Abstract
In contrast to cervical cancer, integration of human papillomavirus (HPV) DNA into the host genome has been considered a rare event in cancer precursor lesions (cervical intraepithelial neoplasia [CIN]). With our new real-time PCR method, we demonstrated that integrated HPV type 16 (HPV16) is already present in CIN lesions. The physical state of HPV16 and the viral load were simultaneously detected. A unique region of the E2 open reading frame (ORF) that is most often deleted during HPV16 integration is targeted by one set of PCR primers and a probe, and another set targets the E6 ORF. In episomal form, both targets should be equivalent, while in integrated form, the copy numbers of E2 would be less than those of E6. The method was tested with DNAs from 31 cervical lesions (non-CIN to CINIII) from 24 women prospectively followed up for 10 years. This report presents viral load and integration results from the largest series of CIN lesions described to date. Only one sample contained exclusively episomal HPV16 DNA, and this lesion regressed spontaneously. Samples from another patient, with only integrated HPV16, rapidly progressed from CINI to CINIII in 2 years. In all other patients, episomal and integrated forms of HPV16 DNA were found to coexist. Rapid progression of the CIN lesions was closely associated with a heavy load of integrated HPV16. Thus, the method described here is a very sensitive tool with which to assess the physical state of HPV, which is useful in predicting disease progression.
Human papillomaviruses (HPVs) comprise more than 120 putative virus types, of which 85 types have been fully sequenced, and approximately 40 types are associated with lesions of the anogenital tract. Infection with high-risk (oncogenic) types of HPV (HPV16 [HPV type 16], -18, -31, -33, -35, -39, -45, -52, -56, -58, and -68) is a well-established risk factor for the development of cervical carcinoma (34, 42), which is the second most common female malignancy worldwide. The most common oncogenic HPV type in cervical cancer is HPV16, which is detectable in more than 50% of the cases (2). Premalignant cervical lesions are commonly staged according to increasing severity, as cervical intraepithelial neoplasia I (CINI), CINII, and CINIII, where CINIII represents severe dysplasia or cancer in situ. NCIN here denotes HPV-infected lesions without signs of neoplasia. Previous studies suggested that benign HPV lesions and low-grade intraepithelial lesions (CINI) mostly contain the viral sequences only as episomes (4, 13, 15, 30, 32, 41). In contrast, viral DNA is integrated into the host genome in virtually all cases of cervical carcinomas and their derivate cell lines (3, 6-9, 14, 22, 36). No previous studies are available in which the physical state and viral load of HPV in premalignant cervical lesions (CIN) have been assessed with new sensitive and quantitative methods such as real-time PCR.
Viral DNA integration into host cell DNA usually disrupts the E1 and E2 open reading frames (ORFs). In contrast, the E6 and E7 ORFs and LCR (long control region) generally remain intact (7, 22, 28, 34, 43). Kalantari and coworkers mapped deletions and disruptions of the E1and E2 ORFs in a large series of HPV16-positive carcinomas and found multiple patterns of deletions, the most common being in the region of the E2 ORF corresponding to the protein “hinge” region (18). A refined method was recently devised with which to assay for integration against a background of episomal HPV based on restriction enzyme cleavage, religation, and inverse PCR (rliPCR) (19).
Elimination of HPV16 E2 protein expression due to integration results in up-regulation of the transcription of the E6 and E7 oncogenes, possibly providing a selective growth advantage for the infected cell (15, 16). Continuous expression of these oncogenic proteins is most likely required for maintenance of the malignant state (12, 16, 20, 27, 28). The E6 and E7 oncoproteins interfere with the normal cell cycle by targeting the p53 and pRb tumor suppressor proteins, respectively (11, 38). In addition to integration, heavy viral loads in CIN lesions have recently been shown to constitute an at least 60-fold increased risk of carcinoma development in situ (17).
We developed a novel real-time PCR method that we used in this study to provide data indicating that HPV16 is integrated at early stages of cervical carcinogenesis, i.e., in low-grade CIN lesions. With our new technique, we were able to calculate viral copy numbers for both episomal and integrated forms separately. In a series of 31 cases of HPV16-positive CIN lesions from women included in the Finnish prospective cohort study (1981 to 1998), integration of HPV16 was present in all of the cases except one. Progression of CIN lesions was closely associated with heavy loads of integrated HPV16.
MATERIALS AND METHODS
Samples.
The SiHa cell line was used to establish the method. This cell line contains one or two copies of HPV16, which are integrated into chromosome 13q14-32 (10). The clinical material used in the present study consisted of 31 cervical biopsy specimens (4 NCIN, 4 CINI, 10 CINII, and 13 CINIII lesions) derived from 24 women included in the Kuopio cohort study of 1981 to 1998 (34). HPV16 positivity was confirmed by amplification of the E6, E7, E1, and E2 ORFs and the LCR (21). There were two patient groups; one consisted of 12 followed-up patients who were not treated for their cervical disease unless the lesion progressed to CINIII (designated F in Table 2 and Fig. 1), and the other consisted of 9 patients treated for their cervical disease with cryosurgery, conization, or interferon (designated T in Table 2 and Fig. 1). However, the biopsy samples available for this study were always taken before treatment was begun. Three patients in this study were included in the double-blind, placebo-controlled interferon trial and treated with vaginal cream containing leukocyte alpha interferon at 1.5 × 106 IU/g or a placebo (40). Patients T2b-88 and T3-87 belonged to the placebo group.
