Abstract
Susceptibilities to macrolides were evaluated in 267 Streptococcus pneumoniae isolates, of which 182 were from patients with invasive diseases and 85 were from healthy carriers. Of the 98 resistant isolates, 20 strains showed an M phenotype and carried mef. Strains that carried both mef(A) and mef(E) were found: 17 strains carried mef(A) and 3 carried mef(E). The characteristics of the strains carrying the mef genes and the properties of the mef-containing elements were studied. Strains carrying mef(A) belonged to serotype 14, were susceptible to all the antibiotics tested except erythromycin, and appeared to be clonally related by pulsed-field gel electrophoresis (PFGE). The three mef(E) strains belonged to different serotypes, showed different susceptibility profiles, and did not appear to be related by PFGE. The sequences of a fragment of the mef-containing element, which encompassed mef and the msr(A) homolog, were identical among the three mef(E)-positive strains and among the three mef(A)-positive strains, although there were differences between the sequences for the two variants at 168 positions. In all mef(A)-positive strains, the mef element was inserted in celB, which led to impairment of the competence of the strains. In line with insertion of the mef(E) element at a different site, the competence of the mef(E)-positive strains was maintained. Transfer of erythromycin resistance by conjugation was obtained from two of three mef(A) strains but from none of three mef(E) strains. Due to the important different characteristics of the strains carrying mef(A) or mef(E), we suggest that the distinction between the two genes be maintained.
Macrolide resistance in Streptococcus pneumoniae is typically due to acquisition of the erm(B) gene, which mediates ribosomal modification (10), or the mef gene, which encodes a drug efflux pump (28). Recently, mutations in the 23S rRNA or ribosomal proteins of S. pneumoniae have been found to confer erythromycin (ERY) resistance in some clinical isolates (30).
The Mef pump confers a low to moderate level of resistance to 14- and 15-membered macrolides but not to lincosamide or streptogramin B antibiotics (M phenotype). Of the two variants of the mef gene, mef(A) was originally found in Streptococcus pyogenes (3) and mef(E) was originally found in S. pneumoniae (29). mef(A) and mef(E) are 90% identical at the nucleotide level and were assigned to the same class of macrolide resistance determinants (22). In most subsequent studies, mef was detected by a PCR assay that did not distinguish between the two variants (27). However, the two variants were considered species specific; therefore, if a mef gene was found in S. pneumoniae, it was generally assumed to be mef(E) (9, 16, 26). However, mef(A) was shown to be present in macrolide-resistant Italian isolates of S. pneumoniae (18).
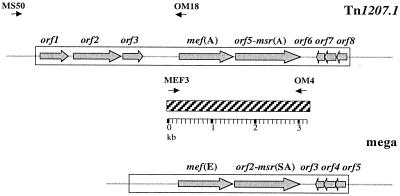
Genetic elements carrying mef genes in S. pneumoniae were recently detected and characterized. The mef(A)-carrying element is a 7.2-kb defective transposon (Tn1207.1) that contains eight open reading frames (ORFs), one of which is a putative site-specific recombinase (23). The element that contains mef(E) (macrolide efflux genetic assembly [the mega element]) is approximately 5.5 kb and contains five ORFs but no putative transposase or recombinase (7). Interestingly, both elements contain an ORF adjacent to mef, designated ORF5 in Tn1207.1 and mel in the mega element, which has homology with the msr(A) gene of Staphylococcus aureus, which coded for a protein of the ABC transporter superfamily involved in macrolide efflux (7, 23).
The aim of this study was to identify the mef genes in a large collection of S. pneumoniae strains from Italy and to characterize the properties of the strains carrying them.
MATERIALS AND METHODS
Bacterial strains.
Two-hundred sixty-seven S. pneumoniae clinical isolates were examined: 182 strains were from patients with invasive diseases (blood or cerebrospinal fluid) isolated in different areas of Italy over 4 years [19]; unpublished data) and 85 strains from the nasopharynges of healthy children attending day-care centers in Rome (20a).
Susceptibilities to ERY, penicillin (PEN), clindamycin, tetracycline (TET), and chloramphenicol (CHL) were assayed by the Etest (AB Biodisk). The breakpoints for resistance were those suggested by NCCLS (17). Isolates were serotyped by capsular swelling in antisera prepared at the Statens Seruminstitut, Copenhagen, Denmark.
Detection of mef genes and distinction between mef(A) and mef(E).
