Abstract
The distributions of the antibiotic resistance patterns in a population of Staphylococcus aureus isolates from a teaching hospital were studied over a 9-year period. The results indicate the existence of successive major epidemic methicillin-resistant strains and the emergence of a methicillin-susceptible strain with an unusual resistance pattern. Our findings suggest that this methicillin-susceptible S. aureus strain could be derived from the dominant gentamicin-susceptible methicillin-resistant S. aureus strain with the loss of a 40-kb DNA fragment.
Methicillin-resistant Staphylococcus aureus (MRSA) has long been known to spread through hospitals. Most epidemiological studies of MRSA are not longitudinal studies but are cross-sectional studies and often involve several hospitals (14). There are a few reports of long-term surveys of MRSA in one hospital (8). We report on the computerized surveillance of MRSA in a French teaching hospital over a 9-year period and provide arguments for the reason for the instability of methicillin resistance in one of the most epidemic clones of MRSA in France.
The University Hospital of Rennes is a 1,800-bed teaching hospital with five facilities. For the 9-year period from 1992 to 2000, the rate of methicillin resistance among S. aureus isolates was 36.5%. During this period, the Committee for Nosocomial Infections Control had published general recommendations for avoidance of the spread of multidrug-resistant bacteria, but policies relating specifically to the prevention of transmission of MRSA were not in place.
Between 1992 and 2000, 13,321 S. aureus isolates were recovered from samples from patients hospitalized in four facilities. Isolates were identified by production of acid on Chapman agar and the presence of catalase and coagulase. Antimicrobial susceptibility was tested by the agar diffusion method (Diagnostics Pasteur, Marnes-la-Coquette, France) on Mueller-Hinton agar (Oxoid, Dardilly, France), according to the recommendations of the Antibiogram Committee of the French Microbiology Society, except that isolates with fosfomycin inhibition zone diameters of >14 and ≤23 mm were categorized as intermediate to this antibiotic. Patient information (sex, age, sample) and antibiogram results were collected from the Laboratory Information System and stored in a specific database. If several isolates with the same antibiograms were recovered at different times from a patient, only the first one was retained. Consequently, 3,350 MRSA isolates have been retained for further analysis and have been grouped by resistance pattern by use of EPILOG software (Saric International, Nanterre, France). A resistance pattern was characterized by combinations of results (susceptible and intermediate or resistant) for oxacillin and the following antibiotics: tobramycin, gentamicin, erythromycin, lincomycin, sulfamethox- azole, pefloxacin, rifampin, and fosfomycin. The combinations of patterns of resistance to all these antibiotics except the glycopeptides were assumed to reflect the patterns of resistance to the most important antibiotics used for the treatment of staphylococcal infections (Table 1). The finding of a resistance pattern in 10 or more oxacillin-resistant S. aureus (MRSA) isolates for 1 year was suspected to represent an epidemic.
TABLE 1.
Descriptions of the five major epidemic MRSA patterns and the MSSA ΔVII pattern
| Pattern | Resistance toa: |
|---|---|
| I | OXA, TOB, GEN, LIN, SUL, PEF, RIF, FOF |
| II | OXA, TOB, GEN, ERY, LIN, PEF |
| III | OXA, TOB, GEN, ERY, PEF |
| VI | OXA, TOB, GEN, ERY, LIN, SUL, PEF, RIF |
| VII | OXA, TOB, ERY, LIN, PEF, FOF |
| ΔVII | ERY, LIN, PEF, FOF |
OXA, oxacillin; TOB, tobramycin; GEN, gentamicin; LIN, lincomycin; ERY, erythromycin; SUL, sulfamethoxazol; PEF, pefloxacin, RIF, rifampin; FOF, fosfomycin.
The clonal nature of epidemic MRSA strains was assessed for randomly selected isolates with each major pattern by two typing methods: the Euclidian distances method and DNA polymorphism after SmaI restriction and pulsed-field gel electrophoresis (PFGE). The diameters of the zones of inhibition for seven antibiotics (tobramycin, gentamicin, erythromycin, lincomycin, sulfamethoxazole, rifampin, and fosfomycin) were collected and the Euclidian distances were calculated by use of ITCF software (Institut Technique des Céréales et Fourrages, Paris, France). As described by Blanc et al. (1), the Euclidian distance method is a means of multivariate analysis of inhibition zone diameters (x) around discs of n antibiotics in which the similarity between two strains, j and k, is calculated by the formula
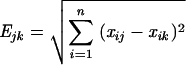 |
where E is Euclidian distance and i is rank of test (from 1 to n).