TABLE 2.
Physical state and viral load of HPV16 in relation to clinical data of cervical preneoplastic lesions
| Patient no. | Sample | Viral load
|
Integration
|
Clinical data
|
||||
|---|---|---|---|---|---|---|---|---|
| E6/50 ng of DNA | E6/SiHa E6 | E2/E6 | E2/integrated E6 | Status | Lesion grade | Clinical outcome | ||
| 1 | F2-85 | 3045992 | 18 | 0.1 | 0.11 | Mixed | CINIII | Progression to CINIII |
| 2 | F3-85 | 45453 | 0.3 | 0.41 | 0.7 | Mixed | NCIN | Regression to normalcy |
| 3 | F4-85 | 22584 | 0.1 | 0.79 | 3.7 | Mixed | CINIII | Progression to CINIII |
| 4 | F5-85 | 1353360 | 7.8 | 0.24 | 0.31 | Mixed | CINII | Regression to normalcy |
| 5 | F6A-91 | 76812 | 0.4 | 0.48 | 0.93 | Mixed | CINII | Progression to CINIII |
| F6B-91 | 166598 | 1 | 0.55 | 1.21 | Mixed | CINIII | ||
| 6 | F7A-85 | 151010 | 0.9 | 0.37 | 0.68 | Mixed | NCIN | Progression to CINIII |
| F7B-85 | 1474 | 0.009 | 0.23 | 0.3 | Mixed | CINI | ||
| F7C-86 | 390265 | 2.3 | 0.34 | 0.52 | Mixed | CINII | ||
| F7D-86 | 699006 | 4 | 0.41 | 0.7 | Mixed | CINII | ||
| F7E-87 | 315849 | 1.8 | 0.34 | 0.51 | Mixed | CINIII | ||
| 7 | F8-85 | 5159863 | 30 | 0.48 | 0.91 | Mixed | CINII | Progression to CINIII |
| 8 | F9-86 | 273723 | 1.6 | 0.48 | 0.93 | Mixed | CINIII | Progression to CINIII |
| 9 | F10-85 | 28754 | 0.2 | 0.38 | 0.62 | Mixed | CINIII | Progression to CINIII |
| 10 | F11b-92 | 1030368 | 6 | 0.46 | 0.85 | Mixed | CINIII | Progression to CINIII |
| 11 | F12A-86 | 86330 | 0.5 | No E2 | No E2 | Integrated | CINII | Progression to CINIII |
| F12B-86 | 94690 | 0.6 | No E2 | No E2 | Integrated | CINII | ||
| F12C-87 | 95856 | 0.6 | No E2 | No E2 | Integrated | CINIII | ||
| 12 | F13-85 | 23352 | 0.1 | 0.8 | 14.03 | Mixed | NCIN | Regression to normalcy |
| 13 | F14-85 | 73936 | 0.4 | 1.02 | No integration | Episomal | CINII | Regression to normalcy |
| 14 | F15-87 | 6764 | 0.04 | 0.34 | 0.51 | Mixed | NCIN | Regression to normalcy |
| 15 | F16-87 | 32130 | 0.2 | 0.79 | 3.83 | Mixed | CINI | Persistence |
| 16 | T1-86 | 25056 | 0.2 | 0.56 | 1.28 | Mixed | CINI | Cure by cryotherapy |
| 17 | T2b-88 | 1355901 | 7.8 | 0.56 | 1.28 | Mixed | CINII | Progression to CINIII, curea |
| 18 | T3-87 | 4687719 | 27 | 0.38 | 0.62 | Mixed | CINII | Progression to CINIII, curea |
| 19 | T4-87 | 276756 | 1.6 | 0.43 | 0.77 | Mixed | CINI | Cure by cryotherapy |
| 20 | T5-87 | 41152 | 0.2 | 0.14 | 0.17 | Mixed | CINIII | Progression CINIII, curea |
| 21 | T6-87 | 1031560 | 6 | 0.79 | 3.84 | Mixed | CINIII | Progression to CINIII, curea |
| 22 | T9-89 | 2497333 | 14 | 0.39 | 0.64 | Mixed | CINIII | Progression to CINIII, curea |
| 23 | T11-90 | 10621 | 0.06 | 0.69 | 2.26 | Mixed | CINIII | Progression to CINIII, curea |
| 24 | T12-90 | 3462483 | 20 | 0.77 | 3.43 | Mixed | CINII | Cure by interferon |
| SiHa | 172991 | 1 | No E2 | No E2 | Integrated | |||
After conization the patient remained disease free.
