Abstract
To better understand the epidemiology and population structure of Cryptococcus neoformans, we determined mating types for 358 C. neoformans strains isolated through the active surveillance program from 1992 to 1994 in four geographic areas in the United States: San Francisco, California; Georgia; Texas; and Alabama. Two assays were used to determine mating types: (i) crossing with standard laboratory tester strains JEC20 and JEC21 on V8 agar medium; and (ii) PCR with the mating type α allele-specific primer of the STE12 gene and with serotype (A and D)- and mating type (a and α)-specific primers of the STE20 gene. Using these two methods, we found that this sample consisted of the following: (i) 324 serotype A, mating type (MAT) α (Aα) strains; (ii) 12 serotype D, α (Dα) strains; (iii) 14 serotype AD strains with mating type alleles Aa and Dα (AaDα); (iv) 2 serotype AD strains with mating type alleles Aα and Da (AαDa); (v) 3 serotype B, α (Bα) strains; and (vi) 3 serotype AD strains but with only one mating type allele. No strain with MATa was found within serotype A, B, or D in this collection. Interestingly, 14 of the 19 serotype AD strains contained the Aa allele at the STE20 locus; 13 of these 14 were from San Francisco. Our results suggest that the environment in San Francisco might contain Aa strains capable of mating with Dα strains. In addition, our result demonstrate that the sample from San Francisco had a significantly higher proportion of self-fertile strains than those from the other three areas.
Cryptococcus neoformans is a basidiomycetous yeast that can cause significant morbidity and mortality in both immunocompromised patients and healthy hosts (3). In patients with human immunodeficiency virus, lifelong suppressive antifungal therapy is usually needed to reduce the likelihood of recurrent cryptococcal infections. However, long-term drug treatments can result in the development of drug resistance in C. neoformans (25, 26, 34). A better understanding of the epidemiology, population structure, and evolution of C. neoformans will enhance our ability to develop appropriate prevention and treatment strategies.
In response to commercial monoclonal antibodies, strains of C. neoformans manifest five distinct serotypes: A, B, C, D, and AD (14). These serotypes differ in their ecological, molecular, and morphological characteristics; epidemiology; pathogenicity; physiology; and geographic distribution (3, 32). A small number of strains do not react with any of the four serotype-specific antibodies. Further classification of serotype groups into higher taxonomic groups remains controversial. However, because serotype has been used as the key feature in all currently proposed classification systems, to avoid confusion, we will restrict the strain affiliations to serotypes.
The mating system of C. neoformans is controlled by one locus with two alternative functional alleles, MATa and MATα. As in many other basidiomycete species, the mating type locus in C. neoformans is highly complex and contains a variety of genes essential for mating and morphogenesis, including the pheromone gene and STE11, STE12, STE20, and others (5, 21, 24). The mating type locus in C. neoformans has also been identified as a potential virulence factor. In a mouse tail vein injection model of systemic cryptococcosis, a MATα strain was found to be more virulent than the congenic MATa strain (16). In addition, mating type genes can play important roles in the epidemiology and evolution of pathogens. For example, in an oomycetous fungus, Phytophthora infestans, which causes late blight of potato and tomato, non-Mexican populations of this species were dominated by a single clonal lineage of the A1 mating type before 1980. However, with the appearance of a compatible A2 mating type, the population became sexual, resulting in new genotypes displacing old clonal lineages and genotypes in only a few years (10).
