Abstract
Of 115 methicillin-resistant Staphylococcus strains collected from sputum specimens, 34 strains reduced susceptibility to vancomycin, 9 of which emerged as heterogeneous vancomycin-resistant strains (hetero-VRS), with various degrees of vancomycin resistance at a frequency of 10−6 or higher. Seventy-six percent (19 of 25) of non-hetero-VRS and 100% (9 of 9) of hetero-VRS were susceptible to synergistic treatment with vancomycin and imipenem. Clinical clearance between 9 hetero-VRS and 25 non-hetero-VRS had an obvious statistical significance (P = 0.001). The hetero-VRS may play an important role in vancomycin therapy failure.
Vancomycin has been a successful treatment for methicillin-resistant Staphylococcus aureus (MRSA) infections in China since the 1980s. The mortality rate of MRSA infections is still high. It reached 43.75% in 1998 in the Guangzhou district of China (18). In Japan, therapeutic failure occurred in 21.3% of cases, and 35.8% of patients continued to develop MRSA infections in the lower respiratory tract after administration of vancomycin. After the first report on heterogeneous vancomycin-resistant Staphylococcus aureus (hetero-VRSA) Mu3 in Japan, heterogeneous vancomycin-resistant strains of Staphylococcus (hetero-VRS) were also successively found in the United States, France, and Italy (1, 2, 14, 15). In China, VRS were not found in the national survey of 1998, and whether there were hetero-VRS strains is unknown. Infections with methicillin-resistant Staphylococcus strains (MRS) were often complicated by the presence of multiple pathogens (17, 18). Therefore, it is necessary to know what broad-spectrum antibiotics, mainly β-lactams, can be used in combination with vancomycin to treat complex infections. Based on the antagonism of vancomycin and β-lactams against hetero-VRSA (5, 6, 7), this will raise a question of how to treat complex pathogens once nosocomial hetero-VRSA infections occur. Recently it was reported that no antagonisms between vancomycin and β-lactams against hetero-VRSA exist (2, 3, 16). Since most of the complex pathogens, such as Pseudomonas aeruginosa, are resistant to many antibiotics and only respond to imipenem, it is very important to understand whether imipenem and vancomycin in combination has final synergism against hetero-VRSA or hetero-VRS.
A total of 115 MRS were clinically isolated from the sputum specimens of patients suffering from lower respiratory tract infections from January 1997 to February 1999. MRS were screened according to the method recommended by the National Committee for Clinical Laboratory Standards (MRSA for which the oxacillin MIC was 4 mg/liter or more and methicillin-resistant coagulase-negative staphylococci for which the oxacillin MIC was 0.5 mg/liter or more on Mueller-Hinton agar [MHA]) (12). Heterogenously resistant bacteria are subpopulations of bacteria with various degrees of vancomycin resistance, demonstrating natural heterogeneity or variability in susceptibility to vancomycin; therefore, a resistant subclone may be selected on the medium with vancomycin. Hetero-VRS was defined as its resistant subclone at a frequency of 10−6 colonies or higher (8). The screening of hetero-VRS was as follows. Overnight cultures of bacteria in tryptic soy broth (Difco) were adjusted to a McFarland standard of 0.5 (about 108 CFU/ml). The bacterial suspension (10 μl) was inoculated onto a brain heart infusion agar (BHIA; Difco) plate containing 4 mg of vancomycin (Sigma Co.) per liter and then was incubated at 37°C for 48 h. Hetero-VRS was confirmed if the strain produced a subclone with a vancomycin MIC of 8 mg/liter or greater on