Abstract
An H1N2 influenza A virus was isolated from a pig in the United States for the first time in 1999 (A. I. Karasin, G. A. Anderson, and C. W. Olsen, J. Clin. Microbiol. 38:2453-2456, 2000). H1N2 viruses have been isolated subsequently from pigs in many states. Phylogenetic analyses of eight such viruses isolated from pigs in Indiana, Illinois, Minnesota, Ohio, Iowa, and North Carolina during 2000 to 2001 showed that these viruses are all of the same reassortant genotype as that of the initial H1N2 isolate from 1999.
H3N2 influenza A viruses have caused widespread outbreaks of respiratory disease and, in some cases, abortion among pigs throughout the major swine-producing regions of the United States since 1998. The most common genotype of H3N2 virus isolated to date is a triple reassortant containing hemagglutinin (HA), neuraminidase (NA), and PB1 polymerase genes of human influenza virus origin, matrix (M), nonstructural (NS), and nucleoprotein (NP) genes of classical swine influenza virus origin, and PA and PB2 polymerase genes of avian influenza virus origin (8, 15, 17). The emergence of these viruses was remarkable because influenza among American pigs had historically been due almost exclusively to infection with classical H1N1 swine influenza viruses (3, 5, 11). Subsequent to the appearance of these H3N2 viruses, an H1N2 virus (A/Swine/Indiana/9K035/99 [Sw/IN/99]) was isolated from a pig in Indiana during a 6-week-long outbreak of influenza-like illness and abortions on the farm of origin in November 1999. Phylogenetic analyses demonstrated that Sw/IN/99 had resulted from reassortment between a classical H1N1 swine influenza virus, which supplied its HA gene segment, and one of the triple-reassortant swine H3N2 viruses, which supplied all of the remaining RNA segments (7).
The Sw/IN/99 virus was the first H1N2 influenza virus isolated from a pig in the United States. However, H1N2 viruses were isolated previously from pigs in France in 1987 and 1988 (4), in Japan in 1978 to 1980 and 1989 to 1992 (6, 9, 10, 12, 13, 16), and in the United Kingdom in 1994 and thereafter (1, 2). The viruses from Japan and France were shown to be products of reassortment between classical (Japan) or avian-like (France) swine H1N1 viruses and human-lineage H3N2 viruses (4, 6, 9, 10, 12, 13, 16). In contrast, the H1N2 viruses in the United Kingdom resulted from multiple reassortment events. Reassortment between human H1N1 and H3N2 viruses is hypothesized to have initially created an H1N2 subtype virus, which subsequently acquired all six of its internal viral protein genes through reassortment with a wholly avian virus (2). The H1N2 viruses in France did not spread beyond their initial farms of origin (4), but the H1N2 viruses in Japan and the United Kingdom caused large-scale outbreaks of disease and spread widely in the swine populations of these countries (1, 2, 6, 12). Most recently it has been shown that H1N2 viruses with HA proteins antigenically related to those of the H1N2 viruses from the United Kingdom are circulating among pigs in Belgium (14).
In this report, we present evidence that H1N2 viruses of the same genotype as that of the initially isolated Sw/IN/99 virus have been isolated from pigs in at least six states in the United States, suggesting that H1N2 viruses are now cocirculating, along with both classical H1N1 and triple-reassortant H3N2 viruses, in the American swine population.
Clinical histories and virus isolation.
Brief clinical histories and isolation information for the viruses analyzed in this study are presented in Table 1. All of the viruses were isolated in mammalian cell culture from either Madin-Darby canine kidney (MDCK) cells or porcine embryonic testicle cells (PT-1; American BioResearch, Sevierville, Tenn.). Following initial isolation, the viruses were passaged twice in MDCK cells to obtain sufficient virus stocks for analyses. Clinical histories and additional diagnostic results differed among the isolates, reflecting the fact that these cases were investigated by different clinical veterinarians and different regional diagnostic laboratories. Nonetheless, it can be seen that the H1N2 viruses were isolated in association with respiratory disease in pigs across a wide range of ages, from 2-week-old nursery pigs through 7-month-old finisher pigs. Many of the infected pigs were, however, also harboring additional infectious agents that may have contributed to the clinical signs observed. It will, therefore, be necessary to conduct experimental infections under controlled conditions to define the pathogenic potential of the H1N2 viruses. Finally, it is of interest to note that, unlike the initially reported Sw/IN/99 H1N2 virus (7), the H1N2 virus infections described herein were not associated with abortions.
