dyskaryosis), a slightly higher frequency of multiple infections (27%) was found. In addition, biopsy specimens showing cervical intraepithelial neoplasia (CIN I (n = 11), II (n = 20), and III (n = 30) and invasive cervical carcinoma (n = 39) showed one or more HR-HPV types in RLB, with HPV type 16 being the predominant HPV type (Table 4). However, considerably fewer multiple HPV infections were detected in these specimens than in the scraping specimens, ranging from 9% for CIN I to 3% for invasive carcinomas.
FIG. 2.
Schematic outline of HPV detection and typing approaches for clinical specimens.
TABLE 4.
Detection of HPV genotypes by RLB in cervical specimens prescreened by HR- and LR-HPV GP5+/6+ PCR-EIA
| Category | No. of the following specimens in each category (typea [no. of times found]):
|
|||||
|---|---|---|---|---|---|---|
| Cervical scraping
|
Biopsy
|
|||||
| Normal | Dyskaryosis | CIN I | CIN II | CIN III | Carcinomas | |
| HR-HPV positiveb | 113 | 30 | 11 | 20 | 30 | 39 |
| LR-HPV LR positiveb | 34 | 1 | ||||
| LR-HPV and HR-HPV positive | 16 | 2 | ||||
| HPV single infections (type) | 127 | 24 | 10 | 18 | 30 | 38 |
| 16 | 43 | 13 | 8 | 8 | 16 | 30 |
| 18 | 7 | 1 | 1 | 1 | ||
| 31 | 9 | 3 | 2 | 5 | 2 | |
| 33 | 6 | 2 | 1 | 2 | 2 | |
| 35 | 2 | 2 | 1 | 3 | ||
| 39 | 4 | 1 | 1 | |||
| 45 | 3 | 1 | 1 | |||
| 51 | 4 | 1 | 1 | |||
| 52 | 5 | 1 | 1 | 1 | ||
| 56 | 3 | 1 | ||||
| 58 | 4 | 1 | ||||
| 59 | 1 | |||||
| 66 | 3 | 1 | 1 | 1 | ||
| 68 | 2 | 1 | ||||
| LR-HPV | 31 (HPV 6 [2], 26, 40, 42 [11], 43 [3], 53, 54 [2], 55, 70 [3], 72, 73, 82MM4 [2], CP6108, 81) | 1 (HPV 6) | ||||
| HPV double infections | 27 (HPV 6/73, 16/18 [2], 16/31 [3], 16/39, 16/42, [2], 16/52, 16/53, 16/70, 16/73, 18/42, 18/82MM4, 31/33, 31/52, 31/56, 31/59, 31/66, 33/51, 39/43, 42/81, 42/73, 56/40, 51/42, 56/81) | 7 (HPV 16/18, 16/31, 16/42, 16/52, 31/45, 33/52, 56/40) | 1 (16/31) | 2 (31/51, 33/39) | 1 (16/31) | 1 (16/59) |
| HPV at least triple infections | 9 (HPV 16/31/51, 18/31/51, 18/31/56, 18/39/42, 31/51/42, 39/52/56, 52/6/11, 56/66/55, 16/59/42/83) | 2 (16/33/52, 18/31/58) | ||||
Type designations indicate the types found in the multiple infections; e.g., 6/73 means that types 6 and 73 were both found in double infections.
Including double infections.
FIG. 3.
Representative example of RLB typing of GP5+/6+ PCR products derived from cervical scraping specimens. Forty cervical scraping specimens (top panel) were typed for 37 HPV types (left panel: OLIGO). HPV types found are indicated (top panel: HPV typing). In addition, GP5+/6+ PCR products for HPV-negative cervical scraping specimens were included to confirm the specificity of RLB (−). It is noteworthy that differences in signal intensity reflect different copy numbers for the HPV types present in the specimens.
(ii) Comparison of RLB typing with EIA typing for HR-HPV types.