the BHIA plate and had stable resistance to vancomycin for more than 9 days in a drug-free medium (8). Bacterial subclones can grow on BHIA containing more than 16 mg of vancomycin/liter at subculture. Vancomycin and imipenem susceptibilities were tested on MHA and BHIA to detect the effects of different nutrient media on Staphylococcus resistance. The plates were supplemented with vancomycin concentrations in the range of 1 to 32 mg/liter and imipenem concentrations in the range of 1 to 256 mg/liter (double dilution). The MHA and BHIA plates were inoculated with 10 μl of a suspension of the testing isolate equivalent to 108 CFU/liter. Test plates were incubated in ambient air at 35°C and were observed after 24 and 48 h. Vancomycin MIC breakpoints were no more than 4 mg/liter (susceptible), 8 to 16 mg/liter (intermediate), and 32 mg/liter or more (resistant), and imipenem MIC breakpoints were no more than 4 mg/liter (susceptible), 8 mg/liter (intermediate), and 16 mg/liter or more (resistant) in reference to National Committee for Clinical Laboratory Standards guidelines (12). The resistant subpopulation analysis of hetero-VRS was done by spreading 50 μl of the initial cell suspension and its serial 10-fold dilutions over BHIA plates containing increments (2 mg/liter) of vancomycin, and suspensions were incubated at 37°C for 48 h. The number of surviving bacteria were counted and demonstrated on a semilogarithmic graph. Vancomycin Etest (AB Biodisk, Solna, Sweden) directly detected the hetero-VRS susceptibilities in the course of subcultures. The fraction of inhibited concentrations (FIC) of vancomycin and imipenem (Merck Sharp & Dohme Ltd.) were determined on BHIA by a checkerboard method. First, the MICs of the two drugs alone for the 34 strains of Staphylococcus were determined by a multiloop inoculator. Fifteen times the highest MIC of each drug was selected for the final concentration of the initial solution, and the concentration was doubled before combination with other solutions. The MICs of the drug solutions alone or in combination were tested by the double dilution method. Procedures for the drug combination were as follows: 18 sterile large test tubes were evenly divided into two rows; 6 ml of saline was put into each tube and 6 ml of the initial drug solution was sucked into the first tube in each row, and then serial double dilution was performed; 121 plates were lined up like a checkerboard, with 11 plates in each column and each row; 6 ml of the drug dilution solution from the first tube in the first row was evenly put into every plate (about 0.5 ml) in the first row and was analogized in order, with the last row of free drug as the control; the other drug was likewise put into every plate (0.5 ml) in the column. The best concentration of drug A or B was expressed as MICA or MICB, respectively. Every tube along an angle bisector from the zero point was equivalent to the center tube. MICA and MICB were read on the axes of X and Y corresponding to the tube with the best MIC. FICA is the MIC of drug A combined with drug B divided by the MIC of drug A alone, and FICB is the MIC of drug B combined with drug A divided by the MIC of drug B alone. The FIC index is FICA plus FICB. Drug A and drug B combined have a synergism with an FIC index of no more than 0.5, an additive index of 0.5 to 1.0, indifference of 1.0 to 2.0, and antagonism of more than 2.0. Staphylococcus species were identified by using the API Staph system (Biomerieux, Marcy l'Etiole, France) based on the manufacture's guidelines. ATCC 29213 acted as a quality control.