TABLE 1.
Brief clinical histories and virus isolation methods for the swine H1N2 viruses analyzed in this study
| Virus | Clinical history | Virus isolation and propagation methods | GenBank reference no. |
|---|---|---|---|
| A/Swine/Minnesota/55551/00 | Unthriftiness and inconsistent growth rates among nursery and grower pigs (∼2-10 wk old); concurrent A. pp. infectionb; PCV2c and PRRSVd positive and M. hyo.e negative | MDCK cell culture isolation from lung tissue from nursery pigs and MDCK cell propagation with TPCK-trypsina | AF455678, AF455686, AF455694, AF455702, AF455710, AF455718, AF455726, AF455734 |
| A/Swine/Indiana/P12439/00 | Acute respiratory disease with coughing and lethargy among 30 of 300 grower pigs (∼9-10 wk old) with 2 deaths; M. hyo. positiveg and PRRSV negative | PT-1 cellsf isolation from pooled lung tissues and MDCK cell propagation with TPCK-trypsin | AF455680, AF455688, AF455696, AF455704, AF455712, AF455720, AF455728, AF455736 |
| A/Swine/Ohio/891/01 | Prolonged outbreak (6 months) of disease on a farm with 700 sows weaning ∼300 piglets/week; high fevers, coughing and diarrheal disease (100% morbidity), and enhanced mortality in pigs 3 to 6 wk old | MDCK cell culture isolation from lung tissue and MDCK cell propagation with TPCK-trypsin | AF455675, AF455683, AF455691, AF455699, AF455707, AF455715, AF455723, AF455731 |
| A/Swine/North Carolina/93523/01 | Acute respiratory disease, increased (twofold) mortality, and reduced rates of weight gain in nursery to grower pigs (∼8 wk old) | MDCK cell culture isolation from lung tissue and MDCK cell propagation with TPCK-trypsin | AF455677, AF455685, AF455693, AF455701, AF455709, AF455717, AF455725, AF455733 |
| A/Swine/North Carolina/98225/01 | Acute respiratory disease and enhanced (0.5-1.0% increase) mortality among grower to finisher pigs (∼12 wk old) | MDCK cell culture isolation from lung tissue and MDCK cell propagation with TPCK-trypsin | AF455676, AF455684, AF455692, AF455700, AF455708, AF455716, AF455724, AF455732 |
| A/Swine/Illinois/100084/01 | Acute respiratory disease with coughing among 5 of 75 finisher pigs (∼7 mo old), with 2 deaths | MDCK cell culture isolation from lung tissue and MDCK cell propagation with TPCK-trypsin | AF455682, AF455690, AF455698, AF455706, AF455714, AF455722, AF455730, AF455738 |
| A/Swine/Illinois/100085a/01 | Coughing of several weeks duration (no improvement with oral antibiotics) among 200 of 1000 young grower pigs (∼5 wk old) | MDCK cell culture isolation from lung tissue and MDCK cell propagation with TPCK-trypsin | AF455681, AF455689, AF455697, AF455705, AF455713, AF455721, AF455729, AF455737 |
| A/Swine/Iowa/930/01 | Sneezing and coughing, progressing to severe dyspnea among 700 of 700 9-12-wk-old pigs | MDCK cell culture isolation from lung tissue and MDCK cell propagation with TPCK-trypsin | AF455679, AF455687, AF455695, AF455703, AF455711, AF455719, AF455727, AF455735 |
MDCK, Madin-Darby canine kidney cells; TPCK, tosylsulfonyl phenylalanyl chloromethyl ketone-treated trypsin.
Actinobacillus pleuropneumoniae (A. pp.) by bacterial culture of lung tissue.
Porcine circovirus type 2 (PCV2) by PCR assay on lung tissue.
Porcine reproductive and respiratory syndrome virus (PRRSV) by virus isolation on MARC-145 cells.
Mycoplasma hyopneumoniae (M. hyo.) by PCR assay on lung tissue.
Porcine embryonic testicle cells (American BioResearch).
Mycoplasma hyopneumoniae by fluorescent antibody assay on lung tissue.