In order to compare RLB and EIA for identifying individual HR-HPV genotypes, we analyzed that 210 cervical scraping specimens previously scored HR-HPV positive in the HR-HPV GP5+/6+ PCR-EIA analysis. Results are shown in Table 5. The same GP5+/6+ PCR products were used for EIA and RLB typing. The typing procedures showed an excellent overall agreement rate of 96.5% (kappa value, 0.77) when the individual typing results were taken into account. A total of 175 out of 178 single infections (97.7%) detected by one or both methods showed identical typing results (Table 5). Complete agreement for all HPV types detected was found for 19 out of 27 double infections (70.4%) and for 3 out of 5 triple infections (60.0%). When partial agreement was allowed, i.e., at least one identical HPV type in double or triple infections, the typing results obtained by EIA and RLB were almost identical (Table 5). Among the mixed infections detected by one or both assays, RLB detected more HPV types than EIA in eight cases, whereas EIA detected more HPV types in two cases. In general, the additional HPV types detected by RLB showed weak RLB spots, indicating relatively low viral loads for those types.
TABLE 5.
Comparison of typing of HPV in cervical specimens by RLB and EIA
| Infection | Identical EIA and RLB results
|
Discrepant results
|
||||
|---|---|---|---|---|---|---|
|
n
|
HPV type found by:
|
|||||
| HPV type | n | EIA | RLB | Type-specific PCRa | ||
| Single | 16 | 60 | 3 | 35 | 35 | |
| 18 | 18 | 59 | 59 | |||
| 31 | 27 | 45 | 45 | |||
| 33 | 6 | |||||
| 35 | 4 | |||||
| 39 | 9 | |||||
| 45 | 9 | |||||
| 51 | 9 | |||||
| 52 | 14 | |||||
| 56 | 10 | |||||
| 58 | 3 | |||||
| 59 | 3 | |||||
| 66 | 2 | |||||
| 68 | 1 | |||||
| Total | 175 | |||||
| Doubleb | 16/33 | 1 | 8 | 45/52 | 52 | 45/52 |
| 16/45 | 1 | 31 | 31/68 | 31 | ||
| 16/51 | 1 | 66 | 56/66 | 56/66 | ||
| 16/52 | 1 | 39 | 39/68 | 39/68 | ||
| 16/56 | 1 | 31 | 31/56 | 31/56 | ||
| 16/58 | 1 | 18 | 18/35 | 18/35 | ||
| 18/31 | 1 | 35/59 | 35 | 35/59 | ||
| 18/33 | 1 | 35 | 35/68 | 35/68 | ||
| 18/58 | 1 | |||||
| 33/58 | 1 | |||||
| 39/56 | 2 | |||||
| 39/58 | 1 | |||||
| 39/66 | 1 | |||||
| 45/51 | 1 | |||||
| 51/52 | 1 | |||||
| 58/66 | 3 | |||||
| Total | 19 | |||||
| Tripleb | 16/33/45 | 1 | 2 | 16/39 | 16/39/51 | 16/39 |
| 16/39/56 | 1 | 18/39 | 18/39/66 | 18/39/66 | ||
| 16/39/52 | 1 | |||||
| Total | 3 | |||||
Type-specific PCR was performed for HPV types found by either EIA or RLB.
See Table 4, footnote a, for an explanation of type designations.
Discrepant cases of EIA and RLB typing were subjected to HPV type-specific PCR. The results are shown in Table 5. With respect to single HPV infections, type-specific PCR confirmed the EIA typing result in the three discrepant cases for which the RLB scored negative. With respect to the 10 discrepant cases in double and triple HPV infections, RLB typing results were confirmed in 6 cases and EIA typing results were confirmed in 4 cases. Ultimately, when all 13 discrepant cases of single and multiple infections were taken into account, RLB typing results were confirmed in 6 cases and EIA typing results were confirmed in 7 cases.
DISCUSSION
In this study, we showed that GP5+/6+ PCR-RLB is an attractive alternative for easy, rapid, and high-throughput identification of individual HPV genotypes. The sensitivities of RLB were identical to those of EIA, and the intensities of EIA signals (optical densities) always corresponded to the intensities of spots in RLB. Despite the fact that for RLB, 20-mer oligoprobes that sometimes differed by no more than two nucleotides from related HPV types were used, the method appeared to be highly specific; no cross-reactivity was found between closely related HPV types, even when high concentrations of PCR products and long ECL exposure times were used.
