Abstract
Chronic periodontitis is a common infectious disease in the adult population. The etiology is clearly bacterial, and a small number of bacterial species have been consistently associated with periodontitis, including Bacteroides forsythus and Porphyromonas gingivalis. Comparatively little attention has been paid to the identification of health-associated and potentially beneficial bacterial species that may reside in the gingival sulcus. The purpose of the present study was to examine the relationship of the presence of B. forsythus and a newly identified Bacteroides phylotype, oral clone BU063, to periodontal health status. The study was accomplished with a set of samples that were collected from subjects with periodontitis and healthy controls. These samples had previously been analyzed for the presence of P. gingivalis. An oral sampling strategy that included every tooth and a PCR-based detection method were used to maximize detection sensitivity. The presence of B. forsythus in the oral cavity was strongly associated with periodontitis, and its nearest genetic neighbor, oral clone BU063, was associated with oral health (P < 0.0001 for both). Colonization with P. gingivalis was independent of the presence of either Bacteroides species, but the two Bacteroides species were found together less often than would be expected by chance (P < 0.0001). This suggests the presence of a specific exclusionary mechanism between the two Bacteroides species. Comparisons between these two organisms may prove useful for studies that determine how B. forsythus functions in the disease process. In addition, oral clone BU063 deserves further study as a possible preventive or therapeutic intervention for periodontitis.
Chronic periodontitis is a common infectious disease in the adult population, with the majority of individuals affected to some degree (2). The etiology is clearly bacterial, although host factors are thought to be important in the disease process. The oral flora is complex, with a recent estimate, based on 16S ribosomal DNA (rDNA) sequence diversity, indicating that approximately 500 species of bacteria are commonly found in the human oral cavity (11). Less than half of these have been cultured, including the as yet uncultivated oral clone BU063 studied in the present investigation. Paster et al. (11) previously identified this phylotype by 16S rDNA sequencing from a subgingival sample.
A unique bacterial cause of chronic periodontitis has not been identified, but, rather, a number of bacterial species have been associated with the disease. A small number of species have been consistently associated with periodontitis, including Bacteroides forsythus. In several recent clinical studies, levels of B. forsythus have been shown to be associated with periodontitis (7, 8) and with progression of the disease in human adults (15, 16). These studies were designed to detect if a species is found at elevated levels in sites of disease activity and if it is more widely distributed throughout the dentition in individuals with periodontitis than in healthy individuals. An alternative and straightforward approach, determining if the presence of B. forsythus in the oral cavity is correlated with periodontal health status, has not been reported. This approach would be helpful because it addresses the question of whether B. forsythus is a commensal opportunist or a pathogen in the classic sense, and this has implications for risk prediction and prevention of periodontitis.
Comparatively little attention has been paid to the identification of health-associated and potentially beneficial bacterial species that may reside in the gingival sulcus. Health-associated bacterial species have been recognized on a number of other mucosal surfaces in the body and have been used to treat or prevent disease (1, 3, 9, 13). Symbiotic organisms may benefit the host by various biological mechanisms, such as through the exclusion of pathogenic species or the promotion of antiallergenic processes.
The purpose of the present study was to examine the relationship of the presence of B. forsythus and a recently identified phylotype, oral clone BU063 (11), to periodontal health status.
MATERIALS AND METHODS
Study population.
Samples from 293 of 311 subjects collected for a previous study were available for reanalysis for the present study (5). As described previously, subjects for this institutionally approved study were recruited from the clinics of the Ohio State University College of Dentistry. Briefly, they were selected for participation if they met criteria for periodontal health or disease designed to identify approximately the most and least healthy one-fourth of the population. The healthy group was matched to the diseased group by age. All teeth present were sampled with endodontic paper points, and samples from each individual were pooled.
Detection of B. forsythus and oral clone BU063.