Genetic and phylogenetic analyses.
To allow determination of the subtype and overall genotype of each virus, the full-length protein coding region sequences of all eight viral RNA segments were determined by cycle sequencing (ABI Big Dye; PE Applied Biosystems, Foster City, Calif.), following amplification by reverse transcription-PCR. Amplifications were conducted using avian myeloblastosis virus reverse transcriptase (Promega Corporation, Madison, Wis.) and Pfu polymerase (Stratagene, La Jolla, Calif.) as previously described (7). The subtypes and genotypes of the viruses were then defined by pairwise comparisons of each gene segment to the sequences of reference influenza viruses available in GenBank using DNASTAR software (version 5.0 for Windows 32).
The reverse transcription-PCRs for the HA and NA genes amplified only H1 and N2 subtype sequences from each virus and, specifically, no H3 or N1 sequences could be recovered. Thus, each virus was of the H1N2 subtype and there was no evidence for mixtures of viruses. For each gene segment, the viruses were highly related to one another and to the original (7) Sw/IN/99 isolate (Table 2). Thus, all of the H1N2 viruses isolated in 2000 to 2001 are of the same overall genotype as that of the original Sw/IN/99 H1N2 virus (7), which includes HA, NP, M, and NS genes of the classical swine influenza virus lineage, NA and PB1 polymerase genes of the human influenza virus lineage, and PA and PB2 polymerase genes of the North American avian influenza virus lineage.
TABLE 2.
Sequence homologies among the swine H1N2 viruses analyzed in this studya
| Gene segment | % Nucleotide sequence identity |
|---|---|
| PB2 | 97.3-99.3 |
| PB1 | 98.0-99.7 |
| PA | 96.9-99.1 |
| HA | 91.9-99.2 |
| NP | 96.3-99.7 |
| NA | 96.7-99.6 |
| M | 96.1-99.8 |
| NS | 97.4-99.6 |
The viruses included in these analyses were the eight swine H1N2 viruses newly described in this paper as well as the original swine H1N2 virus described previously (7).