The presence of the mef gene was detected by PCR with the primer pair designed by Sutcliffe et al. (27). In order to discriminate between mef(A) and mef(E), a PCR-restriction fragment length polymorphism analysis was performed, as suggested by Oster et al. (18). The primers pair used were MEF3 (5′-GCGTTTAAGATAAGCTGGCA-3′) and MEF4 (5′-CCTGCACCATTTGCTCCTAC-3′), both of which were derived from the work of Tait-Kamradt et al. (29), to generate a 1,743-bp PCR product. The amplicon was digested with the BamHI or the DraI restriction enzyme. In mef(A) there is one BamHI site, so restriction generates two fragments of 1,340 and 403 bp, respectively, while in mef(E) there are no BamHI restriction sites. Restriction of mef(A) with DraI yields two fragments of 1,493 and 250 bp, respectively, while restriction of mef(E) yields three fragments of 782, 711, and 250 bp, respectively.
PFGE.
The relatedness among mef(A)- and mef(E)-carrying strains was examined by pulsed-field gel electrophoresis (PFGE) by published methods (19). Genomic DNAs were digested with SmaI prior to electrophoresis with a CHEF-Mapper system (Bio-Rad Laboratories, Milan, Italy). Strains that differed by one to six bands were considered clonally related (31).
Sequencing of mef and orf5 genes.
A 3,201-bp fragment that encompassed the mef gene and orf5-msr(A) homolog was obtained by amplification with primers MEF3 (29) and OM4 (5′-AGGAGCAGTTCGATTTACTG-3′), designed on the basis of the Tn1207.1 sequence (23) (Fig. 1). Sequencing was performed with a Perkin-Elmer ABI 377 DNA sequencer and an ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit (PE Applied Biosystems).
FIG. 1.
Schematic representation of mef(A)-carrying element Tn1207.1 (22) and the mef(E)-carrying mega element (7). The hatched bar represents the fragment amplified and sequenced with primers MEF3 and OM4. Primers MS50 and OM18, used to amplify the chromosomal insertion of the mef(A)-carrying element, are indicated above Tn1207.1. The positions of the primers are indicated by arrows.
As the mef(A) element of S. pneumoniae is inserted into chromosomal gene celB (23), a PCR was performed to amplify a segment spanning a portion of celB (upstream of the putative insertion) and a portion of the inserted mef element. The primers used were MS50 (5′-GCTTATGCTTTTATCCTGACCATG-3′), which anneals upstream of the Tn1207.1 integration site in celB (20, 23), and OM18 (5′-TGCTTGCCCTGCCCATATT-3′), which is designed on the basis of a consensus sequence internal to the mef genes (Fig. 1). In order to detect the chromosomal insertion of the mef(E) element, PCRs were performed with primers specific for the sequences flanking the insertion sites found by Gay and Stephens (7).
Transformation assay.
The assay used to determine the competence of the strains was carried out as described previously (21). The clinical isolates were tested along with a noncompetent control strain, strain MF4 (23), and a competent control strain, strain Rx1 (21). A competence curve was obtained for mef(E)-carrying strain 713. Briefly, transforming DNA and competence-stimulating peptide were added to pneumococcal cells harvested at 15-min intervals during the exponential phase of growth in competence medium (21). Since the transforming DNA contained a novobiocin resistance marker, the number of transformants was determined by plating the cells on selective plates containing novobiocin at 10 μg/ml. The transformation frequency was expressed as the number of CFU of the transformants divided by the number of CFU of the recipients.
Mating experiments.
Transfer of mef by conjugation was tested. The recipient strain was FP10, a streptomycin-resistant derivative of strain Rx1 (rough type 2) (21) in which the comC gene that encodes the competence-stimulating peptide was deleted and replaced by a CHL resistance cassette, making the strain not spontaneously transformable (F. Iannelli et al., unpublished data). Donor and recipient bacteria, grown separately at 37°C in tryptic soy broth until the end of the log phase, were mixed at a 1:10 ratio, plated onto tryptic soy agar plates with 5% horse blood, and incubated in 5% CO2 at 37°C for 4 h. Matings were performed in the presence of DNase at 10 μg/ml. After incubation, the cells were harvested, diluted, and plated. Scoring of transconjugants on multilayer plates was performed as described by Shoemaker et al. (25). In the overlay of the selection plates, ERY was added at 1 μg/ml, streptomycin was added at 500 μg/ml, and CHL was added at 3 μg/ml. The presence of mef genes in the transconjugants was confirmed by PCR.