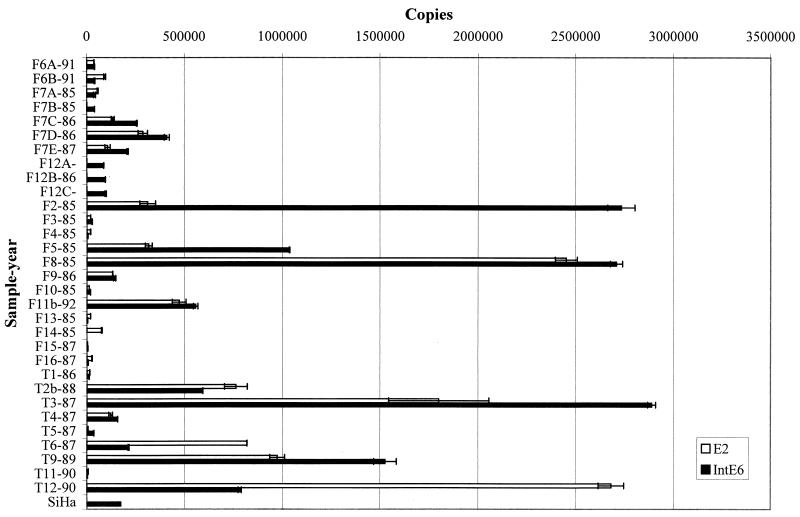
FIG. 1.
Copy numbers of HPV16 E2 and E6 in DNA from clinical samples and the SiHa cell line. The integrated copy numbers (IntE6) were calculated by subtraction of the number of E2 (episomal) copies from the total number of E6 (episomal and integrated) copies. The sample names consist of a unique identifier number preceded by an F for follow-up and T for treated patients plus the sampling year after a hyphen. All biopsy samples were taken before treatment.
DNA extraction.
DNA was extracted with the Miller high-salt method as described by Kurvinen et al. (21). Samples were lysed in 1 ml of 10 mM Tris (pH 8.3)-400 mM NaCl-1% sodium dodecyl sulfate-2 mM EDTA-proteinase K at 300 μg/ml overnight at 37°C. Subsequent protein precipitation was carried out by adding 300 μl of saturated NaCl. After centrifugation, the supernatant was removed and DNA was precipitated with ice-cold absolute ethanol. The DNA pellet was dissolved in sterile water and kept frozen at −70°C.
Real-time PCR.
Real-time PCR was performed with the ABI Prism 7700 Sequence Detection System and the TaqMan Universal PCR Master Mix (PE Applied Biosystems, Perkin-Elmer). The amplification conditions were 2 min at 50°C, 10 min at 95°C, and a two-step cycle of 95°C for 15 s and 60°C for 60 s for a total of 40 cycles. The primers and probe (Table 1) were designed with the aid of the Primer Express program, 1.0b6 (PE Applied Biosystems), for specific amplification of the E2 and E6 ORFs in this study (see Fig. 1 and Table 1). The sizes of the E2 and E6 amplimers were 76 and 81 bp, respectively. The E6 probe was labeled with 6-carboxyfluorescein at the 5′ end and Dark Quencher (Scandinavian Gene Synthesis AB, Koping, Sweden) at the 3′ end. The E2 probe was labeled with BODIPY R6G (the BODIPY dyes used are under license to Scandinavian Gene Synthesis from Molecular Probes Inc.) at the 5′ end and Dark Quencher at the 3′ end. The final primer and probe concentrations, in a total volume of 50 μl, were 0.3 and 0.1 μM, respectively. Fifty nanograms of target DNA from biopsy specimens was added to the reaction mixture. Two standard curves were obtained by amplification of a dilution series of 50 million to 500 copies of a clone of HPV16 in pBR322 (kindly provided by H. zur Hausen, Cancer Research Center, Heidelberg, Germany). There was a linear relationship between the threshold cycle values plotted against the log of the copy number over the entire range of dilutions (data not shown). At least three no-template control reaction mixtures were included in each run. All experiments were performed twice in duplicate with similar ratios.
TABLE 1.
Primers and probes used for real-time PCRa
| Physical state | Name | Sequence (5′→3′) | Tm (°C) | Amplimer length (bp) |
|---|---|---|---|---|
| Episomal | Probe 16E2PRO | (BODIPYR6G)-CACCCCGCCGCGACCCATA-(DQ) | 70 | |
| Primer 1, 16E2F | AACGAAGTATCCTCTCCTGAAATTATTAG | 59 | ||
| Primer 2, 16E2R | CCAAGGCGACGGCTTTG | 60 | 82 | |
| Integrated + episomal | Probe 16E6PRO | (6-FAM)-CAGGAGCGACCCAGAAAGTTACCACAGTT-(DQ) | 69 | |
| Primer 1, 16E6F | GAGAACTGCAATGTTTCAGGACC | 59 | ||
| Primer 2, 16E6R | TGTATAGTTGTTTGCAGCTCTGTGC | 60 | 81 |
For the episomal state, E2 primers and a probe for the episomal viral load were used. For the integrated and episomal states together, E6 primers and a probe for the total viral load were used. F, forward primer; R, reverse primer; DQ, Dark Quencher; 6-FAM, 6-carboxyfluorescein. The melting temperature (Tm) was determined with the Primer Express program, 1.0b6 (PE Biosystems).