One of the most significant findings in epidemiological surveys of C. neoformans was that there were far more MATα strains (∼95%) than MATa strains (∼5%) (12, 17, 23). Mating types in C. neoformans have been traditionally determined by crossing with standard tester strains (3, 12, 17, 23). In this method, cells of strains to be tested are mixed with those of individual tester strains (typically the congenic pair JEC20 [MATa] and JEC21 [MATα]) on medium under incubation conditions conducive for mating. Strains with different mating types can mate to form dikaryotic hyphae, which can undergo nuclear fusion and meiosis. The meiotic process can then restore haploidy in sexual progeny (i.e., basidiospores). Traditionally, the production of hyphae from mating mixtures was used as an indicator of compatible mating types. It should be emphasized, though, that hyphal production does not imply either the successful formation of stable dikaryons, normal meiosis, or viable offspring from these crosses. This method has a few additional problems. First, the MATα tester strain JEC21 and other MATα strains could occasionally undergo haploid filamentation and fruiting, so mating types could be erroneously assigned (30). Second, mating is sensitive to a variety of environmental conditions, including temperature, nutrient availability, and moisture level (3). Third, many strains were unable to mate with either tester strain even after extensive manipulations of environmental conditions (22). Despite these caveats for the traditional crossing method, to be consistent with previous literature, the formation of hyphae from crosses will be considered compatible mating.
Recently, new methods for determining mating type alleles of C. neoformans have been developed. One such method uses mating type-specific primers in PCR (4, 12, 20). Several mating type-specific primers have been used to determine mating types. For example, the mating types of strains of serotype B were recently distinguished by two α mating type-specific primers for a population of C. neoformans from Australia (12). A PCR-restriction fragment length polymorphism method which permits the analysis of the mating type and ploidy based on the sequence divergence of C. neoformans within the mating pheromone gene has also been developed (4). In addition, Lengeler et al. recently developed four pairs of serotype- and mating type-specific primers according to the different sequences within the STE20 locus (20).
To augment our understanding of the epidemiology and evolution of C. neoformans, the objective of this study was to determine mating type allele distributions using a combination of mating assay and the PCR method. We analyzed 358 strains obtained from the Centers for Disease Controls and Prevention (CDC), Atlanta, Ga. This sample is one of the world's most comprehensive epidemiological collections of C. neoformans. It was obtained during the Cryptococcal Disease Active Surveillance program in four geographic areas in the United States between 1992 and 1994 (1, 2). We were specifically interested in the frequency and distribution of the serotype A, MATa (Aa) allele at the STE20 gene locus. Our analyses demonstrated that the Aa allele was widespread among strains of serotype AD.
MATERIALS AND METHODS
Strains.
The 358 isolates used in this study were collected by CDC from four regions in the United States from 1992 through 1994 (1, 2) and were generously provided by Mary E. Brandt. Strains were originally obtained through the Cryptococcal Disease Active Surveillance program in Alabama, Georgia, Texas, and San Francisco, California. Serotype and molecular subtypes based on multilocus enzyme electrophoresis for these strains were determined and described in earlier studies (1, 2).
Seven representative strains with different serotypes (A, B, C, and D) and mating types were tested to confirm the specificity of these primers (see below). These seven strains were E275, E312, B3181, B3184, JEC20, JEC21, and H99 (see Fig. 1). Strains E275 and E312 were obtained from Dee Carter of the University of Sydney, Sydney, Australia. Strains B3181 and B3184 were obtained from Mary E. Brandt of the CDC, Atlanta, Ga. Strains JEC20 and JEC21 were originally constructed by June Kwon-Chung of National Institutes of Health, Bethesda, Md., but were obtained from Joe Heitman of Duke University, Durham, North Carolina. Strain H99 was obtained from John Perfect of Duke University.
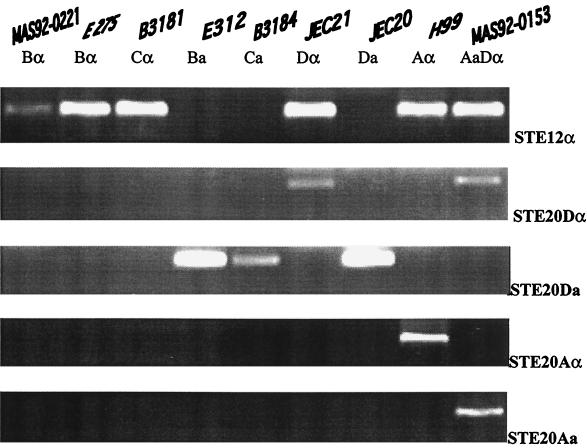
FIG. 1.