The phylogenetic relationships among the H1N2 viruses in this study, the original Sw/IN/99 H1N2 virus isolate, and selected reference strains were estimated from the nucleotide sequences of each RNA segment by the method of maximum parsimony (PAUP software, v.4.0b6; David Swofford, Smithsonian Institution) by using a heuristic search algorithm with the MULTREES option in effect. Confidence levels for the phylogram topologies were determined by bootstrap analysis with 500 replications. To optimize the validity of the phylograms produced, potential reference viruses for which only short sequence fragments were available were not included. Thus, the HA phylogram was based upon full-length HA1 segment reference virus sequences. Likewise, all of the reference virus sequences included in the analyses of the NA, NP, M, NS, PA, PB1, and PB2 gene segments were at least 50% of full length, and the vast majority were full length. The GenBank accession numbers for the reference virus sequences presented in Fig. 1 to 3 are listed in Table 3.
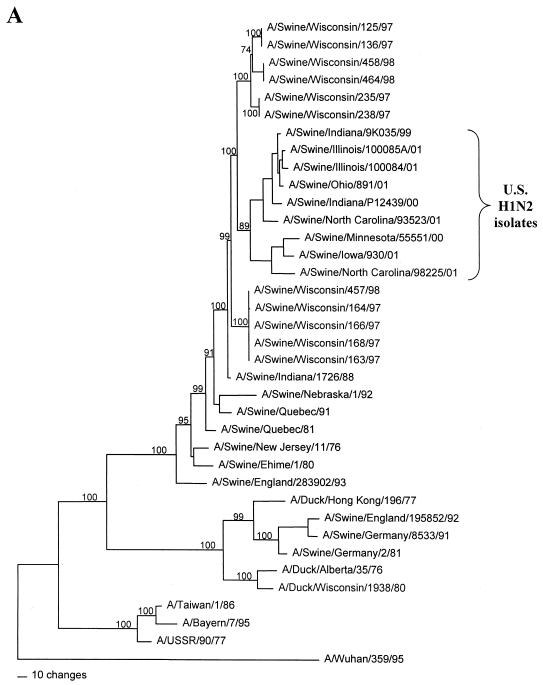
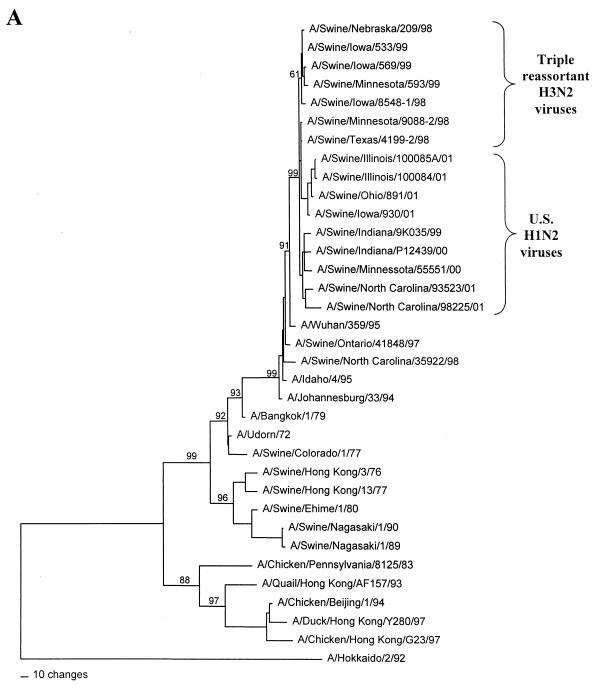
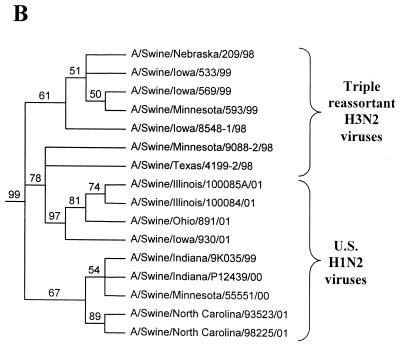
FIG. 1.
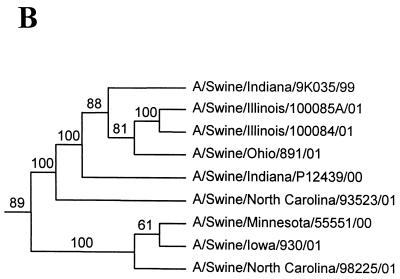
Nucleotide phylograms for the HA genes of swine H1N2 and reference influenza viruses. The evolutionary relationships among these viruses were estimated by the method of maximum parsimony (PAUP software, v.4.0b6; David Swofford, Smithsonian Institution) by using a heuristic search algorithm with the MULTREES option in effect. The numbers at the nodes of the phylograms represent confidence levels for the phylogram topologies as determined by bootstrap analysis with 500 replications. (A) Overall phylogram. Horizontal-line distances are proportional to the minimum numbers of nucleotide changes needed to join nodes and gene sequences. The vertical lines are present simply for spacing the branches and labels. The complete phylogram shown represents the best (score = 1,523) of 64,280 rearrangements generated. (B) Detailed depiction of only the H1N2 viruses isolated recently from pigs in the United States in order to demonstrate the bootstrap values within this cluster of viruses.
FIG. 3.