Despite the advantage of its high specificity, the high stringency of RLB requires that all variants of a certain HPV type show a perfect match with the probe, since a single mismatch may result in the failure of detection. Although the selected RLB probes match perfectly the sequences of all HPV variants currently known (18, 31, 32), the possibility cannot be excluded that variants showing subtle sequence variations in the probe region exist. Therefore, the probes may be subject to subtle changes when more sequence information for HPV variants becomes available in the future. On the other hand, this RLB system may be suitable for the identification of variants when probes are selected in regions where variants differ from each other.
A comparison of RLB and EIA typing of HR-HPV types in patient samples revealed excellent agreement, in line with the identical sensitivities determined for the assays. The fact that for the discrepant cases type-specific PCR confirmed about half of the RLB results and half of the EIA results further supports the notion that discordant results are not due to possible differences in the sensitivities of the assays but rather are due to sampling errors. Generally, the distribution of HPV genotypes found in this study by RLB is in line with data from previous studies (1, 13, 14, 20, 21, 29). Although we classified HPV types 26, 34, 54, 70, and 73 as LR-HPVs, it is noteworthy that their oncogenic potential is still unknown. These types have been reported to be rarely present in invasive cervical carcinomas (1). Indeed, HPV type 26 was identified by RLB for a rare case of cervical carcinoma that previously tested HR-HPV negative but LR-HPV positive by routine HPV diagnostic tests in our laboratory (data not shown). We found HPV types 26, 54, 70, and 73 in a substantial number of normal smears (7%), and their clinical significance in CIN development will be studied in ongoing follow-up investigations.
Multiple HPV infections were found in 22% of the HPV-positive normal scraping specimens by RLB (Table 4). Interestingly, these mixed infections appeared to be age dependent, being most frequent (50%) in younger women (data not shown), most likely reflecting greater sexual activity. Although multiple HPV infections were rather uncommon in biopsy specimens for CIN and invasive carcinoma, a substantial proportion of cervical smears showing dyskaryosis were found associated with multiple infections by RLB. This result suggests that additional, nonrelevant HPV types which are not present in the underlying lesion are detectable in cervical scraping specimens. Similar observations were made in recent studies with SPF10 PCR (13), although these studies revealed an overall higher frequency of multiple HPV infections in cervical smears and biopsy specimens (13, 22). Hence, the biological relevance of multiple HPV infections for CIN disease and cervical cancer is yet unknown; further studies on this issue deserve attention if the presence of multiple HPV types in these lesions can be confirmed by in situ hybridization techniques.
The RLB method developed in this study has the potential for large-scale analysis since it is more user-friendly and less time-consuming than conventional methods. For example, the use of two miniblotters allows the typing of up to 84 samples in less than 1 day, whereas conventional typing by EIA with individual probes takes 1 week for one full-time technician. Typing may be even more rapid when a miniblotter with a larger number of channels becomes available. The identification of HPV types on exposed X-ray films can be facilitated using a special raster (containing both the order of the HPV oligoprobes and the sample numbers) photocopied on a transparent sheet. Alternatively, RLB images can be digitally recorded and processed using specific software packages. Also, because the RLB method uses a nonprecipitating ECL substrate, it has the advantage that nylon filters can be stripped and successfully reused at least 15 times. In contrast, other systems use color substrates and allow only single analysis. Furthermore, owing to the use of negatively charged filters rather than the commonly used positively charged filters, the background of RLB is minimal. By use of other labels, such as fluorophores, or detection systems, such as enzyme-linked chemifluorescence (9), signals can be directly detected by a fluorescent scanner and subsequently processed.