Of the 115 MRS, 29.57% (34 of 115) had reduced susceptibility to vancomycin on BHIA. They showed higher resistance to imipenem (MICs of 8 to 128 mg/liter) and to vancomycin (MICs of 8 to 32 mg/liter) on BHIA than on MHA (imipenem MICs from 4 to 64 mg/liter and vancomycin MICs from 4 to 16 mg/liter) (Table 1).
TABLE 1.
General characteristics of 34 Staphylococcus strains with reduced susceptibility to vancomycin
| Strain no. | Bacterium | Underlying disease or conditiona | MIC (mg/liter) of:
|
|||
|---|---|---|---|---|---|---|
| Vancomycinb
|
Imipenem
|
|||||
| MHA | BHIA | MHA | BHIA | |||
| 2 | S. haemolyticus | COPD | 8 (8) | 32 (24) | 64 | 128 |
| 3 | S. aureus | COPD | 8 | 16 | 16 | 64 |
| 4 | S. aureus | CPHD | 4 | 16 | 32 | 64 |
| 5 | S. hominis | Viral hepatitis B | 4 | 8 | 4 | 8 |
| 6 | S. epidermidis | Uremia | 4 | 16 | 4 | 8 |
| 8 | S. aureus | COPD | 8 | 16 | 32 | 64 |
| 9 | S. sciuri | Bronchiectasis | 4 | 16 | 16 | 64 |
| 10 | S. sciuri | CHD | 8 | 16 | 64 | 128 |
| 11 | S. sciuri | COPD | 4 | 8 | 8 | 16 |
| 12 | S. aureus | COPD | 8 | 8 | 64 | 128 |
| 13 | S. haemolyticus | COPD | 16 (16) | 32 (32) | 8 | 32 |
| 19 | S. aureus | CPHD | 4 | 8 | 32 | 64 |
| 16 | S. aureus | IPF | 8 | 16 | 64 | 128 |
| 18 | S. aureus | Lung cancer | 4 | 8 | 32 | 64 |
| 24 | S. hominis | Uremia | 8 | 16 | 16 | 64 |
| 29 | S. hominis | Cirrhosis of the liver | 4 | 8 | 64 | 128 |
| 31 | S. haemolyticus | Viral hepatitis B | 8 (12) | 32 (32) | 16 | 64 |
| 36 | S. sciuri | Viral hepatitis B | 4 | 8 | 32 | 64 |
| 60 | S. haemolyticus | Chronic respiratory failure | 8 (16) | 32 (48) | 16 | 64 |
| 61 | S. aureus | COPD | 8 | 8 | 64 | 64 |
| 62 | S. chromogenes | Diabetes mellitus | 8 | 16 | 8 | 16 |
| 63 | S. aureus | Acute renal failure | 8 | 16 | 4 | 16 |
| 70 | S. aureus | CHD | 4 | 8 | 64 | 128 |
| 71 | S. aureus | AMI | 16 | 16 | 64 | 128 |
| 73 | S. aureus | Uremia | 8 (24) | 32 (96) | 16 | 64 |
| 94 | S. haemolyticus | Diabetes mellitus | 8 | 16 | 4 | 8 |
| 96 | S. sciuri | Stroke | 4 | 16 | 32 | 64 |
| 97 | S. aureus | Uremia | 8 (32) | 32 (64) | 4 | 8 |
| 99 | S. epidermidis | COPD | 4 | 8 | 4 | 16 |
| 102 | S. aureus | Viral hepatitis B | 16 (24) | 32 (96) | 4 | 8 |
| 108 | S. haemolyticus | COPD | 8 (16) | 32 (32) | 16 | 64 |
| 109 | S. haemolyticus | COPD | 16 (32) | 32 (96) | 8 | 64 |
| 112 | S. sciuri | Diabetes mellitus | 4 | 8 | 32 | 64 |
| 113 | S. sciuri | CHD | 8 | 16 | 8 | 8 |
| ATCC 29213 | 2 | 2 | 0.06 | 0.06 | ||
COPD, chronic obstructive pulmonary disease; CPHD, chronic pulmonary heart disease; IPF, interstitial pulmonary fibrosis; CHD, coronary heart disease; AMI, acute myocardial infarction.
Numbers in parentheses are Etest results.
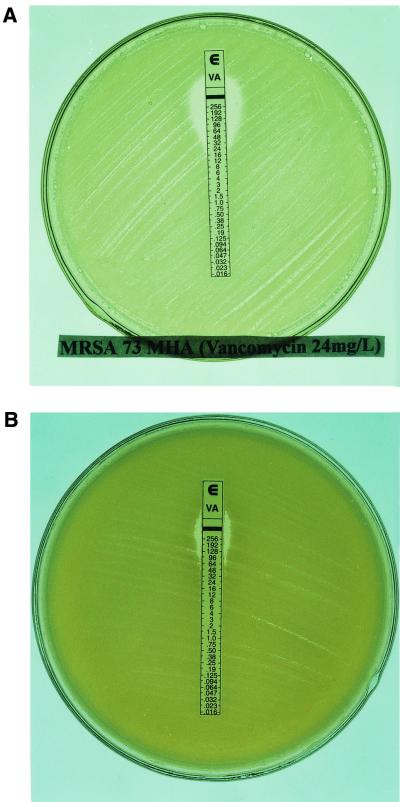
Of 34 Staphylococcus strains with reduced susceptibility to vancomycin, 9 strains picked up from the BHIA plate with vancomycin (MIC of 16 mg/liter) showed highly heterogeneous resistance to vancomycin on BHIA or MHA by Etest at subculture (Table 1 and Fig. 1).
FIG. 1.
MRS 73 picked from the BHIA plate containing 16 mg of vancomycin/liter showed high heterogeneous resistance to vancomycin, with a vancomycin MIC of 24 mg/liter on MHA (A) and 96 mg/liter on BHIA (B). The vancomycin MIC for ATCC 29213 was 2 mg/liter.
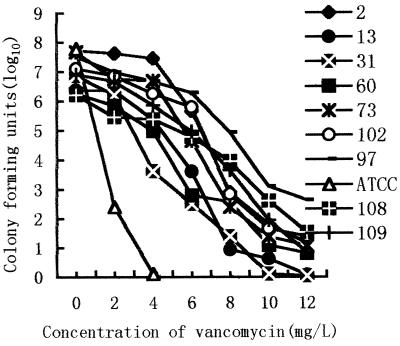
Subpopulation analysis of VRS showed that 9 strains of hetero-VRS from the colony in the BHIA plate containing 8 mg of vancomycin/liter indicated various resistances to vancomycin. Seven strains of Staphylococcus, except the 13th and 31st strains, still grew on the BHIA plates with 10 mg of vancomycin/liter. However, all of them expressed medium-level resistance to vancomycin (MIC ≥8 mg/liter) at a frequency of 10−3 to 10−6 (Fig. 2).