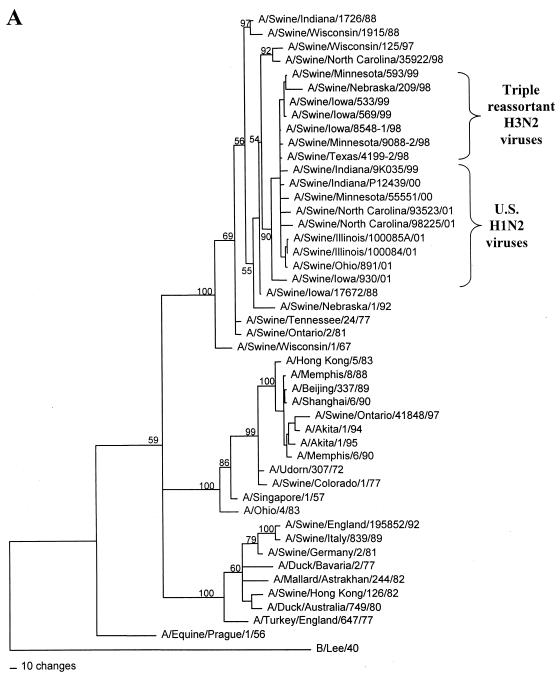
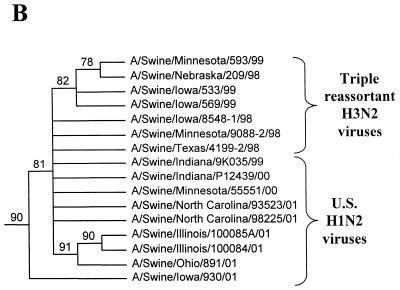
Nucleotide phylograms for the NP genes of swine H1N2 and reference influenza viruses. The evolutionary relationships among these viruses were estimated by the method of maximum parsimony (PAUP software, v.4.0b6; David Swofford, Smithsonian Institution), using a heuristic search algorithm with the MULTREES option in effect. The numbers at the nodes of the phylograms represent confidence levels for the phylogram topologies as determined by bootstrap analysis with 500 replications. (A) Overall phylogram. Horizontal-line distances are proportional to the minimum numbers of nucleotide changes needed to join nodes and gene sequences. The vertical lines are present simply for spacing the branches and labels. The phylogram shown represents the best (score = 2,011) of 2,685,101 rearrangements generated. (B) Detailed depiction of only the H1N2 and triple-reassortant H3N2 viruses isolated recently from pigs in the United States in order to demonstrate the bootstrap values within this cluster of viruses.
TABLE 3.
GenBank accession numbers for the reference virus sequences included in the phylogenetic analyses presented in Figs. 1 to 3
| Virus | HA gene | NA gene | NP gene | Virus | HA gene | NA gene | NP gene | |
|---|---|---|---|---|---|---|---|---|
| A/Swine/Wisconsin/125/97 (H1N1) | AF222026 | AF222768 | ||||||
| A/Swine/Wisconsin/136/97 (H1N1) | AF222027 | |||||||
| A/Swine/Wisconsin/163/97 (H1N1) | AF222028 | |||||||
| A/Swine/Wisconsin/164/97 (H1N1) | AF222029 | |||||||
| A/Swine/Wisconsin/166/97 (H1N1) | AF222030 | |||||||
| A/Swine/Wisconsin/168/97 (H1N1) | AF222031 | |||||||
| A/Swine/Wisconsin/235/97 (H1N1) | AF222032 | |||||||
| A/Swine/Wisconsin/238/97 (H1N1) | AF222033 | |||||||
| A/Swine/Wisconsin/457/98 (H1N1) | AF222034 | |||||||
| A/Swine/Wisconsin/458/98 (H1N1) | AF222035 | |||||||
| A/Swine/Wisconsin/464/98 (H1N1) | AF222036 | |||||||
| A/Swine/Indiana/1726/88 (H1N1) | M81707 | L46849 | ||||||
| A/Swine/Indiana/9K035/99 (H1N2) | AF250124 | AF205126 | AF250127 | |||||
| A/Swine/Nebraska/1/92 (H1N1) | L09063 | L11164 | ||||||
| A/Swine/Quebec/91 (H1N1) | U03719 | |||||||
| A/Swine/Quebec/81 (H1N1) | I03720 | |||||||
| A/Swine/New Jersey/11/76 (H1N1) | K00992 | |||||||
| A/Swine/Ehime/1/80 (H1N2) | X57494 | D00713 | ||||||
| A/Swine/England/283902/93 (H1N1) | U72668 | |||||||
| A/Taiwan/1/86 (H1N1) | D00407 | |||||||
| A/Bayern/7/95 (H1N1) | —a | |||||||
| A/USSR/90/77 (H1N1) | K01330 | |||||||
| A/Duck/Hong Kong/196/77 (H1) | D00839 | |||||||
| A/Swine/England/195852/92 (H1N1) | U72667 | L40332 | ||||||
| A/Swine/Germany/8533/91 (H1N1) | Z46434 | |||||||