Another advantage is that in combination with GP5+/6+ PCR, which amplifies a fragment of approximately 150 bp, this RLB system provides an ideal compromise between efficient small-fragment amplification and specific HPV typing. Although the SPF10 system amplifies an even shorter fragment of 65 bp, typing by a line probe assay is more complicated since it requires the interpretation of specific hybridization patterns rather than single-probe hybridization results for the typing of 9 of the 25 HPV types used (13). This situation is due to the small intervening region in the PCR product left for the selection of specific probes, so that the interpretation of typing results is not facilitated, especially for multiple HPV infections. Although the MY09/11 reverse line blot assay uses individual probes for the identification of types, this method has the disadvantage that relatively large fragments (i.e., about 450 bp) are amplified. Based on the fact that the GP5+/6+ primers are located within the MY09/11 PCR amplicon, our RLB system has the further advantage that it also can be used for the typing of MY09/11 PCR products.
In principle, different scenarios are possible for the use of the RLB system, i.e., a direct typing approach or typing of samples that were prescreened by HPV group-specific assays, such as HR-HPV or LR-HPV GP5+/6+ PCR-EIA or Hybrid Capture 2. We favor a prescreening approach (Fig. 2), particularly for testing samples in which a rather low HPV prevalence is expected. This approach would directly couple clinically validated HPV detection methods with a high-throughput typing procedure suitable for mass screening. Moreover, HPV group-specific detection can still be done in an even less laborious way with a microtiter plate format, and the prescreening approach will prevent redundant typing of HPV-negative samples. As a prescreening method, HR-HPV or LR-HPV GP5+/6+ PCR-EIA is practically most convenient, since the resulting PCR products can be used directly for RLB typing and no additional PCR is required.
In conclusion, the application of GP5+/6+ PCR-RLB can be of great value for studies which require reliable high-throughput HPV typing. The method can be conveniently applied as a second step after GP5+/6+ PCR and HR-HPV or LR-HPV detection by EIA, which is at present one of the most frequently used noncommercial HPV detection systems.
Acknowledgments
We thank A. Splinter for excellent technical assistance, I. van de Pol for technical support, M. Jacobs for helpful discussions, and P. Stern for critically reading the manuscript.
REFERENCES
- 1.Bosch, F. X., M. M. Manos, N. Muñoz, M. Sherman, A. M. Jansen, J. Peto, M. H. Schiffman, V. Moreno, R. Kurman, K. V. Shah, et al. 1995. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J. Natl. Cancer Inst. 87:796-802. [DOI] [PubMed]
- 2.Coutlée, F., P. Gravitt, H. Richardson, C. Hankins, E. Franco, N. Lapointe, H. Voyer, and the Canadian Women's HIV Study Group. 1999. Nonisotopic detection and typing of human papillomavirus DNA in genital samples by the line blot assay. J. Clin. Microbiol. 37:1852-1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Roda Husman, A. M., J. M. M. Walboomers, A. J. C. van den Brule, C. J. L. M. Meijer, and P. J. F. Snijders. 1995. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by polymerase chain reaction. J. Gen. Virol. 76:1057-1062. [DOI] [PubMed] [Google Scholar]
- 4.Forslund, O., B. G. Hansson, and B. Bjerre. 1994. Typing of human papillomaviruses by consensus polymerase chain reaction and a non-radioactive reverse dot blot hybridization. J. Virol. Methods 49:129-140. [DOI] [PubMed] [Google Scholar]