For PFGE, S. aureus DNA preparation and SmaI restriction were done as described previously (3). For epidemiological surveillance, PFGE was performed with a Gene Navigator apparatus (Amersham Pharmacia, Orsay, France), and for comparison of MRSA and methicillin-susceptible S. aureus (MSSA) strains, PFGE was performed with a CHEF-II apparatus (Bio-Rad, Ivry-sur-Seine, France). After electrophoresis, the gels were stained with ethidium bromide and the DNA fragments were visualized with a UV light box. Gel Compar software (Applied Maths, Sint-Martens-Latem, Belgium) was used to calculate the Dice similarity indices and to perform cluster analysis by unweighted pair group matching analysis (tolerance, 2.0%).
PCR was used to amplify the sequences from two different target sites: the mecA gene encoding PBP 2a and the coa gene encoding staphylococcal coagulase. The two sets of primers used have been described previously (9, 19). Positive and negative controls (an MRSA isolate and S. aureus ATCC 25923, respectively) were included with each run for mecA amplification. The thermocycling conditions were as follows: 94°C for 5 min for 1 cycle and then 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min for 35 cycles on a Perkin-Elmer thermocycler. After electrophoresis, the gel was stained with ethidium bromide and the amplicons were visualized. Amplicons produced by coa amplification were restricted with AluI, and the restriction fragments were resolved by 3% agarose gel electrophoresis at 110 V for 45 min. The presence of PBP 2a was investigated by a slide agglutination test with latex particles coated with a monoclonal antibody directed toward the protein (22). The test was performed according to the manufacturer's recommendations (Servibio, Meudon, France) with an MRSA isolate as a positive control and S. aureus ATCC 25923 as a negative control.
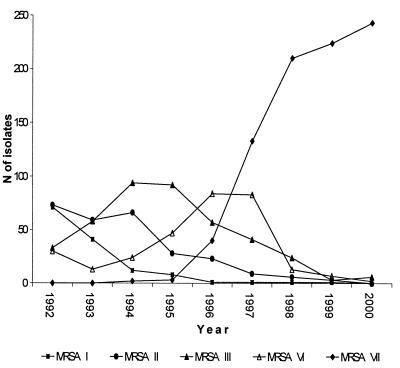
We were able to distinguish 106 different MRSA resistance patterns for the period from 1992 to 2000, and among these, 12 fit the definition of an epidemic pattern (>10 isolates with a single pattern recovered in 1 year). Five of them (designated patterns I, II, III, VI, and VII) included more than 70 strains, and we considered strains with these patterns to be major epidemic MRSA strains, with the number of isolates per year ranging from 0 to 243 (Table 1 and Fig. 1). The seven remaining patterns (patterns IV, V, VIII, IX, X, XI, and XII) included less than 40 strains, and we considered strains with these patterns to be minor epidemic MRSA strains. The isolates which belonged to a minor or a major epidemic pattern accounted for 77.56% ± 4.25% of all MRSA isolates by year.
FIG. 1.
Distributions of major epidemic MRSA isolates between 1992 and 2000.
The distributions of the different major epidemic MRSA strains argue strongly for clonal dissemination (Fig. 1). One or two dominant strains could be identified in each year: isolates with patterns I and II in 1992; isolates with patterns II and III in 1993; isolates with pattern III in 1994 and 1995; isolates with pattern VI in 1996; and isolates with pattern VII in 1997, 1998, 1999, and 2000. The more striking feature is the emergence of MRSA isolates with pattern VII, which consisted of gentamicin-susceptible strains which had replaced almost all the previous gentamicin-resistant, major epidemic MRSA strains. Before 1996 the successive epidemic strains were dominant for more than 1 year (MRSA pattern I isolates have been epidemic in our hospital since 1989), and replacement of one dominant strain by another one was progressive. Conversely, MRSA pattern VI isolates, the next to the last epidemic strains, were dominant only in 1996, and their numbers decreased dramatically in 1998. Thereafter, as previous major epidemic MRSA strains (those with patterns I to VI) have disappeared and no new major epidemic strain has emerged, about 60% of MRSA strains isolated between 1998 and 2000 have MRSA pattern VII (Fig. 1).