| A/Swine/Germany/2/81 (H1N1) | Z30276 | M22579 | ||||||
| A/Duck/Alberta/35/76 (H1N1) | D10477 | |||||||
| A/Duck/Wisconsin/1938/80 (H1N1) | L25071 | |||||||
| A/Wuhan/359/95 (H3N2) | AF008722 | U51246 | ||||||
| A/Swine/Nebraska/209/98 (H3N2) | AF251404 | AF251407 | ||||||
| A/Swine/Iowa/533/99 (H3N2) | AF251412 | AF251415 | ||||||
| A/Swine/Iowa/569/99 (H3N2) | AF251420 | AF251423 | ||||||
| A/Swine/Minnesota/593/99 (H3N2) | AF251428 | AF251431 | ||||||
| A/Swine/Iowa/8548-1/98 (H3N2) | AF153239 | AF153255 | ||||||
| A/Swine/Minnesota/9088-2/98 (H3N2) | AF153238 | AF153254 | ||||||
| A/Swine/Texas/4199-2/98 (H3N2) | AF153237 | AF153253 | ||||||
| A/Swine/Ontario/41848/97 (H3N2) | AF251396 | AF251399 | ||||||
| A/Swine/North Carolina/35922/98 (H3N2) | AF153236 | AF153252 | ||||||
| A/Idaho/4/95 (H3N2) | U51247 | |||||||
| A/Johannesburg/33/94 (H3N2) | U43425 | |||||||
| A/Bangkok/1/79 (H3N2) | K01150 | |||||||
| A/Udorn/72 (H3N2) | J02168 | |||||||
| A/Swine/Colorado/1/77 (H3N2) | AF251389 | AF251392 | ||||||
| A/Swine/Hong Kong/3/76 (H3N2) | D00715 | |||||||
| A/Swine/Hong Kong/13/77 (H3N2) | D21190 | |||||||
| A/Swine/Nagasaki/1/90 (H1N2) | D29659 | |||||||
| A/Swine/Nagasaki/1/89 (H1N2) | D29658 | |||||||
| A/Chicken/Pennsylvania/8125/83 (H5N2) | M11925 | |||||||
| A/Quail/Hong Kong/AF157/93 (H9N2) | AF156399 | |||||||
| A/Chicken/Beijing/1/94 (H9N2) | AF156398 | |||||||
| A/Duck/Hong Kong/Y280/97 (H9N2) | AF156394 | |||||||
| A/Chicken/Hong Kong/G23/97 (H9N2) | AF156392 | |||||||
| A/Hokkaido/2/92 (H1N1) | AF250126 | |||||||
| A/Swine/Wisconsin/1915/88 (H1N1) | M76608 | |||||||
| A/Swine/Iowa/17672/88 (H1N1) | M63768 | |||||||
| A/Swine/Tennessee/24/77 (H1N1) | M30748 | |||||||
| A/Swine/Ontario/2/81 (H1N1) | M63767 | |||||||
| A/Swine/Wisconsin/1/67 (H1N1) | M76607 | |||||||
| A/Hong Kong/5/83 (H3N2) | M22577 | |||||||
| A/Memphis/8/88 (H3N2) | L07370 | |||||||
| A/Beijing/337/89 (H3N2) | L07374 | |||||||
| A/Shanghai/6/90 (H3N2) | L07357 | |||||||
| A/Akita/1/94 (H3N2) | U71144 | |||||||
| A/Akita/1/95 (H3N2) | U71145 | |||||||
| A/Memphis/6/90 (H3N2) | L07356 | |||||||
| A/Udorn/307/72 (H3N2) | M14922 | |||||||
| A/Singapore/1/57 (H2N2) | M63752 | |||||||
| A/Ohio/4/83 (H1N1) | M59334 | |||||||
| A/Swine/Italy/839/89 (H1N1) | M63772 | |||||||
| A/Duck/Bavaria/2/77 (H1N1) | M22574 | |||||||
| A/Mallard/Astrakhan/244/82 (H14N6) | M30764 | |||||||
| A/Swine/Hong Kong/126/82 (H3N2) | M63771 | |||||||
| A/Duck/Australia/749/80 (H1N1) | M63783 | |||||||
| A/Turkey/England/647/77 (H1N1) | M76603 | |||||||
| A/Equine/Prague/1/56 (H7N7) | M63748 | |||||||
| B/Lee/40 | K01395 |
Sequence kindly provided by the Influenza Virus Branch of the Centers for Disease Control and Prevention.