The results were recorded as copy numbers in 50 ng of cellular DNA. The integrated E6 was calculated by subtracting the copy numbers of E2 (episomal) from the total copy numbers of E6 (episomal and integrated). Ratios of E2 to E6 of less than 1 indicate the presence of both integrated and episomal forms. The ratio of E2 to integrated E6 represents the amount of the episomal form in relation to the integrated form. Values of greater than 1 indicate predominance of the episomal form. The relative viral load can be estimated by calculating the ratio of copies of E6 to SiHa cell E6.
RESULTS
Development of the new real-time PCR method.
In the present study, a new real-time PCR (TaqMan) method was developed. The method is based on measurement of the absolute values of the E2 and E6 ORFs in HPV16-positive DNA samples. The positions of the designed E6 and E2 primers and probes are shown in Table 1. E2 primers and probe locations were selected to recognize the E2 hinge region, which is the part of the E2 ORF that is most often deleted upon HPV16 viral integration into the host genome in cervical carcinomas (18).
HPV DNA analysis.
We reanalyzed the physical status of HPV16 in 31 biopsies derived from different grades of cervical cancer precursor lesions (NCIN to CINIII) by using our new real-time PCR method. HPV16 had been previously detected in these biopsies by PCR amplification using primer pairs targeting the E6 and E7 regions of the HPV16 genome; also, the LCR from all of the samples used in this study was previously amplified and sequenced. The E1 ORF was amplified in all samples, whereas no E2 amplification was detected in three samples obtained from the same women, indicating complete viral integration into the host genome (21).
The results of the physical state and the copy numbers of HPV16 E2 and E6 are summarized in Table 2 and Fig. 1. The absolute copy numbers of E2 and E6 in 50 ng of sample DNA are shown. Integrated E6 was calculated by subtracting the values of E2 from those of E6. The viral load per 50 ng of DNA from different samples varied over the very wide range (1 × 103 to 5 × 106 copies). The real-time PCR method is characterized by a dynamic range of several orders of magnitude, which is suitable for such variability.
The episomal form results in remission.
When only the episomal form is present, equivalent copy numbers of both E2 and E6 should be detected. This was found in one sample (F14-85) only. This CINII lesion with only the episomal form regressed to normalcy in 9 years and was the first of the four (F14-86, CINI; F14-87, NCIN; F14-94, normal) follow-up samples from this woman. The relative viral load in the biopsy sample was 40% of that found in 50 ng of SiHa cells (one or two copies per cell).
The integrated form predisposes to rapid progression.
Another patient, whose lesion progressed from CINI to CINIII during the follow-up, showed only integrated HPV16 in her three consecutive samples diagnosed as CINII to CINIII (Table 2 and Fig. 1; samples F12A-86, F12B-86, and F12C-87). Thus, no E2 amplification was detected in any of these samples. The viral load remained at the same level in all of the samples and was 50 to 60% of that in SiHa cells. This lesion progressed from CINI to CINIII in 3 years.
Coexistent episomal and integrated forms of HPV16 are most frequent.
The remaining 22 patients, excluding the 2 described above, had both integrated and episomal forms of HPV DNA in their biopsy samples. Three patients had cervical HPV infections with no CIN lesion (HPV-NCIN), which regressed spontaneously in 2 to 10 years of follow-up (F3-85, F13-85, and F15-87). In all of these samples, a light viral load was detected, with total E6 absolute copy numbers of 45,453, 23,352, and 6,764, respectively. In comparison, the absolute SiHa E6 copy number was 172,991. The episomal form was predominant in one of these samples (F13-85), while two others had more integrated than episomal HPV DNA. Interestingly, spontaneous regression from CINII was found in one patient (F5-85), despite the heavy viral load and predominance of the integrated form of HPV16 DNA.
Two patients showed a rapid progression from an NCIN lesion (F2-85) or CINII (F8-85) to CINIII in 1 year. High copy numbers of HPV16 were detected in both. In F2-85, the copy numbers were 18 times that in SiHa cells and in F8-85, they were 30 times that in SiHa cells. In F2-85, the E2/E6 ratio was low (0.1), indicating a high degree of integration (Table 2 and Fig. 1). In F8-85, there were as many integrated copies as episomal ones.