Nucleotide sequence accession number.
The nucleotide sequence of the 3,152-bp fragment containing mef(E) and the msr(A)-like gene of strain PN150 was assigned GenBank accession no. AF376746.
RESULTS
Characteristics of mef(A)- and mef(E)-positive isolates.
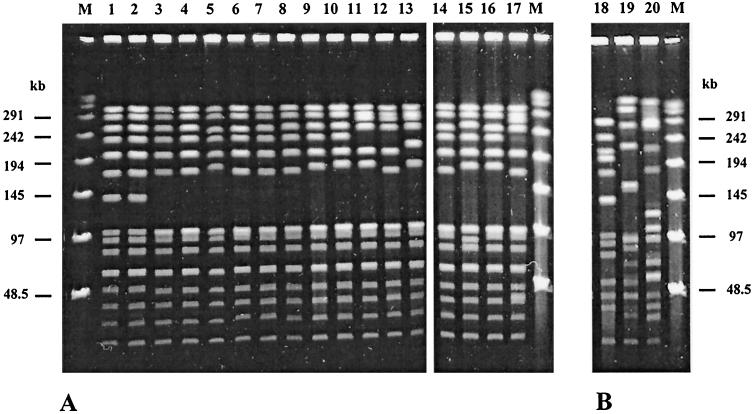
Among a sample of 267 isolates of S. pneumoniae, 98 pneumococcal strains (36.7%) were resistant to macrolides. Twenty of these strains displayed the M phenotype and carried a mef gene. Of the mef-positive strains, 17 carried mef(A) and 3 carried mef(E). The 17 mef(A)-carrying isolates belonged to serotype 14. By PFGE their profiles appeared to be very similar (Fig. 2A). Six isolates shared an identical profile (indicated profile A1); the other isolates showed six different patterns (profiles A2 to A7) that differed from the principal profile by two to six bands. These differences are compatible with a clonal origin of the isolates (31), although they were not related in terms of times of isolation, geographical area of isolation in Italy, or the characteristics of the patients from which they were isolated (Table 1).
FIG. 2.
PFGE of SmaI-digested chromosomal DNA of S. pneumoniae. (A) mef(A)-carrying serotype 14 strains; (B) mef(E)-carrying strains. Lane 1, PNS06; lane 2, PNS07; lane 3, 1514; lane 4, 1711; lane 5, PN67; lane 6, PN151; lane 7, PN83; lane 8, PN98; lane 9, PN88; lane 10, PN137; lane 11, PN165; lane 12, PN17; lane 13, 1044; lane 14, PN138; lane 15, PN139; lane 16, PN92; lane 17, PNS11; lane 18, PN150; lane 19, PN34; lane 20, 713; lanes M, bacteriophage lambda ladder molecular mass marker. See Table 1 for details about the strains.
TABLE 1.
Characteristics of 20 clinical isolates carrying the mef gene
| Strain | Place (yr) of isolation | Source | Patient | Serotype | Resistance pattern | mef gene | PFGE profile |
|---|---|---|---|---|---|---|---|
| PN17 | Cuneo (1997) | CSFa | Adult | 14 | ERY | mef(A) | A5 |
| PN67 | Rome (1997) | CSF | Child | 14 | ERY | mef(A) | A3 |
| PN83 | Verona (1998) | CSF | Child | 14 | ERY | mef(A) | A1 |
| PN88 | Reggio Calabria (1998) | CSF | Child | 14 | ERY | mef(A) | A3 |
| PN92 | Monza (1998) | CSF | Child | 14 | ERY | mef(A) | A3 |
| PN98 | Rome (1998) | CSF | Child | 14 | ERY | mef(A) | A1 |
| PN137 | Arezzo (1999) | CSF | Adult | 14 | ERY | mef(A) | A3 |
| PN138 | Parma (1999) | CSF | Adult | 14 | ERY | mef(A) | A1 |
| PN139 | Arezzo (1999) | CSF | Adult | 14 | ERY | mef(A) | A7 |
| PN151 | Mantova (1999) | CSF | Adult | 14 | ERY | mef(A) | A1 |
| PN165 | Palermo (1999) | CSF | Child | 14 | ERY | mef(A) | A4 |
| PNS06 | Rome (1998) | Blood | Adult | 14 | ERY | mef(A) | A2 |
| PNS07 | Rome (1998) | Blood | Adult | 14 | ERY | mef(A) | A2 |
| PNS11 | Rome (1998) | Blood | Adult | 14 | ERY | mef(A) | A5 |
| 1514 | Rome (1999) | Nasopharynx | Child | 14 | ERY | mef(A) | A1 |
| 1711 | Rome (1999) | Nasopharynx | Child | 14 | ERY | mef(A) | A1 |
| 1044 | Rome (1999) | Nasopharynx | Child | 14 | ERY | mef(A) | A6 |
| PN34 | Trento (1997) | CSF | Adult | 6A | ERY, TET, CHL | mef(E) | B |
| PN150 | Bologna (1999) | CSF | Adult | 19F | ERY, PEN, TET | mef(E) | C |
| 713 | Rome (1999) | Nasopharynx | Child | 6A | ERY, TET | mef(E) | D |
CSF, cerebrospinal fluid.
