Abstract
Campylobacter lari is an infrequent cause of intestinal and extraintestinal infection in humans. We report a case of C. lari prosthetic joint infection and bacteremia in an 81-year-old immunocompetent man. The infection was associated with septic shock and fatal outcome. C. lari may cause severe disease, even in an immunocompetent host.
Bacteria of the genus Campylobacter are important causes of enteritis and extraintestinal infection in humans. Although the majority of documented Campylobacter infections are caused by C. jejuni, C. coli, and C. fetus, other species are being increasingly recognized as human pathogens.
Campylobacter lari is infrequently isolated from humans, but has been associated with enteritis (3, 5, 12, 15, 16), bacteremia (5, 6, 8-11, 14, 15, 16), permanent pacemaker infection (10), purulent pleurisy (4), and urinary tract infection (2). We report a case of C. lari prosthetic joint infection and bacteremia in an immunocompetent patient, which had a fatal outcome. To our knowledge, this is the first report to document infection of an orthopedic prosthesis with this microorganism.
An 81-year-old male was admitted to the hospital with a 1-day history of severe pain in his right prosthetic hip joint. He had experienced pain after flexion in the same joint 2 weeks earlier, but this had resolved completely following physiotherapy. The patient had also felt generally unwell for about 3 weeks prior to admission, with dizzy spells, feverishness, and chills. He did not experience diarrhea at any time. The right total hip prosthesis was inserted 4 years previously for osteoarthritis. His past medical history also included coronary artery disease, hypertension, and atrial fibrillation, for which he was being treated with cilazapril, amiodarone, and felodipine.
At the time of admission to hospital, he had a temperature of 38.1°C, was hemodynamically stable, but was unable to stand due to severe pain in the right hip, particularly on flexion and rotation. Blood tests at admission included a leukocyte count of 15.5 × 109/liter, a neutrophil count of 13.9 × 109/liter, and a C-reactive protein of 143 mg/liter (normal range, <10 mg/liter). An aspirate of the right hip showed numerous leukocytes, but Gram staining revealed no organisms and was reported as negative.
A diagnosis of prosthetic joint infection was made, and the patient was commenced on intravenous penicillin, flucloxacillin, and gentamicin, while waiting for the results of blood and hip aspirate cultures. A washout of the affected joint was performed 1 day after admission, and aspirated purulent material and tissue were sent for microscopy and culture.
During surgery, the patient became hypotensive, developed ischemic electrocardiogram changes, and was transferred to the Intensive Care Unit for continued ventilation and hemodynamic support. In the Intensive Care Unit, his initial mean arterial pressure was 55 mmHg (low), the cardiac index was 3.5 liter/min/m2 (elevated), and the pulmonary artery occlusion pressure was 20 mmHg (high). These measurements suggested significant myocardial dysfunction in the presence of distributive (septic) shock. Over the next few hours, he required escalating doses of norepinephrine and epinephrine. During this time, there was progressive acidosis and multiple organ dysfunction. Troponin T increased from 0.2 to 0.75 μg/liter (normal, <0.1 μg/l) over the next day, and the creatinine kinase had increased from 105 to 278 IU/liter (normal, 25 to 175 IU/liter). The raised creatinine kinase was consistent with his postoperative state, and the rise in the troponin T level could be explained by acute renal failure. However, an acute myocardial infarction could not be excluded. In view of his failure to respond to these measures, supportive therapies were withdrawn. The patient died on hospital day 2, 1 day after his admission to the Intensive Care Unit.
Blood cultures taken on the day of admission were incubated in the automated BacT/ALERT system (Organon Teknika Corporation, Durham, N.C.). The anaerobic BacT/ALERT bottle became positive after 26 h of incubation with a faintly staining curved gram-negative bacillus. The aerobic bottle remained negative. The organism was subcultured onto 5% sheep blood agar, MacConkey agar, and chocolate agar and was incubated at 36°C in a 5% CO2 environment. Inoculated sheep blood agar plates were also incubated in an anaerobic atmosphere and at 42°C in a microaerophilic environment; the latter was prompted by the Gram stain appearance suggestive of Campylobacter species. Growth appeared on the microaerophilic plate after 24 h and on the anaerobic plate after 2 days. In view of these findings, the original Gram stains of the hip joint aspirates were reviewed, and bacteria with similar Gram stain morphology were seen. The original report was amended appropriately. The same microorganism was subsequently cultured from both blood and joint aspirates and was presumptively identified as C. lari based on phenotypic characteristics. Key test results include growth at 42°C; growth in 1% glycine; and no growth in 3.5% NaCl; positive tests for catalase production and nitrate reduction; negative tests for hippurate hydrolysis, indoxyl acetate hydrolysis, and H2S production; and resistance to both nalidixic acid and cephalothin.