In addition to the patient with only the integrated form (F12), there were two others with several follow-up samples available. From patient F6-91, two samples, one from a CINII lesion and the other from a CINIII lesion, were taken at 6-month intervals. Five years earlier, the lesion was HPV-NCIN, progressing finally to CINIII in 5 years from the first diagnosis. We found that in 6 months, the viral load more than doubled and reached the same level as in SiHa cells. The copy numbers of the integrated and episomal forms were nearly equivalent. Five samples were available from the patient F7. A CINII lesion was diagnosed 1 year before the first biopsy was taken for this study, and the disease progressed to CINIII in 11 years. In the lesion without CIN (HPV-NCIN), equal amounts of the integrated and episomal forms were detected and the copy numbers were equivalent to that in SiHa cells. In the next biopsy with CINI, only few copies were present, with predominance of the integrated form. From the CINII lesion onward, 10- to 20-times heavier viral loads were detected, along with predominance of integrated form.
Patients T1-86 and T4-87 were treated with cryotherapy, and both were cured. All CINIII lesions were treated with conization. In these patients, a variation of the viral load from 0.06 to 20 times the level in SiHa cells was disclosed (Table 2). Two further patients, who belonged to the interferon trial but were treated with placebo cream (T2b-88 and T3-87) showed rapid progression from CINII to CINIII. The third patient (T12-90) was treated with interferon, and she was cured and remained disease free during the 5-year follow-up. Prior to treatment, the lesion contained both episomal and integrated forms of HPV DNA, with a heavy viral load (20 times the level in SiHa cells) and 3.4 times more episomal than integrated HPV DNA.
DISCUSSION
In this study, we have established a new quantitative real-time PCR (TaqMan) method by which to detect the physical state of HPV16 DNA in clinical samples. This new technique can also be used to determine the total viral load in these samples. It is known that CIN lesions and cervical cancer can be attributed largely to HPV infections. To support their own persistence and replication, high-risk HPVs interfere with normal cellular control mechanisms, leading to abnormal growth, genetic alterations, and transformation. The most important viral oncogenes involved in cellular immortalization are E6 and E7. Continuous expression of E6 and E7 is required to induce and maintain the neoplastic phenotype of cervical cancer cells (37). Integration of HPV into the human genome usually disrupts the E2 ORF of the HPV genome, and this may cause the up-regulation of E6 and E7 (31). E2 controls E6 and E7 expression by binding to and repressing the viral promoter directing expression of E6 and E7 (14). It was also recently shown that expression of exogenous E2 results in cellular growth arrest and cellular senescence (39). Integration of HPV16 DNA into the human genome also results in increased stability of the E6 and E7 mRNAs (16) and correlates with a selective growth advantage of cells (15). Thus, detection of integrated HPV might be a promising marker for cervical disease at progression. As far as we are aware, the present study presents the largest series of cervical precancerous lesions in which both the viral load and the physical state of HPV16 have been characterized.
The method described here is very sensitive, and we can find low copy numbers of integrated HPV DNA even in the presence of excess episomal DNA. The remarkable range of copy numbers obtained in this study and calculated for each sample per 50 ng of input DNA is in a good agreement with the data of Swan and coworkers (33).
In some previous studies, in which Southern blotting, PCR, and two-dimensional electrophoresis have been used, high-level episomal forms can mask the presence of low-level integrated HPV forms (5, 6, 26). This new method is also simpler to perform than the rliPCR procedure recently described by Kalantari et al. (19). A minor disadvantage of the present method is that only a small, although representative, region of E2 is assayed for deletion. This is a region deleted in HPV16 in SiHa cells and was also deleted in a majority of cervical cancer samples analyzed by rliPCR (19).
In order to minimize the material and labor costs, we also tested a multiplex configuration that amplifies the E6 and E2 targets in the same tube. In the preliminary experiments, the multiplexing was functional but the data obtained were less reliable than those obtained when the reaction was run in two separate tubes. Therefore, only the two-tube results are presented here. In the future, an optimized multiplex configuration currently under testing in our laboratory could include a real-time PCR assay of a frequently deleted region of E1.
Even the early studies in the mid-1980's and early 1990's (1, 6, 23, 25, 32) on the physical stage of HPV in cervical disease suggested that the integration of HPV16 already occurs in the precancerous stage, and both episomal and integrated forms were detected simultaneously (23, 32). During the past 10 years, however, integration has been generally regarded as an event that occurs at the invasive cancer stage (42). Our data further support the view that integration of HPV already occurs at an early stage of the disease. Indeed, only one sample (F14-85) contained exclusively the episomal form of HPV16. As could be expected, this CINII lesion regressed to normalcy during the follow-up and the relative viral load was half of that found in 50 ng of SiHa cell DNA (one or two HPV16 copies per cell). A rapid progression from NCIN and CINII lesions to CINIII was found in two patients, F2 and F8, respectively. In both patients, both the integrated and episomal forms of HPV16 were found along with a heavy viral load. Interestingly, the HPV16 found in F2 was of the Asian-American type, which is known to be associated with more aggressive behavior (21, 35).