In the HA gene, the H1N2 viruses isolated in 2000 to 2001 and the original Sw/IN/99 H1N2 virus form a distinct phylogenetic clade with two major branches (Fig. 1). These HA genes are all most closely related to classical swine H1N1 viruses isolated in 1997 to 1998. The two branches within the H1N2 clade suggest that these viruses may have been derived as the result of two separate reassortment events involving different classical swine H1 viruses. However, it will be necessary to examine a larger number of H1N2 viruses over a longer time period to confirm this possibility. In the remaining gene segments, the H1N2 viruses isolated in 2000 to 2001 and the original Sw/IN/99 H1N2 virus demonstrate very close phylogenetic relationships with each other and the triple-reassortant H3N2 influenza viruses from pigs. In the NA and M genes, the H1N2 viruses form definably separate branches from the H3N2 viruses. As an example of this pattern, the NA phylogram is shown in Fig. 2. As in the HA phylogram, there appear to be two clusters of H1N2 viruses on separate branches of the NA phylogram, but this was not the case for any of the other gene segments. In the NP, NS, PB1, PB2, and PA genes, because of the very low overall levels of genetic differences between the H1N2 and H3N2 viruses, it is not possible to define distinctly separate H1N2 and H3N2 lineages. As an example of this pattern, the NP phylogram is shown in Fig. 3.
FIG. 2.
Nucleotide phylograms for the NA genes of swine H1N2 and reference influenza viruses. The evolutionary relationships among these viruses were estimated by the method of maximum parsimony (PAUP software, v.4.0b6; David Swofford, Smithsonian Institution) by using a heuristic search algorithm with the MULTREES option in effect. The numbers at the nodes of the phylograms represent confidence levels for the phylogram topologies as determined by bootstrap analysis with 500 replications. (A) Overall phylogram. Horizontal-line distances are proportional to the minimum numbers of nucleotide changes needed to join nodes and gene sequences. The vertical lines are present simply for spacing the branches and labels. The complete phylogram shown represents the best (score = 1,970) of 1,009,555 rearrangements generated. (B) Detailed depiction of only the H1N2 and triple-reassortant H3N2 viruses isolated recently from pigs in the United States in order to demonstrate the bootstrap values within this cluster of viruses.
Our results indicate that, since the initial isolation of an H1N2 virus from pigs in 1999 (7), H1N2 viruses of the same overall genotype have spread widely within the swine population of the United States. The emergence of these viruses presents two important issues. First, if the overall epidemiologic pattern of swine influenza in the coming years is to be defined, it will be important to subtype all viruses as to both HA and NA subtypes. Otherwise, it will not be possible to distinguish H1N1 from H1N2 viruses (and possibly H3N1 from H3N2 viruses in the future). It will also be important to determine experimentally whether vaccination of pigs with the available H1N1 and H3N2 swine influenza virus vaccines will provide protection against H1N2 virus challenge. Logically this should be the case, but, to our knowledge, this has yet to be proven experimentally.
Nucleotide sequence accession numbers.
The GenBank numbers assigned to the gene sequences determined in this study are listed in Table 1.
Acknowledgments
This work was supported by funding from the USDA National Research Initiative Competitive Grants Program and a grant from the University of Wisconsin School of Veterinary Medicine Food Animal Grant Program.
We thank Gabrielle Landolt from the University of Wisconsin—Madison for reviewing the manuscript and for many helpful discussions.