The identification of the isolates was also confirmed by molecular methods based on a multiplex PCR targeting of the Campylobacter lpxA gene. Whole-cell lysates (WCLs) were prepared by suspending the Campylobacter cells (48 h of growth) in 2 ml of distilled water to a McFarland equivalent of 1. A 1-ml aliquot was transferred to a sterile 1.5-ml Eppendorf tube and boiled for 10 min. Cellular debris was removed after centrifugation (10,000 × g for 10 min), and supernatants were transferred to a second sterile 1.5-ml Eppendorf tube and stored at −20°C until use.
Three primers, recognizing species-specific regions of the Campylobacter jejuni (0121; 5′-ACAACTTGGTGACGATGTTGTA-3′), C. coli (0120; 5′-AGACAAATAAGAGAGAATCAG-3′), or C. lari (0122;, 5′-CTTACCAAATGTTAAAATAGGC-3′) lpxA gene were used in combination with primer KK2(5′-CAATCATGWGCNATATGRCAATANGCC-3′). KK2binds to a conserved region of sequence identified for all of the thermotolerant Campylobacter species. Fifty-microliter reaction mixtures for the lpxA multiplex consisted of 10 pmol of the primers 0120, 0121, and 0122 per reaction; 30 pmol of KK2 per reaction; 200 μM each deoxynucleoside triphosphate (dNTP); 4 mM MgCl2; 1× PCR buffer (Roche); and 10 μl of WCLs. Taq DNA polymerase (2.5 U) was added to the reaction mixtures prior to amplification. In addition, a portion of the 16S rRNA gene was amplified. Primers 16S357F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 16S1087R (5′-CTCGTTGCGGGACTTACCC-3′) (30 pmol per reaction) were substituted for the lpxA primers above, but all other reagents were identical. For all of the PCRs, C. jejuni, C. coli, and C. lari experiments, as well as no-DNA control experiments, were performed.
The conditions used for thermocycling were as follows: DNA was initially denatured at 94°C for 3 min. Thirty-one cycles of the following steps were subsequently performed: denaturation at 94°C for 1 min followed by annealing at 64°C for 1 min and amplification at 72°C for 1 min. A final cycle was included in which denaturation and annealing times and temperatures remained the same, but the extension time was extended to 5 min to permit completion of all initiated products. PCR amplicons were analyzed by electrophoresis through 3% agarose, gels were stained with ethidium bromide, and amplicons were visualized under UV light (254 nm). Images were captured with a Kodak electrophoresis documentation and analysis system 120.
PCR amplicons to be sequenced were first passed through a Qiagen PCR cleanup spin column (Qiagen GmbH, Hilden, Germany) as per the manufacturer's instructions. Amplicons were sent to the University of Waikato DNA Sequencing Centre, Hamilton, New Zealand, and were processed by fluorescence dye chemistry on an ABI3100 (Perkin-Elmer). Electronic readouts were systematically compared to electropherograms, and ambiguities were resolved by using a minimum of twofold sequence redundancy of the target DNA fragments. Edited nucleotide sequences were sent to the BLAST server (National Center for Biotechnology Information) for nucleotide sequence comparisons. Alignments of DNA sequences were performed with the program Clustal X.
This identification system was developed in the laboratory of one of the authors (J.D.K.) and was validated in the following manner. The complete, redundant nucleotide sequences of the lpxA gene from five C. jejuni, three C. coli, and three C. lari isolates, as well as one Campylobacter upsaliensis isolate (the type strain of each species in addition to other phenotypically typical isolates), were determined by Sanger dideoxy DNA sequencing by fluorescent labeling technology. The edited sequence was aligned by the program Clustal X. Regions of maximal intraspecies identity and interspecies diversity were identified, and oligonucleotide sequence primers were developed for each of these species. The multiplex PCR primers were subsequently validated in two assays. In the first assay, purified genomic DNA from 17 Campylobacter and 20 non-Campylobacter species were used as a template in the multiplex PCR. With the exception of Campylobacter jejuni subsp. Doylei (which was identified as a C. jejuni) and Campylobacter hyoilei (which has been shown to be a strain of C. coli), none of the nontarget species resulted in an amplicon from this multiplex PCR. A second assay, in which over 100 environmental and clinical isolates of thermotolerant Campylobacter were tested, also showed a 100% correlation with the multiplex PCR. These isolates were previously confirmed as either C. jejuni, C. coli, or C. lari based on classic phenotypic tests as well as species-specific PCR and pulsed-field gel electrophoresis.