The mef(E)-carrying strains belonged to two different serotypes, and their macrorestriction profiles were different (Fig. 2B). All mef(A)-carrying strains were resistant to ERY and sensitive to other antibiotics, including PEN, TET, and CHL, while the mef(E)-carrying strains were resistant to ERY and to other drugs (Table 1).
mef and msr(A)-like genes.
The 3,201-bp DNA fragment containing the mef and msr(A)-like genes was obtained by PCR with primers designed on the basis of the sequence of mef(A)-carrying genetic element Tn1207.1 (Fig. 1). PCR products of identical sizes were obtained from all 20 mef strains, and sequencing was performed for the 3 mef(E)-carrying strains and for 3 randomly chosen mef(A)-carrying strains (strains PN83, PN92, and PN137; Table 1). The nucleotide sequences of the mef(A)-carrying strains were identical to that of Tn1207.1, while the nucleotide sequences of the mef(E)-carrying strains were identical to each other but differed from that of Tn1207.1 at 168 positions. In the mef(E)-carrying strains, the sequences encoding mef(E) and the msr(A) homolog were identical to the corresponding sequences published by Gay and Stephens (7). However, a 16-bp deletion and six single-base mismatches were found in the noncoding region upstream of mef(E), and one mismatch was found in the noncoding region downstream of the msr(A) homolog.
Chromosomal insertion site of the mef element.
Since mef(A)-carrying genetic element Tn1207.1 was found to be integrated into the pneumococcal celB gene (23), PCR primer pair MS50-OM18 was designed on the basis of celB and mef sequences (Fig. 1) to investigate the insertion sites of the mef-carrying genetic elements. Identical 3.9-kb fragments could be amplified from all the 17 mef(A) strains examined in this study, indicating integration in celB. Sequencing of the insertion site in one isolate showed that it was coincident at the nucleotide level with the insertion site previously described in transformant MF4 (23). No PCR products were obtained from the three mef(E) strains, suggesting integration elsewhere. In strain 713, the insertion site was found to correspond to class I of Gay and Stephens (7). In the other two mef(E) strains the insertion sites were different from those described by Gay and Stephens (7).
Transformability of mef-carrying pneumococci.
Since insertional inactivation of celB should lead to impairment of natural transformation (20), our strains were tested for competence, as was noncompetent control strain MF4 (23) and competent control strain Rx1 (21). As expected, the three mef(A)-carrying strains (strains PN83, PN92, and PN137) did not become competent, while the mef(E) strains were transformed, confirming that the mef(E) elements are not integrated in celB. A competence curve was obtained for mef(E)-carrying S. pneumoniae strain 713 by using as a positive control competent strain Rx1. The transformation frequency was calculated at each time point. The maximum activity was at 45 min for both strains, with a value of 4.2 × 10−4 transformants per recipient in strain 713 and 3 × 10−2 transformants per recipient in strain Rx1.
Conjugal transfer of mef.
The three mef(E)-carrying strains and the three mef(A)-carrying strains were used as donors in conjugation experiments in which rough strain FP10 was the recipient. mef(A) was transferred at a frequency of 1.5 × 10−3 transconjugants per donor from PN92 and at a frequency of 1.8 × 10−4 transconjugants per donor from PN137, while no transfer (<5 × 10−8 transconjugants per donor) was detected when PN83 was the donor. In the two transconjugants, the mef(A) element was found to be inserted in celB. No mef(E)-carrying strain was able to transfer macrolide resistance by conjugation.