- 5.Gravitt, P. E., C. L. Peyton, R. J. Apple, and C. M. Wheeler. 1998. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. J. Clin. Microbiol. 36:3020-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gravitt, P. E., C. L. Peyton, T. Q. Alessi, C. M. Wheeler, F. Coutlée, A. Hildesheim, M. H. Schiffman, D. R. Scott, and R. J. Apple. 2000. Improved amplification of genital human papillomaviruses. J. Clin. Microbiol. 38:357-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacobs, M. V., A. M. de Roda Husman, A. J. C. van den Brule, P. J. F. Snijders, C. J. L. M. Meijer, and J. M. M. Walboomers. 1995. Group-specific differentiation between high- and low-risk human papillomavirus genotypes by general primer-mediated PCR and two cocktails of oligonucleotide probes. J. Clin. Microbiol. 33:901-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobs, M. V., P. J. F. Snijders, A. J. C. van den Brule, T. J. M. Helmerhorst, C. J. L. M. Meijer, and J. M. M. Walboomers. 1997. A general primer GP5+/6(+)-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J. Clin. Microbiol. 35:791-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jordens, J. Z., S. Lanham, M. A. Pickett, S. Amarasekara, I. Abeywickrema, and P. J. Watt. 2000. Amplification with molecular beacon primers and reverse line blotting for the detection and typing of human papillomavirus. J. Virol. Methods 89:29-37. [DOI] [PubMed] [Google Scholar]
- 10.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaufhold, A., A. Podbielski, G. Baumgarten, M. Blokpoel, J. Top, and L. Schouls. 1994. Rapid typing of group A streptococci by the use of DNA amplification and nonradioactive allele specific oligonucleotide probes. FEMS Microbiol. Lett. 119:19-26. [DOI] [PubMed] [Google Scholar]
- 12.Kleter, B., L. J. van Doorn, J. ter Schegget, L. Schrauwen, K. van Krimpen, M. Burger, B. ter Harmsel, and W. Quint. 1998. Novel short-fragment PCR assay for highly sensitive broad-spectrum detection of anogenital human papillomaviruses. Am. J. Pathol. 153:1731-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleter, B., L. J. van Doorn, L. Schrauwen, A. Molijn, S. Sastrowijoto, J. ter Schegget, J. Lindeman, B. ter Harmsel, M. Burger, and W. Quint. 1999. Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J. Clin. Microbiol. 37:2508-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krüger Kjaer, S., A. J. C. van den Brule, J. E. Bock, P. A. Poll, G. Engholm, M. E. Sherman, J. M. M. Walboomers, and C. J. L. M. Meijer. 1996. Human papillomavirus—the most significant risk determinant of cervical intraepithelial neoplasia. Int. J. Cancer 65:601-606. [DOI] [PubMed] [Google Scholar]
- 15.Liaw, K.-L., A. G. Glass, M. M. Manos, C. E. Greer, D. R. Scott, M. Sherman, R. D. Burk, R. J. Kurman, S. Wacholder, B. B. Rush, D. M. Cadell, P. Lawler, D. Tabor, and M. Schiffman. 1999. Detection of human papillomavirus DNA in cytologically normal women and subsequent cervical squamous intraepithelial lesions. J. Natl. Cancer Inst. 91:954-960. [DOI] [PubMed] [Google Scholar]
- 16.Manos, M. M., Y. Ting, D. K. Wright, A. J. Lewis, T. R. Broker, and S. M. Wolinsky. 1989. Use of polymerase chain reaction amplification for the detection of genital human papillomaviruses. Cancer Cells 7:209-214. [Google Scholar]
- 17.Meijer, C. J. L. M., L. Rozendaal, J. C. van der Linden, T. J. M. Helmerhorst, F. J. Voorhorst, and J. M. M. Walboomers. 1997. Human papillomavirus testing for primary cervical cancer screening, p. 338-349. In E. Franco and J. Monsonego (ed.), New developments in cervical cancer screening and prevention. Blackwell Science Ltd., Oxford, England.
- 18.Meyers, G., C. Baker, K. Münger, F. Svedrup, A. McBride, H.-U. Bernard, and J. Meissner (ed.). 1997. Human papillomaviruses 1997. A compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics Group T-10, Los Alamos National Laboratory, Los Alamos, N. Mex.