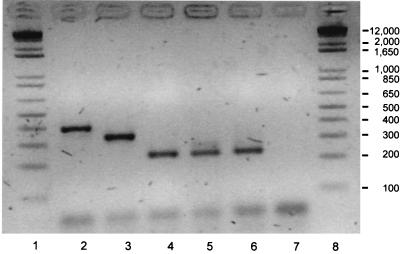
The WCLs of two independently collected Campylobacter species isolates from the present case (hip aspirate and blood) subjected to lpxA multiplex PCR were consistent with an identification of C. lari (Fig. 1). In order to confirm this result, an approximately 700-bp fragment of the small subunit rRNA gene was amplified, sequenced by methods described elsewhere (7), and compared to similar sequences from other thermotolerant and nonthermotolerant Campylobacter species. A comparison of the C. lari nucleotide sequence from the patient to the nucleotide sequence contained in the GenBank database revealed less than 1% divergence.
FIG. 1.
lpxA PCR analysis of Campylobacter spp. isolated from the patient in this study. Lanes: 1 and 8; Gibco-BRL 1 kb Plus; 2, Campylobacter coli control (isolate WA27); 3, Campylobacter jejuni control (isolate ANR0697); 4, Campylobacter lari control (isolate DRI879); 5, Campylobacter spp., hip aspirate; 6, Campylobacter spp., blood isolate; 7, no-DNA control.
C. lari infection was first described in 1980 by Skirrow and Benjamin, who isolated the microorganism from seagulls of the genus Larus (13). The organism was named Campylobacter laridis in 1983 (1), and the name was changed to C. lari in 1990 (18). The main reservoir for C. lari is gulls, chickens, other birds, and some domestic mammals, although it does not appear to cause overt infection in these animals.
C. lari belongs to the group of thermophilic campylobacters that grow optimally at 42°C. Other members of this group are C. jejuni and C. coli, which are both well-recognized human pathogens. Useful tests to distinguish C. lari from other Campylobacter species include resistance to nalidixic acid, demonstration of anaerobic growth in the presence of trimethylamine-N-oxide, susceptibility to triphenyl tetrazolium chloride, hydrolysis of indoxyl acetate, and the absence of hippurate hydrolysis. It may be difficult to distinguish C. lari from C. jejuni based on phenotypic properties alone, especially since both hippurate-negative and nalidixic acid-resistant C. jejuni strains have been described (1, 17). Molecular testing is useful in this situation to confirm identification.
Generally, Campylobacter species are not easily visualized with the safranin counterstain commonly used in Gram staining. In the present case, no microorganisms were seen when the Gram stains of the hip aspirates were initially examined. When reviewed after bacteria were detected in blood cultures, faintly staining curved gram-negative bacilli were seen in all hip aspirate samples and the tissue. This highlights the difficulty in identifying Campylobacter in clinical samples, especially from a site at which Campylobacter is infrequently or unexpectedly isolated.
Our case is unusual for several reasons. It is the first reported case of a prosthetic joint infection due to C. lari. C. lari had been the causative agent of a permanent pacemaker infection (10), raising the question as to whether C. lari could have a predilection for prosthetic devices. Second, our patient was not known to be immunocompromised. Most reported cases of C. lari bacteremia have occurred in people with significant medical conditions, usually associated with immune suppression (9). Third, our patient had a severe illness associated with septic shock and fatal outcome. Only two other fatal cases of C. lari bacteremia had been described before, both occurring in immunosuppressed patients: one patient with multiple myeloma and chronic renal failure, the other with AIDS (6, 11). Although our patient may have had a small perioperative myocardial infarction, his general condition and hemodynamic measurements were more in keeping with a picture of septic shock. Although we suspect bloodstream invasion arose from an intestinal focus, the patient experienced no antecedent gastrointestinal symptoms. This is in keeping with the findings from a review of eight other cases of C. lari bacteremia; only two cases were associated with gastroenteritis (9).
C. lari is an infrequent, possibly underrecognized, cause of human disease. Although usually having a favorable outcome, C. lari may cause severe disease, even in an immunocompetent host.
Acknowledgments
We thank Lois Seaward from the Microbiology Unit, Canterbury Health Laboratories, and the staff of the Institute of Environmental Science & Research, Porirua, New Zealand, for assistance with identification of the microorganism and helpful discussion.