DISCUSSION
The high rate of macrolide resistance observed in this study is not unprecedented among isolates from Italy. In the last decade a steep increase in the rate of resistance to macrolides has been observed, from 5% in 1993 (12) to over 30% in 1998 to 1999 (14, 19). In our study, ERY resistance was mediated by the drug efflux mechanism in only 20 of 98 isolates (20%). This confirms that in Italy, as in other European countries, the mef gene is relatively uncommon among macrolide-resistant S. pneumoniae isolates (2, 5, 13, 19), while in North America it is more frequent than the erm(B) gene (4, 6, 9). Although carriage of mef(E) has been considered typical of S. pneumoniae, we have found 17 strains that carry mef(A) and only 3 strains that carry mef(E).
As both the element carrying mef(A) and the element carrying mef(E) contain another putative efflux gene, a homolog to msr(A), we amplified and sequenced a fragment that included mef and the msr(A) homolog in six Italian isolates. The sequences were identical for the three mef(A) strains and the sequences were identical for the three mef(E) strains, while the sequences of the two variants were divergent at a number of positions. This suggests that the elements have recently emerged from a common ancestor and evolved in different hosts. It is possible that the original host of the mef(A) element is S. pyogenes (M. Santagati, F. Iannelli, C. Messina, M. R. Oggioni, S. Stefani, and G. Pozzi, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. 2014, 2001), while mef(E) is carried by viridans group streptococci (1). In the nasopharynx there are opportunities for these elements to be transferred from both species to S. pneumoniae. The presence of mef(A) in Italian isolates of S. pneumoniae might be the consequence of the high prevalence of macrolide-resistant mef-carrying S. pyogenes isolates in Italy (8).
An important characteristic that is different between the two elements is the insertion site: while the mef(A) element was found to be integrated at a single specific chromosomal site (celB) in all the strains examined, including transformant MF4 and the two transconjugants, the mef(E) element was found to be inserted at different chromosomal locations (7). Site-specific integration of the mef(A) element is a property shared by other mobile DNA elements, including transposons (32) and the pathogenicity islands of enteric pathogens (24).
As the target gene for mef(A) integration (celB) is involved in competence, mef(A)-carrying strains are defective in transformability, whereas mef(E) strains can be transformed.
We found that two of three mef(A)-carrying isolates were able to transfer the mef(A) element by conjugation. Previously described transposon Tn1207.1 appeared to be a defective element and was not transferable (23). Although we did not examine the size of the mef(A) element in the clinical isolates, it is conceivable that the strains able to transfer macrolide resistance possess an element that is larger than Tn1207.1, similar to that described recently in S. pyogenes (Santagati et al., 41st ICAAC), and that represents a complete conjugative transposon. Conversely, the mef(E) element was not transferable by conjugation from our three isolates. Although mef(E) has been found to be transferable from S. pneumoniae in some instances (11), the mega element described by Gay and Stephens (7) has features of a defective transposon that lacks the enzymes required for DNA transposition.
Interestingly, the resistance phenotypes of the strains are different. mef(A)-positive strains are susceptible to all the antibiotics tested other than ERY, while mef(E)-positive strains display resistance to various antibiotics, including PEN. The uniform susceptibility of mef(A) strains might be due to the fact that all the mef(A) isolates belong to a single serotype 14 clone. This finding is intriguing and has also been confirmed in a subsequent large set of mef(A)-carrying invasive isolates (data not shown). As the isolates are not related in terms of times and areas of isolation, it appears that the spread of the mef(A) element in Italy occurred through the expansion of a single PEN-susceptible serotype 14 clone that has acquired mef(A). An alternate explanation is that different strains belonging to a well-established clone have acquired the element independently. On the basis of SmaI PFGE fingerprinting and multilocus sequence typing (data not shown), the mef(A)-positive clone appears to belong to the England14-9 clone, one of the major antibiotic-resistant pneumococcal clones (15). It is noteworthy that clonal expansion has also been noted in ERY-resistant serotype 14 isolates carrying mef(E) (7) or erm(B) (19).
We do not know whether the prevalence of mef(A) in S. pneumoniae is a peculiarity of the Italian situation, as the identity of the mef genes has been investigated in only a few geographical areas. On the other hand, our data and those of Gay and Stephens (7) indicate that the mef(E) element is disseminated by horizontal transfer to different strains and therefore seems to be more adapted for S. pneumoniae.