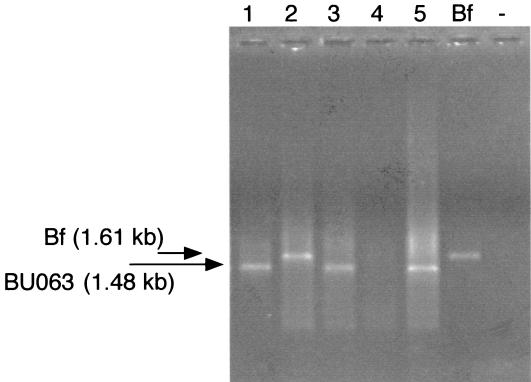
DNA was isolated from the plaque samples and was amplified by PCR with universal prokaryotic primers specific for regions in the 16S and 23S genes, as described previously (10). PCRs were performed with 2.5 U of Taq DNA polymerase in a total volume of 100 μl in buffer containing 50 mM KCl, 10 mM Tris-HCl (pH 8.8), 3 mM MgCl2, 0.1% Triton X-100, 0.2 mM (each) deoxynucleoside triphosphates, and 2 to 4 μl of template DNA. Each sample was subjected to 25 to 27 cycles (denaturation at 94°C for 1 min, annealing at 42°C for 2 min, and primer extension at 72°C for 3 min) in an automated thermal cycler (Perkin-Elmer). In a second, nested amplification, a previously designed oligonucleotide specific for the B. forsythus 16S rRNA gene (TGCGATATAGTGTAAGCTCTACAG) (4) was paired with a second universal 23S primer (GGTACTTAGATGTTTCAGTTC) and used to amplify the intergenic spacer region from B. forsythus and oral clone BU063. Two microliters of product from the first amplification was added as template DNA. The PCR conditions were the same as those for the first amplification except that the annealing step was performed at 52°C and anti-Taq antibody was added to each reaction mixture in an amount sufficient to inhibit the Taq polymerase until denaturation of the template. DNA fragments were separated by electrophoresis in a 1% agarose gel in TBE (0.1 M Tris-borate [pH 8.3], 2 mM EDTA) (see Fig. 1). The gels were stained with ethidium bromide and were photographed under UV light. The samples were scored as positive or negative for the presence of B. forsythus or oral clone BU063 on the basis of the presence of two bands running at 1.61 kb (B. forsythus) and 1.48 kb (oral clone BU063). All second amplifications were repeated, and if the results were not in agreement, they were repeated again. The presence of DNA was confirmed for all negative samples by use of universal prokaryotic primers.
FIG. 1.
PCR amplification of ISR rDNAs of B. forsythus and oral clone BU063. The lane labeled “Bf” contains DNA amplified from B. forsythus strain ATCC 43037, and the last lane labeled with a minus sign contains a negative (no-template) control. The B. forsythus DNA template yielded a fragment of 1.61 kb, and oral clone BU063 yielded a fragment of 1.48 kb. Lanes 1 through 5 contain DNA amplified from clinical samples; lane 2 was scored as positive for B. forsythus, and lanes 1, 3, and 5 were scored as positive for BU063.
Sequencing.
B. forsythus strain ATCC 43037 was provided by Mark Maiden. PCR products were sequenced with an ABI 310 automated DNA sequencer as described previously (12). Sequences were assembled with SeqApp software (http://IUBIO.BIO.INDIANA.EDU) and analyzed with MacVector (version 7.0) software.
Statistical methods.
Chi-square analysis was used to compare the prevalences of B. forsythus, P. gingivalis, and oral clone BU063 in healthy individuals and individuals with periodontitis, to examine differences due to race and sex, and to examine cocolonization with these species. Odds ratios with 95% confidence intervals were calculated for detection of P. gingivalis, B. forsythus, and BU063 in the periodontitis group versus the healthy group and to examine cocolonization with these species or phylotypes.
Nucleotide sequence accession numbers.
The sequences of the 16S and 23S ribosomal intergenic spacer region for both B. forsythus and BU063 are available for electronic retrieval from GenBank (accession numbers AF466818 and AF466819).