REFERENCES
- 1.Benjamin, J., S. Leaper, R. J. Owen, and M. B. Skirrow. 1983. Description of Campylobacter laridis, a new species comprising the nalidixic acid resistant thermophilic campylobacter (NARTC) group. Curr. Microbiol. 8:231-238. [Google Scholar]
- 2.Bézian, M. C., G. Ribou, C. Barberis-Giletti, and F. Mégraud. 1990. Isolation of a urease-positive thermophilic variant of Campylobacter lari from a patient with urinary tract infection. Eur. J. Clin. Microbiol. Infect. Dis. 9:895-897. [DOI] [PubMed] [Google Scholar]
- 3.Broczyk, A., S. Thomson, D. Smith, and H. Lior. 1987. Water-borne outbreak of Campylobacter laridis-associated gastroenteritis. Lancet i:164-165. [DOI] [PubMed] [Google Scholar]
- 4.Bruneau, B., L. Burc, C. Bizet, N. Lambert-Zechovsky, and C. Branger. 1998. Purulent pleurisy caused by Campylobacter lari. Eur. J. Clin. Microbiol. Infect. Dis. 17:185-188. [DOI] [PubMed] [Google Scholar]
- 5.Chiu, C. H., C. Y. Kuo, and J. T. Ou. 1995. Chronic diarrhea and bacteraemia caused by Campylobacter lari in a neonate. Clin. Infect. Dis. 21:700-701. [DOI] [PubMed] [Google Scholar]
- 6.Dionisio, D., D. Milo, D. Mazzotta, and P. Cecile. 1989. Campylobacter laridis bacteraemia in an AIDS patient. Boll. Ist. Sieroter. Milan. 68:199-200. [PubMed] [Google Scholar]
- 7.Godfrey, S. A. C., S. A. Harrow, J. W. Marshall, and J. D. Klena. 2001. Characterization by 16S rRNA sequence analysis of pseudomonads causing blotch disease of cultivated Agaricus bisporus. Appl. Environ. Microbiol. 67:4316-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Godreuil, S., J. Maslin, M. Morillon, E. Sagui, J. J. De Pina, and G. Martet. 2000. Campylobacter lari bacteraemia. Presse Med. 29:1603.. [PubMed] [Google Scholar]
- 9.Martinot, M., B. Jaulhac, R. Moog, S. De Martino, P. Kehrli, H. Monteil, and Y. Piemont. 2001. Campylobacter lari bacteraemia. Clin. Microbiol. Infect. 7:96-97. [DOI] [PubMed] [Google Scholar]
- 10.Morris, C. N., B. Scully, and G. J. Garvey. 1998. Campylobacter lari associated with permanent pacemaker infection and bacteraemia. Clin. Infect. Dis. 27:220-221. [DOI] [PubMed] [Google Scholar]
- 11.Nachamkin, I., C. Stowell, D. Skalina, A. M. Jones, R. M. Hoop, and R. M. Smibert. 1984. Campylobacter laridis causing bacteraemia in an immunosuppressed patient. Ann. Intern. Med. 101:55-57. [DOI] [PubMed] [Google Scholar]
- 12.Simor, A. E., and L. Wilcox. 1987. Enteritis associated with Campylobacter laridis. J. Clin. Microbiol. 25:10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skirrow, M. B., and J. Benjamin. 1980. “1001” campylobacters: cultural characteristics of intestinal campylobacter from man and animals. J. Hyg. 85:427-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skirrow, M. B., D. M. Jones, E. Sutcliffe, and J. Benjamin. 1993. Campylobacter bacteraemia in England and Wales 1981-1991. Epidemiol. Infect. 110:567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soderstrom, C., C. Schalen, and M. Walder. 1991. Septicaemia caused by an unusual Campylobacter species (C. laridis and C. mucosalis). Scand. J. Infect. Dis. 23:369-371. [DOI] [PubMed] [Google Scholar]
- 16.Tauxe, R. V., C. M. Patton, P. Edmonds, T. J. Barrett, D. J. Brenner, and P. A. Blake. 1985. Illness associated with Campylobacter laridis, a newly recognized Campylobacter species. J. Clin. Microbiol. 21:222-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Totten, P. A., C. M. Patton, F. C. Tenover, T. J. Barrett, W. E. Stamm, A. G. Steigerwalt, J. Y. Lin, K. K. Holmes, and D. J. Brenner. 1987. Prevalence and characterization of hippurate-negative Campylobacter jejuni in King County, Washington. J. Clin. Microbiol. 25:1747-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Von Graevenitz, A. 1990. Revised nomenclature of Campylobacter laridis, Enterobacter intermedium, and “Flavobacterium branchiophila.” Int. J. Syst. Bacteriol. 40:211.. [DOI] [PubMed] [Google Scholar]