By the Euclidian distances method and by use of a cutoff value of 18 mm for the inhibition zone diameter, the five major epidemic MRSA patterns were clearly separated from each other (data not shown). Antibiotyping has proved to be a powerful method for the typing of MRSA strains when a quantitative method was used (1, 21). In this study, for a well-defined period and in one hospital, qualitative antibiotyping has worked as well as a quantitative method; this is likely due to the small number of epidemic strains with distinct resistance patterns.
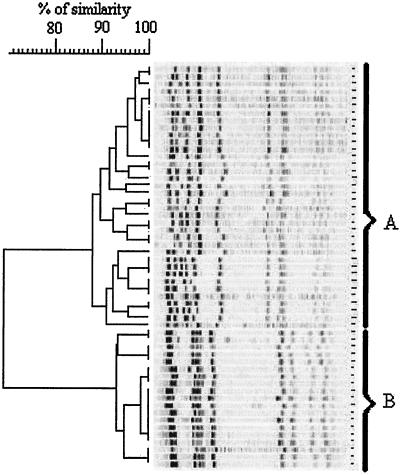
After SmaI restriction and PFGE, all the major epidemic MRSA grouped into two well-separated clones: those with patterns I, VI, and VII and those with patterns II and III (Fig. 2). Each genomic clone was homogeneous, with a high degree of internal similarity among the clone with patterns II and III (index = 94%) but with a rather low degree of similarity among the clone with patterns I, VI, and VII (index = 87.5%). Most gentamicin-susceptible MRSA strains remained resistant to tobramycin by the production of the nucleotidyltransferase ANT4′ but did not produce the bifunctional enzyme APH6′-AAC2", which inactivates gentamicin and netilmicin (6, 7). These strains have spread widely in French hospitals since 1992, and their epidemiological and molecular features have been well described in recent years (2, 7, 17, 18). Our results fully agree with previous findings: all the isolates have the same resistance pattern (pattern 1) and most of them have the same pulsotype (pulsotype A1) as those reported by Lemaítre et al. (18) (Fig. 2). More recently, Laurent and colleagues (15) have suggested that French epidemic strains of MRSA (including an MRSA pattern VII strain from our hospital) have a competitive advantage over gentamicin-resistant clones but are also genetically related to some of them. On the basis of this relatedness, some investigators have suggested that gentamicin-sensitive MRSA clones could have emerged from gentamicin-resistant clones (2, 7, 17). Therefore, we cannot prove an exogenous origin of the MRSA pattern VII clone rather than the local emergence from a gentamicin-resistant MRSA strain, but its presence in many hospitals in the context of the nationwide spread argues well for the first hypothesis. Epidemic MRSA strains have been described for a long time, and there have been many reports of very large outbreaks that are due to a clone and that involve hospitals in a region, a country, or a continent (5, 14).
FIG. 2.
Classification of PFGE restriction patterns of MRSA pattern I, II, III, VI, and VII isolates (55 isolates) by using the Dice coefficient and unweighted pair grouping matching analysis.
MSSA strains are usually susceptible to most antibiotics except benzylpenicillin (to which 88% of isolates are resistant) and, at lower rates, macrolides-lincosamides-streptogramin B (MLSB) (rate of inducible resistance to MLSB, 14.5%) and fluoroquinolones (to which 4.5% of isolates are resistant). Constitutive resistance to MLSB is rare in MSSA isolates (2.5%) but frequent in MRSA isolates (77%). Since 1997 we have identified MSSA isolates with a surprising resistance pattern that consists of constitutive MLSB resistance, resistance to fluoroquinolones, and resistance to fosfomycin. Moreover, these isolates were resistant to spectinomycin, an aminoglycoside antibiotic which we used as a resistance marker for some MRSA strains, especially MRSA strains with pattern VII. Since this resistance pattern was the same except for a lack of resistance to oxacillin and tobramycin, we have assumed that this strain was derived from an MRSA pattern VII strain and have designated it MSSA pattern ΔVII. The number of MSSA pattern ΔVII isolates has progressively increased over 5 years: in 1997, 2 isolates; in 1998, 5 isolates; in 1999, 14 isolates; in 2000, 22 isolates; and from 1 January to 30 June 2001, 24 isolates. Five patients were first infected with an MRSA pattern VII strain and then with a MSSA pattern ΔVII strain a few months later (Table 2).