RESULTS
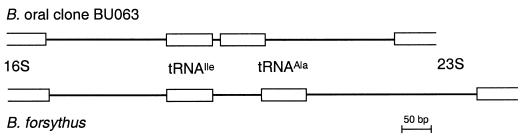
A previously designed oligonucleotide specific for the B. forsythus 16S rRNA gene (4) was used as a PCR primer for detection of B. forsythus in subgingival samples. PCRs with this primer paired with a universal bacterial primer specific for a sequence in the 23S rDNA produced two fragments of different sizes, as seen by agarose gel electrophoresis (Fig. 1). A 1.61-kb PCR product was the same size as a band amplified from B. forsythus strain ATCC 43037 (Fig. 1) as well as several additional laboratory strains of B. forsythus (data not shown). Sequencing of this PCR product confirmed that it was B. forsythus rDNA. The second PCR product gave a band of 1.48 kb and did not match in size any bands from B. forsythus strains. The 3′ end of the 16S rRNA gene from this PCR product was sequenced and, by comparison of the sequence to known 16S rRNA sequences with the BLAST search program, was found to match the 16S rRNA gene of oral clone BU063 (11). This uncharacterized phylotype is the species most closely related to B. forsythus, as determined by 16S rRNA phylogenetic analysis. Both PCR products contained approximately 700 bp of the 16S rRNA gene, 200 bp of 23S DNA, and the entire intergenic spacer region (ISR) between the 16S and 23S rRNA genes. The ISRs were sequenced for both B. forsythus and oral clone BU063 (GenBank accession numbers AF466818 and AF466819). A representation of the two ISRs is shown in Fig. 2. The ISRs differ in length by approximately 130 bp, but both contain sequences homologous to tRNAs for isoleucine and alanine. These tRNAs share homology with each other as well as with the tRNAs found in ISRs from other Bacteroides and Porphyromonas species. The remaining sequences are approximately 50% homologous, in contrast to the 16S genes, which are 91% homologous.
FIG. 2.
Schematic diagram of the rDNA ISR between the 16S and 23S rRNA genes for B. forsythus and oral clone BU063. Genes are shown as boxes, and noncoding regions are shown as lines. The same tRNA genes are present in both organisms, but noncoding regions differ in both size and sequence.
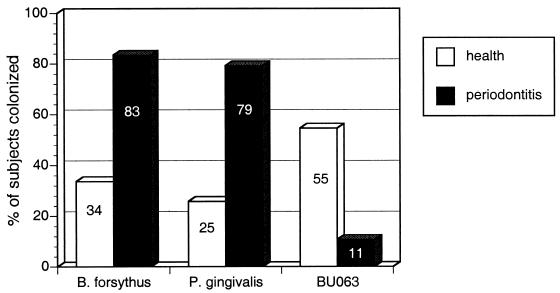
Samples from 293 subjects (172 samples from the healthy group and 121 samples from the group with periodontitis) were analyzed by PCR for the presence of B. forsythus and oral clone BU063. The demographics and disease status of both groups have been described previously (5). The mean ages of the groups were 49.2 and 51.4 years, respectively, and this difference was not significant (5). More females and fewer African-American subjects were included in the healthy group than in the group with periodontitis, but as reported previously, no differences in colonization with Porphyromonas gingivalis were seen with respect to race or sex (5). Similarly, in the present study no significant differences in the presence of B. forsythus or BU063 were seen by sex or race by chi-square analysis (data not shown). The distributions of B. forsythus, oral clone BU063, and P. gingivalis in healthy individuals and individuals with periodontitis are shown in Fig. 3. The P values for chi-square analysis and the odds ratios for being infected with B. forsythus and oral clone BU063 in relation to periodontal health status are shown in Table 1. Previously reported data for P. gingivalis (5) are also shown for comparison. The presence of B. forsythus and P. gingivalis was strongly associated with periodontitis; in contrast, oral clone BU063 was much more common in healthy individuals than in individuals with periodontitis (P < 0.0001). The P values for chi-square analysis and the odds ratios for finding these species or phylotypes together are shown in Table 2. Colonization with P. gingivalis was independent of the presence of either Bacteroides species, but the two Bacteroides species were found together less often than would be expected by chance (P < 0.0001).
FIG. 3.