FIG. 2.
Profile of a vancomycin-resistant subpopulation of methicillin-resistant staphylococci. MRS 2, 13, 108, 60, 31, and 109 were S. haemolyticus, and MRS 73, 97, and 102 were S. aureus. These strains were picked from the colony on the BHIA plate containing 8 mg of vancomycin/liter. ATCC 29213 (ATCC) was used as a control.
The separate imipenem and vancomycin MICs at which 90% of the isolates tested are inhibited were 64 and 8 times as high as the MIC when the drugs were combined, respectively. The two drugs in combination against Staphylococcus strains had synergism, with an FIC index of 0.047 to 0.281 (82.35%; 28 of 34 strains), additive of 0.508 to 0.516 (11.76%; 4 of 34), and indifference of 1.016 to 1.016 (5.88%; 2 of 34) (Table 2). Vancomycin and imipenem combined had better synergism against hetero-VRS (100%; 9 of 9) (FIC index, 0.047 to 0.281) than against non-hetero-VRS (76%; 19 of 25) (FIC index, 0.141 to 0.313).
TABLE 2.
MIC and FIC (mg/liter) of vancomycin and imipenem alone or in combination against 34 Staphylococcus strains
| Antibiotics | MIC and FIC
|
||
|---|---|---|---|
| Range | 50% | 90% | |
| Vancomycin (alone) | 4-32 | 16 | 32 |
| Vancomycin (with imipenem) | 0.25-4 | 4 | 4 |
| Imipenem (alone) | 4-128 | 64 | 128 |
| Imipenem (with vancomycin) | 0.5-4 | 1 | 2 |
Of a total of 34 strains of staphylococci with reduced susceptibility to vancomycin, 26 were cleared after treatment by vancomycin (total clearance, 76.5% [26 of 34]). Three out of 9 strains of hetero-VRS which showed high-level vancomycin resistance at subculture (vancomycin MIC of more than 32 μg/ml on BHIA) were cleared (clearance, 33.3% [3 of 9]). Twenty-three of 25 Staphylococcus strains for which the vancomycin MICs were no more than 8 or 16 μg/ml at subculture were cleared (clearance, 92% [23 of 25]). The hetero-VRS were cleared with more difficulty than non-hetero-VRS (P = 0.001). It is well known that MRS have heterogeneous resistance to β-lactams. In this study, population analysis of 9 strains of hetero-VRS (6 strains of Staphylococcus haemolyticus and 3 strains of S. aureus) showed that the majority of these bacteria could grow on BHIA containing vancomycin concentrations of 8 to 12 mg per liter at a frequency of 10−3 to 10−6. Heterogeneous response to glycopeptides appeared to be a common feature of S. haemolyticus (1). Of 9 Staphylococcus strains, 6 strains of S. haemolyticus had highly heterogeneous resistance to vancomycin, and this suggests that S. haemolyticus may more likely be heterogeneously resistant than S. aureus. The drug MICs for precursors of 8 in 9 hetero-resistant staphylococci were within 4 to 8 mg per liter more than the 3 mg per liter of strain Mu3, and exceptionally, that of strain 97 was 2 mg per liter. The subcultures of these bacteria may extend to above 96 mg per liter. The progenitor of hetero-VRS may emerge heterogeneously resistant to low levels of vancomycin. Conventional MIC tests will not be predictive of the in vivo therapeutic effect of vancomycin. Brilliant nutrient BHI media can promote the growth of MRS, especially hetero-VRS. Vancomycin MICs for hetero-VRS were increased two- to fourfold. For example, the vancomycin MIC for MRSA 73 on the BHIA plate was 96 mg per liter higher than that on the MHA plate (24 mg per liter).
The vancomycin Etest strip may induce hetero-VRS on BHIA plates during subcultures and may directly indicate the MIC. We found that the hetero-VRS produced a double inhibitory zone around vancomycin Etest strips at subculture for more than 48 h. The bacteria within interior zones started to grow after about 24 h with heterogeneous resistance to vancomycin (unpublished observations). Up to now, little has been known about the mechanism of heterogeneously resistant staphylococci. One of the mechanisms may be associated with heterogeneous-to-homogeneous conversion of methicillin resistance (10). The response regulator vraR has recently been thought to be one of the key regulators modulating the level of vancomycin resistance in S. aureus (11). Although prophylactic vancomycin may reduce the morbidity of potential MRSA nosocomial infections (13), it is prudent that vancomycin is widely used as an empirical therapy in MRSA infections at the expense of homogeneous resistance of staphylococci to vancomycin (4, 9).