REFERENCES
- 1.Brown, I. H., P. Chakraverty, P. A. Harris, and D. J. Alexander. 1995. Disease outbreaks in pigs in Great Britain due to an influenza A virus of H1N2 subtype. Vet. Rec. 136:328-329. [DOI] [PubMed] [Google Scholar]
- 2.Brown, I. H., P. A. Harris, J. W. McCauley, and D. J. Alexander. 1998. Multiple genetic reassortment of avian and human influenza A viruses in European pigs, resulting in emergence of an H1N2 virus of novel genotype. J. Gen. Virol. 79:2947-2955. [DOI] [PubMed] [Google Scholar]
- 3.Chambers, T. M., V. S. Hinshaw, Y. Kawaoka, B. C. Easterday, and R. G. Webster. 1991. Influenza viral infection of swine in the United States 1988-1989. Arch. Virol. 116:261-265. [DOI] [PubMed] [Google Scholar]
- 4.Gourreau, J. M., C. Kaiser, M. Valette, A. R. Douglas, J. Labie, and M. Aymard. 1994. Isolation of two H1N2 influenza viruses from swine in France. Arch. Virol. 135:365-382. [DOI] [PubMed] [Google Scholar]
- 5.Hinshaw, V. S., W. J. Bean, Jr., R. G. Webster, and B. C. Easterday. 1978. The prevalence of influenza viruses in swine and the antigenic and genetic relatedness of influenza viruses from man and swine. Virology 84:51-62. [DOI] [PubMed] [Google Scholar]
- 6.Ito, T., Y. Kawaoka, A. Vines, H. Ishikawa, T. Asai, and H. Kida. 1998. Continued circulation of reassortant H1N2 influenza viruses in pigs in Japan. Arch. Virol. 143:1773-1782. [DOI] [PubMed] [Google Scholar]
- 7.Karasin, A. I., G. A. Anderson, and C. W. Olsen. 2000. Genetic characterization of an H1N2 influenza virus isolated from a pig in Indiana. J. Clin. Microbiol. 38:2453-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karasin, A. I., M. M. Schutten, L. A. Cooper, C. B. Smith, K. Subbarao, G. A. Anderson, S. Carman, and C. W. Olsen. 2000. Genetic characterization of H3N2 influenza viruses isolated from pigs in North America, 1977-1999: evidence for wholly human and reassortant virus genotypes. Virus Res. 68:71-85. [DOI] [PubMed] [Google Scholar]
- 9.Nerome, K., M. Ishida, A. Oya, and K. Oda. 1982. The possible origin of H1N1 (Hsw1N1) virus in the swine population of Japan and antigenic analysis of the isolates. J. Gen. Virol. 62:171-175. [DOI] [PubMed] [Google Scholar]
- 10.Nerome, K., Y. Yoshioka, S. Sakamoto, M. Yasuhara, and A. Oya. 1985. Characterization of a 1980-swine recombinant influenza virus possessing H1 haemagglutinin and N2 neuraminidase similar to that of the earliest Hong Kong (H3N2) virus. Arch. Virol. 86:197-211. [DOI] [PubMed] [Google Scholar]
- 11.Olsen, C. W., S. Carey, L. Hinshaw, and A. I. Karasin. 2000. Virologic and serologic surveillance for human, swine and avian influenza virus infections among pigs in the north-central United States. Arch. Virol. 145:1399-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ouchi, A., K. Nerome, Y. Kanegae, M. Ishida, R. Nerome, K. Hayashi, T. Hashimoto, M. Kaji, Y. Kaji, and Y. Inaba. 1996. Large outbreak of swine influenza in southern Japan caused by reassortant (H1N2) influenza viruses: its epizootic background and characterization of the causative viruses. J. Gen. Virol. 77:1751-1759. [DOI] [PubMed] [Google Scholar]
- 13.Sugimura, T., H. Yonemochi, T. Ogawa, K. Tanaka, and T. Kumagai. 1980. Isolation of a recombinant influenza virus (Hsw1N2) from swine in Japan. Arch. Virol. 66:271-274. [DOI] [PubMed] [Google Scholar]
- 14.VanReeth, K., I. H. Brown, and M. Pensaert. 2000. Isolations of H1N2 influenza A virus from pigs in Belgium. Vet. Rec. 146:588-589. [DOI] [PubMed] [Google Scholar]
- 15.Webby, R. J., S. L. Swenson, S. L. Krauss, P. J. Gerrish, S. M. Goyal, and R. G. Webster. 2000. Evolution of swine H3N2 influenza viruses in the United States. J. Virol. 74:8243-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yasuhara, M., T. Hirahara, M. Nakai, N. Sasaki, J. Kata, T. Watanabe, and M. Morikawa. 1983. Further isolation of a recombinant virus (H1N2), formerly (Hsw1N2), from a pig in Japan in 1980. Microbiol. Immunol. 27:43-50. [DOI] [PubMed] [Google Scholar]
- 17.Zhou, N. N., D. A. Senne, J. S. Landgraf, S. L. Swenson, G. Erickson, K. Rossow, L. Liu, K.-J. Yoon, S. Krauss, and R. G. Webster. 1999. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J. Virol. 73:8851-8856. [DOI] [PMC free article] [PubMed] [Google Scholar]