In conclusion, as the mef(A) and the mef(E) elements are endowed with important genetic differences and confer distinctive characteristics to the strains, it might be appropriate to distinguish between them by referring to them as mef(A), subclass mef(A), or subclass mef(E). When describing the epidemiology of macrolide resistance in S. pneumoniae in different parts of the world, it might be important to distinguish these two genes, as this could contribute to an understanding of the spread of ERY resistance in S. pneumoniae.
Acknowledgments
We are indebted to Kathryn Gay and David S. Stephens for making the sequence of the mega element available to us before publication. We thank Fabio D'Ambrosio for experienced technical assistance.
This work was supported in part by grants from Ministero della Sanità (Programmi per la Ricerca Finalizzata 1999 and Progetti di ricerca finalizzata IRCCS, ICS 120.5/RF 97.99) and from MURST (COFIN 2000, 22010709/16010190).
REFERENCES
- 1.Arpin, C., M. H. Canron, J. Maugein, and C. Quentin. 1999. Incidence of mefA and mefE genes in viridans group streptococci. Antimicrob. Agents Chemother. 43:2335-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baquero, F., J. A. Garcia-Rodriguez, J. G. De Lomas, L. Aguilar, and the Spanish Surveillance Group for Respiratory Pathogens. 1999. Antimicrobial resistance of 1,113 Streptococcus pneumoniae isolates from patients with respiratory tract infections in Spain: results of a 1-year (1996-1997) multicenter surveillance study. Antimicrob. Agents Chemother. 43:357-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clancy, J., J. Pepitpas, F. Dib-Hajj, W. Yuan, M. Cronan, A. V. Kamath, J. Bergeron, and A. Retsema. 1996. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol. Microbiol. 22:867-879. [DOI] [PubMed] [Google Scholar]
- 4.Doern, G. V., A. B. Brueggemann, H. Huynh, E. Wingert, and P. Rhomberg. 1999. Antimicrobial resistance with Streptococcus pneumoniae in the United States, 1997-98. Emerg. Infect. Dis. 5:757-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fitoussi, F., C. Doit, P. Geslin, N. Brahimi, and E. Bingen. 2001. Mechanism of macrolide resistance in clinical pneumococcal isolates in France. Antimicrob. Agents Chemother. 45:636-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gay, K., W. Baughman, Y. Miller, D. Jackson, C. G. Whitney, A. Schuchat, M. M. Farley, F. Tenover, and D. S. Stephens. 2000. The emergence of Streptococcus pneumoniae resistant to macrolide antimicrobial agents: a 6-year population-based assessment. J. Infect. Dis. 182:1417-1424. [DOI] [PubMed] [Google Scholar]
- 7.Gay, K., and D. S. Stephens. 2001. Structure and dissemination of a chromosomal insertion element encoding macrolide efflux in Streptococcus pneumoniae. J. Infect. Dis. 184:56-65. [DOI] [PubMed] [Google Scholar]
- 8.Giovanetti, E., M. P. Montanari, M. Mingoia, and P. E. Varaldo. 1999. Phenotypes and genotypes of erythromycin-resistant Streptococcus pyogenes strains in Italy and heterogeneity of inducibly resistant strains. Antimicrob. Agents Chemother. 43:1935-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnston, N. J., J. C. De Azavedo, J. D. Kellner, and D. E. Low. 1998. Prevalence and characterization of the mechanisms of macrolide, lincosamide, and streptogramin resistance in isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2425-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leclercq, R., and P. Courvalin. 1991. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob. Agents Chemother. 35:1267-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luna, V. A., P. Coates, E. A. Eady, J. H. Cove, T. T. H. Nguyen, and M. C. Roberts. 1999. A variety of gram-positive bacteria carry mobile mef genes. J. Antimicrob. Chemother. 44:19-25. [DOI] [PubMed] [Google Scholar]
- 12.Marchese, A., E. A. Debbia, A. Arvigo, A. Pesce, and G. C. Schito. 1995. Susceptibility of Streptococcus pneumoniae strains isolated in Italy to penicillin and ten other antibiotics. J. Antimicrob. Chemother. 36:833-837. [DOI] [PubMed] [Google Scholar]
- 13.Marchese, A., E. Tonoli, E. A. Debbia, and G. C. Schito. 1999. Macrolide resistance mechanisms and expression of phenotypes among Streptococcus pneumoniae circulating in Italy. J. Antimicrob. Chemother. 44:461-464. [DOI] [PubMed] [Google Scholar]