TABLE 2.
Samples and dates of recovery of major epidemic MRSA pattern VII and MSSA pattern ΔVII isolates from five patients
| Patient | MRSA pattern VII isolates
|
MSSA pattern ΔVII isolates
|
||
|---|---|---|---|---|
| Sample | Recovery date (day.mo.yr) | Sample | Recovery date (day.mo.yr) | |
| 1 | Surgical wound | 10.10.1998 | Blood culture | 16.02.1999 |
| 2 | Abdominal drainage | 27.03.1999 | Abdominal abcess | 24.04.1999 |
| 3 | Urine | 07.01.2000 | Abcess | 23.02.2000 |
| 4 | Urine | 04.08.2000 | Urine | 30.05.2001 |
| 5 | Urine | 18.01.2001 | Urine | 18.05.2001 |
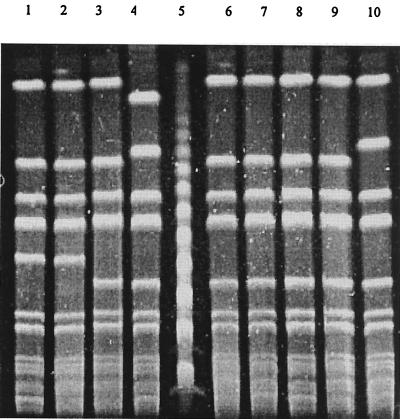
Methicillin susceptibility was confirmed by the absence of expression of PBP 2a (agglutination test negative) and the absence of the mecA gene (PCR negative) in all MSSA pattern ΔVII isolates tested. The AluI polymorphisms of the coa PCR products showed that MRSA pattern VII and MSSA pattern ΔVII strains had identical restriction patterns (data not shown). The PFGE pattern of MSSA pattern ΔVII strains differs from that of major epidemic MRSA pattern VII strains by only one band, corresponding to the loss of a 40-kb fragment from the 200-kb DNA band (Fig. 3). Phenotypic as well as genotypic markers indicated that MSSA pattern ΔVII strains could be derived from major epidemic MRSA strains by the loss of the mecA region. Previous reports have suggested that some MRSA strains possess an unstable mecA DNA region. The instability of mecA was first described in vitro (10, 11) and more recently has been described in clinical situations (4, 12, 16, 20, 23). The role of recently discovered cassette chromosomal recombinases CCRA and CCRB has been underlined by the induction of the precise excision of the mecA DNA region (13). The deletion of the Staphylococcus chromosome cassette (SCC) is responsible for a new resistance phenotype (resistance to erythromycin and spectinomycin only) in MRSA strain N315 (13). Similarly, methicillin susceptibility in MSSA pattern ΔVII strains should be due to the loss of mecA. As the genes mecA and aad (the gene encoding for ANT4′) are localized on the same 185- to 215-kb SmaI restriction fragments in French gentamicin-sensitive MRSA clones (17) and the unique difference between the MRSA pattern VII and the MSSA pattern ΔVII SmaI restriction patterns is found on these fragments, tobramycin susceptibility is likely due to the loss of aad. Excision of SCC implies the deletion of Tn 554, which is localized upstream of mecA, and then the loss of genes for resistance to erythromycin and spectinomycin. According to Katayama and collaborators (13), we assume that the remaining resistance to these two antibiotics is due to the presence of multiple copies of Tn 554.
FIG. 3.