Prevalence of B. forsythus, P. gingivalis, and oral clone BU063, a newly identifed phylotype closely related to B. forsythus, in subjects with periodontitis and healthy subjects. Actual percentages are shown inside the bars. The data for P. gingivalis have been reported previously (5). The total sample size was 293 subjects, with samples obtained from 121 subjects with periodontitis (▪) and 172 healthy, age-matched controls (□). Differences in the prevalences of all three species in orally healthy individuals and individuals with periodontitis were significant by chi-square analysis (P < 0.0001).
TABLE 1.
Presence of bacteria and periodontal health status
| Species or phylotype | P value for chi-squaresa | Odds ratio (95% confidence interval)b |
|---|---|---|
| BU063 | <0.0001 | 0.1 (0.05-0.19) |
| B. forsythus | <0.0001 | 9.9 (5.6-17.6) |
| P. gingivalis | <0.0001 | 10.9 (6.4-18.8) |
Chi-square for distribution of the species or phylotype in subjects with periodontitis and healthy subjects.
Odds of finding the species or phylotype in subjects with periodontitis compared to finding the species or phylotype in healthy subjects.
TABLE 2.
Likelihood of infection with multiple species
| Species or phylotype | Subjects with periodontitis
|
Healthy subjects
|
||
|---|---|---|---|---|
| P value for chi-squaresa | Odds ratio (95% confidence interval)b | P value for chi-squaresa | Odds ratio (95% confidence interval)b | |
| B. forsythus and BU063 | <0.0001 | 0.05 (0.01-0.19) | 0.014 | 0.45 (0.23-0.86) |
| P. gingivalis and BU063 | 0.773 | 0.81 (0.21-3.22) | 0.233 | 1.52 (0.76-3.05) |
| P. gingivalis and B. forsythus | 1.00 | 1.00 (0.30-3.32) | 0.300 | 1.45 (0.72-2.93) |
Chi-square for distribution of the two species or phylotypes in the same subject.
Odds of finding either species when the other is present compared to when the other is not present.
DISCUSSION
An oligonucleotide had been developed as a hybridization probe specific for B. forsythus 16S rDNA sequences (4). When used in combination with a universal 23S primer for PCR amplification of dental samples, two PCR products were generated (Fig. 1). The sequence of the larger product matched that of the PCR product generated with genomic DNA from a reference strain of B. forsythus. The 16S rDNA sequence of the smaller PCR product was greater than 99.5% homologous with the 16S rDNA sequence from oral clone BU063 reported by Paster et al. (11). Sequences complementary to the PCR primer were identical for both B. forsythus and BU063. The BU063 16S rRNA gene differed from the B. forsythus 16S rRNA gene by 9%. Although this difference is greater than that found between some bacterial species, BU063 is termed a “phylotype” rather than a “species” since it has not been cultured and characterized. Yet-to-be cultured clone BU063 is the oral organism most closely related to B. forsythus that has been reported (11), and comparison of all known 16S rDNA sequences failed to identify additional species or phylotypes more closely related to either organism.
For bacterial identification, amplification of ribosomal targets that include the ISR provides two advantages over assays limited to the 16S rRNA gene. First, there are usually enough variations in ISRs, but not 16S rRNA genes, to differentiate strains of a species, as, for example, in P. gingivalis (12). Second, unlike 16S rRNA genes, ISRs show considerable interspecies size variations. So, even closely related bacteria (e.g., B. forsythus and oral clone BU063) generate different-sized PCR products from the same primers. This is useful when amplifying DNA from complex populations such as those found in the oral cavity, where novel species or species considered nonoral may be encountered. If the 16S rRNA gene, but not the ISR, from an unknown species were amplified with a primer targeted to a known species, the two PCR products would be the same size and would therefore not be distinguishable by agarose gel electrophoresis. Similarly, hybridization probes that can bind to closely related species would not discriminate between those species.
The association of B. forsythus and Bacteroides phylotype BU063 with periodontitis was examined by analyzing samples from subjects with periodontitis and a group of healthy, aged-matched controls. Previously published data on the prevalence of P. gingivalis (5) were also included for comparison. By use of a sensitive assay to analyze whole-mouth samples, highly significant differences in the presence of these organisms were found between healthy and diseased subjects (Table 1; Fig. 3).