- 14.Marchese, M., E. Tonoli, G. Balistreri, E. Debbia, and G. C. Schito. 2000. Antibiotic susceptibility patterns and serotypes of antibiotic resistant and/or invasive Streptococcus pneumoniae strains circulating in Italy. Microb. Drug Resist. 6:163-170. [DOI] [PubMed] [Google Scholar]
- 15.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefevre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montanari, M. P., M. Mingoia, E. Giovannetti, and P. E. Varaldo. 2001. Differentiation of resistance phenotypes among erythromycin-resistant pneumococci. J. Clin. Microbiol. 39:1311-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 5th ed. NCCLS document M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 18.Oster, P., A. Zanchi, S. Cresti, M. Lattanzi, F. Montagnani, C. Cellesi, and G. M. Rossolini. 1999. Patterns of macrolide resistance determinants among community-acquired Streptococcus pneumoniae isolates over a 5-year period of decreased macrolide susceptibility rates. Antimicrob. Agents Chemother. 43:2510-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pantosti, A., F. D'Ambrosio, A. Tarasi, S. Recchia, G. Orefici, and P. Mastrantonio. 2000. Antibiotic susceptibility and serotype distribution of Streptococcus pneumoniae causing meningitis in Italy, 1997-1999. Clin. Infect. Dis. 31:1373-1379. [DOI] [PubMed] [Google Scholar]
- 20.Pestova, E. V., and D. A. Morrison. 1998. Isolation and characterization of three Streptococcus pneumoniae transformation-specific loci by use of a lacZ reporter insertion vector. J. Bacteriol. 180:2701-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Petrosillo, N., A. Pantosti, E. Boroli, A. Spanò, M. Del Grosso, B. Tallaride, and G. Ippolito. Prevalence and determinants of Streptococcus pneumoniae nasopharyngeal colonization and molecular epidemiology of the isolates from healthy children in Rome. Eur. J. Clin. Microbiol. Infect. Dis., in press. [DOI] [PubMed]
- 21.Pozzi, G., L. Masala, F. Iannelli, R. Manganelli, L. S. Havarstein, L. Piccoli, D. Simon, and D. A. Morrison. 1996. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J. Bacteriol. 178:6087-6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppala. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santagati, M., F. Iannelli, M. Oggioni, S. Stefani, and G. Pozzi. 2000. Characterization of a genetic element carrying the macrolide efflux gene mef(A) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2585-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt, H., W.-L. Zhang, U. Hemmrich, S. Jelacic, W. Brunder, P. I. Tarr, U. Dobrindt, J. Hacker, and H. Karch. 2001. Identification and characterization of a novel genomic island integrated at selC in locus of enterocyte effacement-negative, Shiga toxin-producing Escherichia coli. Infect. Immun. 69:6863-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shoemaker, N., M. Smith, and W. R. Guild. 1980. DNase-resistant transfer of chromosomal cat and tet insertions by filter mating in pneumococcus. Plasmid 3:80-87. [DOI] [PubMed] [Google Scholar]
- 26.Shortridge, V. D., G. V. Doern, A. B. Brueggemann, J. M. Beyer, and R. K. Flamm. 1999. Prevalence of macrolide resistance mechanisms in Streptococcus pneumoniae isolates from a multicenter antibiotic resistance surveillance study conducted in the United States in 1994-1995. Clin. Infect. Dis. 29:1186-1188. [DOI] [PubMed] [Google Scholar]
- 27.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sutcliffe, J., A. Tait-Kamradt, and L. Wondrack. 1996. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolide but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob. Agents Chemother. 40:1817-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tait-Kamradt, A., J. Clancy, M. Cronan, F. Dib-Hajj, L. Wondrack, W. Yuan, and J. Sutcliffe. 1997. mefE is necessary for the erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:2251-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tait-Kamradt, A., T. Davies, P. C. Appelbaum, F. Depardieu, P. Courvalin, J. Petitpas, L. Wondrack, A. Walker, M. R. Jacobs, and J. Sutcliffe. 2000. Two new mechanisms of macrolide resistance in clinical strains of Streptococcus pneumoniae from Eastern Europe and North America. Antimicrob. Agents Chemother. 44:3395-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vijayakumar, M. N., and S. Ayalew. 1993. Nucleotide sequence analysis of the termini and chromosomal locus involved in site-specific integration of the streptococcal conjugative transposon Tn5252. J. Bacteriol. 175:2713-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]