PFGE restriction patterns of MRSA pattern VII and MSSA pattern ΔVII isolates. Lane 1, MRSA pattern VII strain from patient 1; lane 2, MRSA pattern VII strain from patient 2; lane 3, MSSA pattern ΔVII strain from patient 1; lane 5, DNA ladder (n × 48.5-kb fragments); lane 6, MSSA pattern ΔVII strain from patient 2; lanes 4, 7, 8, 9, and 10, MSSA pattern ΔVII strains from other patients
Wagenvoort and colleagues (23) have indicated that the loss of epidemicity relies on the loss of mecA. On the one hand, the recovery of MSSA pattern ΔVII strains from patients formerly infected with a major epidemic MRSA pattern VII strain suggests that most of these strains are directly derived from MRSA strains. On the other hand, we have recovered MSSA pattern ΔVII strains from premature twins who were hospitalized in the same room of a neonatology intensive care unit and who were infected with such strains but who had not previously had staphylococcal infections. Nose carriage of MSSA pattern ΔVII strains has been detected in medical staff members during investigations of two outbreaks due to major epidemic MRSA pattern VII strains. Moreover, the increasing number of MSSA pattern ΔVII isolates might indicate that such strains are potentially transmissible from person to person, independent of the deletion of mecA.
We have described the evolution of dominant MRSA strains in our hospital and collected data which indicate that the mecA region is unstable in the most epidemic strain. It remains unclear if insertion of mecA and reacquisition of methicillin resistance could occur in such strains. Conversely, it would be of interest to know if the loss of mecA could occur in other epidemic MRSA strains and contribute to the temporal evolution of MRSA in our hospital.
Acknowledgments
We acknowledge Gilles Weidman, Philippe Gautier, Marie-France Travert, and the staff of Laboratoire de Bactériologie-Virologie, CHU of Rennes, for excellent technical assistance.
REFERENCES
- 1.Blanc, D. S., C. Lugeon, A. Wenger, H. H. Siegrist, and P. Francioli. 1994. Quantitative antibiogram typing using inhibition zone diameters compared with ribotyping for epidemiological typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 32:2505-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanc, D. S., P. Francioli, A. Le Coustumier, L. Gazagne, E. Lecaillon, P. Gueudet, F. Vandenesch, and J. Etienne. 2001. Reemergence of gentamicin-susceptible strains of methicillin-resistant Staphylococcus aureus in France: a phylogenetic approach. J. Clin. Microbiol. 139:2287-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carles-Nurit, M. J., B. Christophle, S. Broche, A. Gouby, N. Bouziges, and M. Ramuz. 1992. DNA polymorphisms in methicillin-susceptible and methicillin-resistant strains of Staphylococcus aureus. J. Clin. Microbiol. 30:2092-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deplano, A., P. T. Tassios, Y. Glupczynski, E. Godfroid, and M. J. Struelens. 2000. In vivo deletion of the methicillin resistance mec region from the chromosome of Staphylococcus aureus strains. J. Antimicrob. Chemother. 46:617-619. [DOI] [PubMed] [Google Scholar]
- 5.de Sousa, M. A., I. Santos Sanches, M. L. Ferro, M. J. Vaz, Z. Saraiva, T. Tendeiro, J. Serra, and H. de Lencastre. 1998. Intercontinental spread of a multidrug-resistant methicillin-resistant Staphylococcus aureus clone. J. Clin. Microbiol. 36:2590-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubin, D. T., P. R. Matthews, S. G. Chikramane, and P. R. Stewart. 1991. Physical mapping of the mec region of an American methicillin-resistant Staphylococcus aureus strain. Antimicrob. Agents Chemother. 35:1661-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galdbart, J.-O., A. Morvan, and N. El Solh. 2000. Phenotypic and molecular typing of nosocomial methicillin-resistant Staphylococcus aureus strains susceptible to gentamicin isolated in France from 1995 to 1997. J. Clin. Microbiol. 38:185-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Givney, R., A. Vickery, A. Holliday, M. Pegler, and R. Benn. 1998. Evolution of an endemic methicillin-resistant Staphylococcus aureus population in an Australian hospital from 1967 to 1996. J. Clin. Microbiol. 36:552-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goh, S. H., S. K. Byrne, J. L. Zhang, and A. W. Chow. 1992. Molecular typing of Staphylococcus aureus on the basis of coagulase gene polymorphisms. J. Clin. Microbiol. 30:1642-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grubb, W. B., and D. I. Annear. 1972. Spontaneous loss of methicillin resistance in Staphylococcus aureus at room temperature. Lancet ii:1257. [DOI] [PubMed] [Google Scholar]