The prevalences of B. forsythus and P. gingivalis were high in subjects with periodontitis and low in healthy subjects (Fig. 3). This is consistent with previous reports linking the levels of these organisms to disease (7, 8, 15, 16). In the present study, subjects with periodontitis were 9.9 times more likely than their healthy counterparts to be infected with B. forsythus. For comparison, the odds for the presence of P. gingivalis in the same population were quite similar, at 10.9. (This number differs slightly from the previously published value [5] because only part of the sample from the original study was included in the present investigation.) These data suggest that B. forsythus may not be a normal or desirable member of the oral flora. In a previous report, not all strains of P. gingivalis were associated with periodontitis (6), probably accounting for the presence of the organism in some healthy subjects. A similar phenomenon may explain the presence of B. forsythus in at least some healthy subjects.
In contrast to B. forsythus and P. gingivalis, uncultivated oral clone BU063 was associated with periodontal health. It was found in more than half of healthy subjects but in only 11% of subjects with periodontitis. The odds for finding oral clone BU063 were 0.1 in subjects with periodontitis compared to their healthy counterparts. So, while B. forsythus is highly associated with periodontal disease, its closest known relative, BU063, is associated with periodontal health. Studies comparing these two closely related organisms may prove useful for determining how B. forsythus functions in the disease process. Oral clone BU063 may also prove useful for therapy involving the replacement of pathogenic flora with beneficial species. Similar beneficial relationships have been exploited as therapies for other mucosal surfaces such as the digestive tract and urogenital system (1, 3, 13). Periodontal therapy has almost exclusively been focused on the eradication of pathogenic bacteria, but the oral cavity is not a sterile environment, and establishment of a healthy flora may be essential to stabilization of the ecosystem. Oral clone BU063 will have to be cultured for these studies, but attempts to grow it under standard culture conditions, including conditions with N-acetylmuramic acid, which is required by B. forsythus, have not yet been successful.
The two Bacteroides species, B. forsythus and oral clone BU063, were found together less often than would be expected by chance, suggesting a mechanism of exclusion. In subjects with periodontitis, the odds ratio for finding B. forsythus and BU063 in the same sample indicates that either species was only 0.05 times as likely to be found when the other species was present as when it was not present (Table 2). This difference in cocolonization was highly significant by chi-square analysis (P < 0.0001). To a lesser extent, the two Bacteroides species were also unlikely to be found together in healthy subjects (Table 2). This could mean that BU063 is overgrown or pushed out by B. forsythus in subjects with periodontitis and that BU063 provides some protection from acquisition of B. forsythus. This is consistent with data indicating that oral clone BU063 is common in early childhood and is less prevalent as subjects reach adulthood (X. Chu, A. L. Griffen, and E. J. Leys, unpublished data), and the distribution of B. forsythus with respect to age in children is the opposite, increasing with age (X. Chu, A. L. Griffen, and E. J. Leys, International Association for Dental Research Abstracts, J. Dent. Res. 79:621, 2000).
In contrast to the exclusion seen between B. forsythus and BU063, colonization with either of these bacteria and the periodontal pathogen P. gingivalis was independent, as shown in Table 2; the odds ratio for finding P. gingivalis in the presence or absence of B. forsythus was 1.00, and for oral clone BU063 it was 0.81. This independence of colonization between both Bacteroides species and P. gingivalis helps to rule out more general environmental factors as an explanation for the exclusion between the Bacteroides species. Previous investigators have reported a strong association within subjects for the presence and levels of B. forsythus and P. gingivalis at various oral sites (14), but on the basis of our findings, individual subjects are independently colonized with these organisms.
In conclusion, the presence of B. forsythus in the oral cavity was strongly associated with periodontitis, and the opposite was true for its health-associated, nearest genetic neighbor, oral clone BU063. In addition, a specific exclusionary mechanism between the two Bacteroides species may occur since they were found together significantly less often than was expected. Comparisons between these two organisms may prove useful for studies that determine how B. forsythus functions in the disease process. In addition, further study of oral clone BU063 may provide insight into the natural history of periodontal health and may suggest preventive or therapeutic interventions for periodontitis.