The data presented here would fit into the speculative model of temporal relationships in HPV-induced carcinogenesis presented in Fig. 2. In this model, we emphasize that initial heavy viral loads will increase the probability of integration into chromosomal sites favorably selected to gain a growth advantage. This will explain the high frequency of mixed episomal and integrated forms of the viral DNA, as these precancerous cases could still be in the selection phase. This is further supported by the experiments done with the cell lines established from HPV-infected premalignant genital lesions (15, 29; Peitsaro et al., personal communication). These studies indicate that the integrated form was already present in the original premalignant biopsy sample but the episomal form predominated. Similar patterns of integrated and episomal forms were also detected in early passages of the cultured cells. In one study, Jeon and coworkers (15) were unable to detect differences in the cellular properties of integrated cell populations over extrachromosomal cell populations, other than the capacity for the integrated cell populations to outgrow the extrachromosomal cell populations. The two cell populations were indistinguishable in resistance to the induction of terminal differentiation.
FIG. 2.
Hypothetical temporal multistep events in cervical carcinogenesis. The model explains the frequent occurrence of mixed episomal and integrated forms of HPV DNA in cancer precursors found in this study. These cases will be predicted to be still at the stage at which there is selection for integrated cell clones. In bold text are all of the factors identified as important predictors of cancer progression.
The hypothesis of HPV-induced carcinogenesis presented here is further supported by the model outlined for hamster polyomavirus-induced lymphomas (24). This hypothesis underlines two essential prerequisites for hamster polyomavirus to become lymphomagenous: (i) suppression of the late coding functions of the viral genome and (ii) expression of the viral oncogenes above a threshold level. The amount of viral early RNAs yielded by a single integrated copy was shown to be very similar to that associated with several thousand extrachromosomal copies of the viral genome.
To conclude, we present here a novel method that detects both the viral load and the physical state of HPV16 simultaneously. By using sequential biopsies from prospectively followed-up patients, we were able to show that integration of HPV can be detected in CIN lesions and even in those containing HPV but no CIN. The mixed forms were most prevalent in precancerous lesions. A rapid progression, in 1 to 2 years, from non-CIN lesions or CINII to CINIII was associated with a heavy load of integrated HPV. Thus, simultaneous identification of integrated HPV and measurement of the viral load provides a new potential prognostic tool with which to estimate the risk of cervical cancer in individual patients.
Acknowledgments
We thank H. zur Hausen and L. Gissmann for the kind donation of plasmid pHPV16.
This work was supported by grants from the Academy of Finland (1S30096) and the Cancer Foundation of Finland.
REFERENCES
- 1.Battista, C., J. Hillova, M. Hill, M. Reynes, and G. Mathe. 1988. Presence of human papillomavirus types 16 and 18 in genital warts and cervical neoplasias. Med. Oncol. Tumor Pharmacother. 5:1-9. [DOI] [PubMed] [Google Scholar]
- 2.Bosch, F. X., M. M. Manos, N. Munoz, M. Sherman, A. M. Jansen, J. Peto, M. H. Schiffman, V. Moreno, R. Kurman, K. V. Shah, et al. 1995. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J. Natl. Cancer Inst. 87:796-802. [DOI] [PubMed] [Google Scholar]
- 3.Boshart, M., L. Gissman, H. Ikenberg, A. Kleinheinz, W. Scheurlen, and H. zur Hausen. 1984. A new type of papillomavirus DNA, its presence in genital cancer biopsies and in cell lines derived from cervical cancer. EMBO J. 3:1151-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choo, K. B., C. C. Pan, and S. H. Han. 1987. Integration of human papillomavirus type 16 into cellular DNA of cervical carcinoma: preferential deletion of the E2 gene and invariable retention of the long control region and the E6/E7 open reading frames. Virology 161:259-261. [DOI] [PubMed] [Google Scholar]
- 5.Choo, K. B., C. C. Pan, M. S. Liu, H. T. Ng, C. P. Chen, Y. N. Lee, C. F. Chao, C. L. Meng, M. Y. Yeh, and S. H. Han. 1987. Presence of episomal and integrated human papillomavirus DNA sequence in cervical carcinoma. J. Med. Virol. 21:101-107. [DOI] [PubMed] [Google Scholar]