- 11.Hiramatsu, K., E. Suzuki, H. Takayama, Y. Katayama, and T. Yokota. 1990. Role of penicillinase plasmids in the stability of the mecA gene in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 34:600-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inglis, B., W. El Adhani, and P. R. Stewart. 1993. Methicillin-sensitive and -resistant homologues of Staphylococcus aureus occur together among clinical isolates. J. Infect. Dis. 167:323-328. [DOI] [PubMed] [Google Scholar]
- 13.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, Staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerr, S., G. E. Kerr, C. A. Mackintosh, and R. R. Marples. 1990. A survey of methicillin-resistant Staphylococcus aureus affecting patients in England and Wales. J. Hosp. Infect. 16:35-48. [DOI] [PubMed] [Google Scholar]
- 15.Laurent, F., H. Lelièvre, M. Cornu, F. Vandenesch, G. Carret, J. Etienne, and J.-P. Flandrois. 2001. Fitness and competitive growth advantage of new gentamicin-susceptible MRSA clones spreading in French hospitals. J. Antimicrob. Chemother. 47:277-283. [DOI] [PubMed] [Google Scholar]
- 16.Lawrence, C., M. Cosseron, P. Durand, Y. Costa, and R. Leclercq. 1996. Consecutive isolation of homologous strains of methicillin-resistant and methicillin-susceptible Staphylococcus aureus from a hospitalized child. J. Hosp. Infect. 33:49-53. [DOI] [PubMed] [Google Scholar]
- 17.Lelièvre, H., G. Lina, M. E. Jones, C. Olive, F. Forey, M. Roussel-Delvallez, M.-H. Nicolas-Chanoine, C. M. Bébéar, V. Jarlier, A. Andremont, F. Vandenesch, and J. Etienne. 1999. Emergence and spread in French hospitals of methicillin-resistant Staphylococcus aureus with increasing susceptibility to gentamicin and other antibiotics. J. Clin. Microbiol. 37:3452-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemaítre, N., W. Sougakoff, A. Masmoudi, M.-H. Fievet, R. Bismuth, and V. Jarlier. 1998. Characterization of gentamicin-susceptible strains of methicillin-resistant Staphylococcus aureus involved in nosocomial spread. J. Clin. Microbiol. 136:81-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murakami, K., W. Minamide, K. Wada, E. Nakamura, H. Teraoka, and S. Watanabe. 1991. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J. Clin. Microbiol. 29:2240-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sá-Leão, R., I. Santos Sanches, D. Dias, I. Peres, R. M. Barros, and H. de Lencastre. 1999. Detection of an archaic clone of Staphylococcus aureus with low-level resistance to methicillin in a pediatric hospital in Portugal and in international samples: relics of a formerly widely disseminated strain? J. Clin. Microbiol. 37:1913-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tenover, F. C., R. Arbeit, G. Archer, J. Biddle, S. Byrne, R. Goering, G. Hancock, G. A. Hebert, B. Hill, R. Hollis, W. R. Jarvis, B. Kreiswirth, W. Eisner, J. Maslow, L. K. McDougal, J. M. Miller, M. Mulligan, and M. A. Pfaller. 1994. Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J. Clin. Microbiol. 32:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Leeuwen, W. B., C. Vant Pelt, A. D. Luijensijk, H. A. Verbrugh, and W. H. Goessens. 1999. Rapid detection of methicillin resistance in Staphylococcus aureus isolates by the MRSA-Screen latex agglutination test. J. Clin. Microbiol. 37:3029-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagenvoort, J. H. T., H. M. J. Toenbreker, M. E. O. C. Heck, W. J. van Leeuwen, and W. J. B. Wannet. 2000. Hospital outbreak of methicillin-resistant Staphylococcus aureus followed by an in vivo change to a mecA-negative mutant with loss of epidemicity. Eur. J. Clin. Microbiol. Infect. Dis. 19:976-977. [DOI] [PubMed] [Google Scholar]