- 19.Nelson, J. H., G. A. Hawkins, K. Edlund, M. Evander, L. Kjellberg, G. Wadell, J. Dillner, T. Gerasimova, A. L. Coker, L. Pirisi, D. Petereit, and P. F. Lambert. 2000. A novel and rapid PCR-based method for genotyping human papillomaviruses in clinical samples. J. Clin. Microbiol. 38:688-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nobbenhuis, M. A. E., J. M. M. Walboomers, T. J. M. Helmerhorst, L. Rozendaal, A. J. Remmink, E. K. J. Risse, H. C. van der Linden, F. J. Voorhorst, P. Kenemans, and C. J. L. M. Meijer. 1999. Relation of human papillomavirus status to cervical lesions and consequences for cervical-cancer screening: a prospective study. Lancet 354:20-25. [DOI] [PubMed] [Google Scholar]
- 21.Peyton, C. L., P. E. Gravitt, W. C. Hunt, R. S. Hundley, M. Zhao, R. J. Apple, and C. M. Wheeler. 2001. Determinants of genital human papillomavirus detection in a US population. J. Infect. Dis. 183:1554-1564. [DOI] [PubMed] [Google Scholar]
- 22.Quint, W. G., G. Scholte, L. J. van Doorn, B. Kleter, P. H. Smits, and J. Lindeman. 2001. Comparative analysis of human papillomavirus infections in cervical scrapes and biopsy specimens by general SPF(10) PCR and HPV genotyping. J. Pathol. 194:51-58. [DOI] [PubMed] [Google Scholar]
- 23.Rijpkema, S. G., M. J. Molkenboer, L. M. Schouls, F. Jongejan, and J. F. Schellekens. 1995. Simultaneous detection and genotyping of three genomic groups of Borrelia burgdorferi sensu lato in Dutch Ixodes ricinus ticks by characterization of the amplified intergenic spacer region between 5S and 23S rRNA genes. J. Clin. Microbiol. 33:3091-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snijders, P. J. F., A. J. C. van den Brule, H. F. J. Schrijnemakers, G. Snow, C. J. L. M. Meijer, and J. M. M. Walboomers. 1990. The use of general primers in the polymerase chain reaction permits the detection of a broad spectrum of human papillomavirus genotypes. J. Gen. Virol. 71:173-181. [DOI] [PubMed] [Google Scholar]
- 25.Solomon, D., M. Schiffman, and R. Tarone. 2001. Comparison of three management strategies for patients with atypical squamous cells of undetermined significance: baseline results from a randomized trial. J. Natl. Cancer Inst. 93:293-299. [DOI] [PubMed] [Google Scholar]
- 26.van den Brule, A. J. C., P. J. F. Snijders, R. L. J. Gordijn, O. P. Bleker, C. J. L. M. Meijer, and J. M. M. Walboomers. 1990. General primer mediated polymerase chain reaction permits the detection of sequenced and still unsequenced human papillomavirus genotypes in cervical scrapes and carcinomas. Int. J. Cancer 45:644-649. [DOI] [PubMed] [Google Scholar]
- 27.van den Brule, A. J. C., C. J. L. M. Meijer, V. Bakels, P. Kenemans, and J. M. M. Walboomers. 1990. Rapid detection of human papillomavirus in cervical scrapes by combined general primer-mediated and type-specific polymerase chain reaction. J. Clin. Microbiol. 28:2739-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vernon, S. D., E. R. Unger, and D. Williams. 2000. Comparison of human papillomavirus detection and typing by cycle sequencing, line blotting, and hybrid capture. J. Clin. Microbiol. 38:651-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walboomers, J. M. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, A. Kummer, K. V. Shah, P. J. F. Snijders, J. Peto, C. J. L. M. Meijer, and N. Muñoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]
- 30.Wallin, K. L., F. Wiklund, T. Angstrom, F. Bergman, U. Stendahl, G. Wadell, G. Hallmans, and J. Dillner. 1999. Type-specific persistence of human papillomavirus DNA before the development of invasive cervical cancer. N. Engl. J. Med. 341:1633-1638. [DOI] [PubMed] [Google Scholar]
- 31.Yamada, T., C. M. Wheeler, A. L. Halpern, A.-C. M. Stewart, A. Hildesheim, and S. A. Jenison. 1995. Human papillomavirus type 16 variant lineages in U.S. populations characterized by nucleotide sequence analysis of the E6, L2, and L1 coding segments. J. Virol. 69:7743-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamada, T., M. M. Manos, J. Peto, C. E. Greer, N. Muñoz, F. X. Bosch, and S. M. Wheeler. 1997. Human papillomavirus type 16 sequence variation in cervical cancers: a worldwide perspective. J. Virol. 71:2463-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]