Acknowledgments
This work was supported by NIH grant DE10467.
REFERENCES
- 1.Alvarez-Olmos, M. I., and R. A. Oberhelman. 2001. Probiotic agents and infectious diseases: a modern perspective on a traditional therapy. Clin. Infect. Dis. 32:1567-1576. [DOI] [PubMed] [Google Scholar]
- 2.American Academy of Periodontology. 1996. Position paper: epidemiology of periodontal diseases. J. Periodontol. 67:935-945. [PubMed] [Google Scholar]
- 3.Chan, R. C., G. Reid, R. T. Irvin, A. W. Bruce, and J. W. Costerton. 1985. Competitive exclusion of uropathogens from human uroepithelial cells by Lactobacillus whole cells and cell wall fragments. Infect. Immun. 47:84-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dix, K., S. M. Watanabe, S. McArdle, D. I. Lee, C. Randolph, B. Moncla, and D. E. Schwartz. 1990. Species-specific oligodeoxynucleotide probes for the identification of periodontal bacteria. J. Clin. Microbiol. 28:319-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffen, A. L., M. R. Becker, S. R. Lyons, M. L. Moeschberger, and E. J. Leys. 1998. Prevalence of Porphyromonas gingivalis and periodontal health status. J. Clin. Microbiol. 36:3239-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffen, A. L., S. R. Lyons, M. R. Becker, M. L. Moeschberger, and E. J. Leys. 1999. Porphyromonas gingivalis strain variability and periodontitis. J. Clin. Microbiol. 37:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grossi, S. G., J. J. Zambon, A. W. Ho, G. Koch, R. G. Dunford, E. E. Machtei, O. M. Norderyd, and R. J. Genco. 1994. Assessment of risk for periodontal disease. I. Risk indicators for attachment loss. J. Periodontol. 65:260-267. [DOI] [PubMed] [Google Scholar]
- 8.Haffajee, A. D., M. A. Cugini, A. Tanner, R. P. Pollack, C. Smith, R. L. Kent, Jr., and S. S. Socransky. 1998. Subgingival microbiota in healthy, well-maintained elder and periodontitis subjects. J. Clin. Periodontol. 25:346-353. [DOI] [PubMed] [Google Scholar]
- 9.Kalliomaki, M., S. Salminen, H. Arvilommi, P. Kero, P. Koskinen, and E. Isolauri. 2001. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet 357:1076-1079. [DOI] [PubMed] [Google Scholar]
- 10.McClellan, D. L., A. L. Griffen, and E. J. Leys. 1996. Age and prevalence of Porphyromonas gingivalis in children. J. Clin. Microbiol. 34:2017-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rumpf, R. W., A. L. Griffen, B. G. Wen, and E. J. Leys. 1999. Sequencing of the ribosomal intergenic spacer region for strain identification of Porphyromonas gingivalis. J. Clin. Microbiol. 37:2723-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakamoto, I., M. Igarashi, K. Kimura, A. Takagi, T. Miwa, and Y. Koga. 2001. Suppressive effect of Lactobacillus gasseri OLL 2716 (LG21) on Helicobacter pylori infection in humans. J. Antimicrob. Chemother. 47:709-710. [DOI] [PubMed] [Google Scholar]
- 14.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol 25:134-144. [DOI] [PubMed] [Google Scholar]
- 15.Tanner, A., M. F. Maiden, P. J. Macuch, L. L. Murray, and R. L. Kent, Jr. 1998. Microbiota of health, gingivitis, and initial periodontitis. J. Clin. Periodontol. 25:85-98. [DOI] [PubMed] [Google Scholar]
- 16.Tran, S. D., J. D. Rudney, B. S. Sparks, and J. S. Hodges. 2001. Persistent presence of Bacteroides forsythus as a risk factor for attachment loss in a population with low prevalence and severity of adult periodontitis. J. Periodontol. 72:1-10. [DOI] [PubMed] [Google Scholar]