- 6.Cullen, A. P., R. Reid, M. Campion, and A. T. Lorincs. 1991. Analysis of the physical state of different human papillomavirus DNAs in intraepithelial and invasive cervical neoplasms. J. Virol. 65:606-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daniel, B., A. Rangarajan, G. Mukherjee, E. Vallikad, and S. Krishna. 1997. The link between integration and expression of human papillomavirus type 16 genomes and cellular changes in the evolution of cervical intraepithelial neoplastic lesions. J. Gen. Virol. 78:1095-1101. [DOI] [PubMed] [Google Scholar]
- 8.Das, B. C., J. K. Sharma, V. Gopalakrishna, and U. K. Luhtra. 1992. Analysis by polymerase chain reaction of the physical state of human papillomavirus type 16 DNA in cervical preneoplastic and neoplastic lesions. J. Gen. Virol. 73:2327-2336. [DOI] [PubMed] [Google Scholar]
- 9.Dürst, M., A. Kleinheinz, M. Hotz, and L. Gissmann. 1985. The physical state of human papillomavirus type 16 DNA in benign and malignant genital tumours. J. Gen. Virol. 66:1515-1522. [DOI] [PubMed] [Google Scholar]
- 10.Dürst, M., C. M. Croce, L. Gissmann, E. Schwarz, and K. Huebner. 1987. Papillomavirus sequences integrate near cellular oncogenes in some cervical carcinomas. Proc. Natl. Acad. Sci. USA 84:1070-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyson, N., P. M. Howley, K. Munger, and E. Harlow. 1989. The human papillomavirus-16 E7 oncoprotein is able to bind the retinoblastoma gene product. Science 243:934-937. [DOI] [PubMed] [Google Scholar]
- 12.Francis, D. A., S. I. Schmid, and P. M. Howley. 2000. Repression of the integrated papillomavirus E6/E7 promoter is required for growth suppression of cervical cancer cells. J. Virol. 74:2679-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groff, D. E., and W. D. Lancaster. 1984. Evidence that integration of virus DNA may not be necessary for maintenance of cell transformation. Prog. Med. Virol. 29:218-230. [PubMed] [Google Scholar]
- 14.Howley, P. M. 1985. Presence and expression of human papillomavirus sequences in human cervical carcinoma cell lines. Am. J. Pathol. 119:361-366. [PMC free article] [PubMed] [Google Scholar]
- 15.Jeon, S., B. L. Allen-Hoffmann, and P. F. Lambert. 1995. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 69:2989-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeon, S., and P. F. Lambert. 1995. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc. Natl. Acad. Sci. USA 92:1654-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Josefsson, A. M., P. K. Mangnusson, N. Ylitalo, P. Sörensen, P. Qwarforth-Tubbin, P. K. Andersen, M. Melbye, H. O. Adami, and U. B. Gyllensten. 2000. Viral load of human papillomavirus 16 as a determinant for development of cervical carcinoma in situ: a nested case-control study. Lancet 355:2189-2193. [DOI] [PubMed] [Google Scholar]
- 18.Kalantari, M., F. Karlsen, G. Kristensen, R. Holm, B. Hagmar, and B. Johansson. 1998. Disruption of the E1 and E2 reading frames of HPV 16 in cervical carcinoma is associated with poor prognosis. Int. J. Gynecol. Pathol. 17:146-153. [DOI] [PubMed] [Google Scholar]
- 19.Kalantari, M., E. Blennow, B. Hagmar, and B. Johansson. 2001. Physical state of HPV 16, and chromosomal mapping of the integrated form in cervical carcinomas. Diagn. Mol. Pathol. 10:46-54. [DOI] [PubMed] [Google Scholar]
- 20.Kessis, T. D., D. C. Connolly, L. Hedrick, and K. R. Cho. 1996. Expression of HPV16 E6 or E7 increases integration of foreign DNA. Oncogene 13:427-431. [PubMed]
- 21.Kurvinen, K., M. Yliskoski, S. Saarikoski, K. Syrjänen, and S. Syrjänen. 2000. Variants of the long control region of human papillomavirus 16. Eur. J. Cancer 36:1402-1410. [DOI] [PubMed] [Google Scholar]
- 22.Lazo, P. A. 1997. Papillomavirus integration: prognostic marker in cervical cancer? Am. J. Obstet. Gynecol. 176:1121-1122. [DOI] [PubMed] [Google Scholar]
- 23.Lehn, H., L. L. Villa, F. Marziona, M. Hilgarth, H. G. Hillemans, and G. Sauer. 1988. Physical state and biological activity of human papillomavirus genomes in precancerous lesions of the female genital tract. J. Gen. Virol. 69:187-196. [DOI] [PubMed] [Google Scholar]
- 24.Mazur, S., J. Feunteun, and C. D. Saintandre. 1995. Episomal amplification or chromosomal integration of the viral genome: alternative pathways in hamster polyomavirus-induced lymphomas. J. Virol. 69:3059-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitrani-Rosenbaum, S., D. Gal, M. Friedman, N. Kitron, R. Tsvieli, N. Mordel, and S. O. Anteby. 1998. Papillomaviruses in lesions of the lower genital tract in Israeli patients. Eur. J. Cancer Clin. Oncol. 24:725-731. [DOI] [PubMed] [Google Scholar]