Acknowledgments
We thank W. Peng and C. Chen for providing English language editing and Y. Xi for identification of vancomycin-resistant staphylococci.
This work was supported by the Roche Research Fund for Infectious Diseases for Young Doctors from Roche (China) Pharmaceuticals Company Limited.
REFERENCES
- 1.Biavasco, F., C. Vignardi, R. Lazzarini, P. E. Varaldo. 2000. Glycopeptide susceptibility profiles of Staphylococcus haemolyticus bloodstream isolates. Antimicrob. Agents Chemother. 44:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chesneau, O., A. Morvan, and N. E. Solh. 2000. Retrospective screening for heterogeneous vancomycin resistance in diverse Staphylococcus aureus clones disseminated in French hospitals. J. Antimicrob. Chemother. 45:887-890. [DOI] [PubMed] [Google Scholar]
- 3.Climo, M. W., R. L. Patron, and G. C. Archer. 1999. Combinations of vancomycin and β-lactams are synergistic against staphylococci with reduced susceptibilities to vancomycin. Antimicrob. Agents Chemother. 43:1747-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dagan, O., P. N. Cox, L. Ford-Jones, J. Ponsonby, and D. J. Bohn. 1999. Nosocomial infection following cardiovascular surgery comparison of two periods, 1987 vs 1992. Crit. Care Med. 27:104-108. [DOI] [PubMed] [Google Scholar]
- 5.Hanaki, H., and K. Hiramatsu. 1999. Combination effect of teicoplanin and various antibiotics against hetero-VRSA. Kansenshogaku Zasshi 73:1048-1053. [DOI] [PubMed] [Google Scholar]
- 6.Hanaki, H., Y. Inaba, K. Sasaki, and K. Hiramatsu. 1998. A novel method of detecting Staphylococcus aureus heterogeneously resistant to vancomycin (hetero-VRSA). Jpn. J. Antibiot. 530:51-58. [PubMed] [Google Scholar]
- 7.Haraga, I., S. Nomura, and A. Nagayama. 1999. The effects of vancomycin and beta-lactam antibiotics on vancomycin-resistant staphylococcus aureus. N. Engl. J. Med. 341:1624.. [DOI] [PubMed] [Google Scholar]
- 8.Hiramatsu, K., N. Aritaka, H. Hanaki, S. Kawasaki, Y. Hoaoda, S. Hori, Y. Fukachi, and I. Kobayashi. 1997. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350:1670-1673. [DOI] [PubMed] [Google Scholar]
- 9.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-136. [DOI] [PubMed] [Google Scholar]
- 10.Hiramatsu, K. 1995. Molecular evolution of MRSA. Microbiol. Immunol. 39:531-543. [DOI] [PubMed] [Google Scholar]
- 11.Kurodam, K., A. Kuwahara, and K. Hiramatsu. 2000. Identification of the up-and-down-regulated genes in vancomycin-resistant staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem. Biophys. Res. Commun. 269:485-490. [DOI] [PubMed] [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. 1999. Minimum inhibitory concentration interpretive standards (μg/ml) for Staphylococcus spp. M7-A4 (M100-S9). National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 13.Roghmann, M. C. 2000. Predicting methicillin resistance and the effect of inadequate empiric therapy on survival in patients with Staphylococcus aureus bacteremia. Arch. Intern. Med. 160:1001-1004. [DOI] [PubMed] [Google Scholar]
- 14.Shimada, K., H. Obayashi, K. Sunagawa, T. Inamatsu, and K. Yamagachi. 1995. Clinical summary of intravenous use of vancomycin hydrochloride for severe infection caused by methicillin-resistant Staphylococcus aureus. Jpn. J. Chemother. 43:1048-1061. [Google Scholar]
- 15.Sieradzki, K., P. Villari, and A. Tomasz. 1998. Decreased susceptibility to teicoplanin and vancomycin among coagulase-negative methicillin-resistant clinical isolates of staphylococci. Antimicrob. Agents Chemother. 42:100-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siereszki, K., R. B. Roberts, S. W. Haber, and A. Tomasz. 1999. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N. Engl. J. Med. 340:1624.. [DOI] [PubMed] [Google Scholar]
- 17.Takeda, S., K. Kono, and K. Arakawa. 1997. Relation between candidiasis and nutrition of patients and MRSA infection. Kansenshogaku Zasshi 71:899-902. [DOI] [PubMed] [Google Scholar]
- 18.Wu, B., Y. Tang, and J. Zhu. 2000. High risk factors lead to nosocomial pulmonary infections caused by MRSA. Chin. J. Tuber. Respir. Dis. 23:413-416. [PubMed] [Google Scholar]