- 26.Park, J., E. Hwang, S. Park, H. Ahn, S. Um, C. Kim, S. Kim, and S. Namkoong. 1997. Physical status and expression of HPV genes in cervical cancers. Gynecol. Oncol. 65:121-129. [DOI] [PubMed] [Google Scholar]
- 27.Rohlfs, M., S. Winkenbach, S. Meyer, T. Rupp, and M. Durst. 1991. Viral transcription in human keratinocyte cell lines immortalized by human papillomavirus type-16. Virology 183:331-342. [DOI] [PubMed] [Google Scholar]
- 28.Romanzuk, H., and P. Howley. 1992. Disruption of either the E1 and E2 regulatory gene of human papillomavirus type 16 increases viral immortalization capacity. Proc. Natl. Acad. Sci. USA 89:3159-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sastre-Garau, X., S. Schneider-Maunoury, J. Couturier, and G. Orth. 1990. Human papillomavirus type 16 DNA is integrated into chromosome region 12q14-q15 in a cell line derived from a vulvar intraepithelial neoplasia. Cancer Genet. Cytogenet. 44:243-251. [DOI] [PubMed] [Google Scholar]
- 30.Schneider-Maunoury, S., O. Croissant, and G. Orth. 1987. Integration of human papillomavirus type 16 DNA sequences: a possible early event in the progression of genital tumors. J. Virol. 61:3295-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwarz, E., U. K. Freese, L. Gissmann, W. Mayer, B. Roggenbuck, A. Stremlau, and H. zur Hausen. 1985. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature 314:111-114. [DOI] [PubMed] [Google Scholar]
- 32.Shirasawa, H., A. Tomita, K. Kubota, T. Kasai, S. Sekiya, H. Takamizawa, and B. Simizu. 1986. Detection of human papillomavirus type 16 DNA and evidence for integration into cell DNA in cervical dysplasia. J. Gen. Virol. 67:2011-2015. [DOI] [PubMed] [Google Scholar]
- 33.Swan, D. C., R. A. Tucker, G. Tortolero-Luna, M. F. Mitchell, L. Wideroff, E. R. Unger, R. A. Nisenbaum, W. C. Reeves, and J. P. Icenogle. 1999. Human papillomavirus (HPV) DNA copy number is dependent on grade of cervical disease and HPV type. J. Clin. Microbiol. 37:1030-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Syrjänen, K., and S. Syrjänen. 2000. Papillomavirus infections in human pathology. J. Wiley & Sons, Inc., New York, N.Y.
- 35.Tornesello, M. L., F. M. Buonagura, L. Buonagura, I. Salatiello, E. Beth-Giraldo, and G. Giraldo. 2000. Identification and functional analysis of sequence rearrangements in the long control region of human papillomavirus type 16 Af-1 variants isolated from Ugandan penile carcinomas. J. Gen. Virol. 81:2969-2982. [DOI] [PubMed] [Google Scholar]
- 36.Vernon, S. D., E. R. Unger, D. L. Miller, D. R. Lee, and W. C. Reeves. 1997. Association of human papillomavirus type 16 integration in the E2 gene with poor disease-free survival from cervical cancer. Int. J. Cancer 74:50-56. [DOI] [PubMed] [Google Scholar]
- 37.von Knebel Doeberitz, M., C. Rittmüller, F. Aengeneyndt, P. Jansen-Dürr, and D. Spitkovsky. 1994. Reversible repression of papillomavirus oncogene expression in cervical carcinoma cells: consequences for the phenotype and E6-p53 and E7-pRB interactions. J. Virol. 68:2811-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vousden, K. 1993. Interactions of human papillomavirus transforming proteins with the products of tumor suppressor genes. FASEB J. 7:872-879. [DOI] [PubMed] [Google Scholar]
- 39.Wells, S., D. A. Francis, A. Y. Karpova, J. J. Dowhanick, J. D. Benson, and P. M. Howley. 2000. Papillomavirus E2 induces senescence in HPV-positive cells via pRB- and p21cip-dependent pathways. EMBO J. 19:5762-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yliskoski, M., K. Syrjänen, S. Syrjänen, S. Saarikoski, and A. Nethersell. 1991. Systemic α-interferon (Wellferon) treatment of genital human papillomavirus (HPV) type 6, 11, 16, and 18 infections: double-blind, placebo-controlled trial. Gynecol. Oncol. 43:55-60. [DOI] [PubMed] [Google Scholar]
- 41.Ylitalo, N., A. Josefsson, M. Melbye, P. Sörensen, M. Frisch, P. K. Andersen, P. Sparen, M. Gustafsson, P. Magnusson, J. Ponten, U. Gyllensten, and H. O. Adami. 2000. A prospective study showing long-term infection with human papillomavirus 16 before the development of cervical carcinoma in situ. Cancer Res. 60:6027-6032. [PubMed] [Google Scholar]
- 42.zur Hausen, H. 2000. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J. Natl. Cancer Inst. 92:690-698. [DOI] [PubMed] [Google Scholar]
- 43.zur Hausen, H. 1996. Papillomavirus infections—a major cause of human cancers. Biochim. Biophys. Acta 1288:F55-F78. [DOI] [PubMed] [Google Scholar]