Confirmation of serotype- and mating type-specific primers using seven representative strains of C. neoformans. Two additional strains (MAS92-0221 and MAS92-0153) from this collection were also included in Fig. 1. The following five primer pairs were tested: STE12α, STE20Aa, STE20Aα, STE20Da, and STE20Dα (see Table 1 for details about these primer pairs). Strains MAS92-0221 (serotype B, MATα), E275 (serotype B, MATα), B3181 (serotype C, MATα), E312 (serotype B, MATa), B3184 (serotype C, MATa), JEC21 (serotype D, MATα), JEC20 (serotype D, MATa), H99 (serotype A, MATα strain), and MAS92-0153 (serotype AD, mating type Aa and Dα) are shown.
Mating type determination based on laboratory crosses.
The congenic strains JEC20 (serotype D, MATa) and JEC21 (serotype D, MATα) were used as testers. These two strains differ only at the mating type locus (13). A loopful of 2-day-old yeast cells was mixed evenly with JEC20 or JEC21, respectively, on V8 juice medium (5% V8 juice, 0.05% potassium phosphate [monobasic], 4% agar [pH 7.2]). A negative control of each strain alone was included. Strains JEC20 and JEC21 were mixed as positive controls. Plates were incubated at 25°C for 2 to 5 weeks and observed periodically with the naked eye and with a microscope. Following traditional interpretations, hyphal growth at the edge of the mating mixture was considered to be evidence of compatible mating (12, 17, 23). Self-fertility was inferred if the negative control containing a single inoculum produced hyphae. For strains not able to form hyphae with either tester strain in the first round of the crossing experiment, up to two more rounds of crosses were performed. Positive controls of crosses between JEC20 and JEC21 always produced hyphae.
It should be noted that a compatible mating does not imply that the hyphae contains stable dikaryons or that all dikaryons will go through proper nuclear fusion and meiosis or that all meiotic progenies will be viable and fertile. Compatible mating types are a prerequisite but not sufficient for completing the sexual life cycle of C. neoformans, a multistage process involving cell-cell fusion, formation and maintenance of stable dikaryotic hyphae, nuclear fusion, meiosis, and basidiospore formation in C. neoformans (3, 21).
Mating type determination through PCR.
Genomic DNA was isolated from each strain by the method described by Xu et al. (33). Five pairs of primers were used in this study to determine mating types (Table 1). The STE12α primer pair was MATα specific (Fig. 1). The other four pairs of primers were designed from DNA sequences of the STE20 gene, and they were specific for serotype (A and D) and mating type (a and α) (20). The specificity and selectivity of these primers were originally determined with a random sample of 65 strains of serotypes A, D, and AD by Lengeler et al. (20). In addition, these primers were tested here using seven strains representing all four serotypes A, B, C, and D (see above). Specificity of the STE12α primer pair was confirmed for strains in all four serotypes (Fig. 1). However, cross amplifications were observed for STE20Da (for strains from serotypes B and C, Fig. 1) and occasionally STE20Dα primers (for strains from serotype A, see below).
TABLE 1.
Primers used for identification of serotype- and mating type-specific alleles
| Primer and irection | Sequence (5′→3′) | Annealing temp (°C) | No. of cycles | Expected band size (bp) |
|---|---|---|---|---|
| STE20Aa | 55 | 35 | 865 | |
| Forward | TCCACTGGCAACCCTGCGAG | |||
| Reverse | ATCAGAGACAGAGGAGCAAGAC | |||
| STE20Aα | 55-45 | 38 | 588 | |
| Forward | CCAAAAGCTGATGCTGTGGA | |||
| Reverse | AGGACATCTATAGCAGAT | |||
| STE20Da | 60 | 32 | ∼440 | |
| Forward | GATCTGTCTCAGCAGCCAC | |||
| Reverse | AATATCAGCTGCCCAGGTGA | |||
| STE20Dα | 61 | 28 | 443 | |
| Forward | GATTTATCTCAGCAGCCACG | |||
| Reverse | AAATCGGCTACGGCACGTC | |||
| STE12α | 55 | 35 | 379 | |
| Forward | CTGAGGAATCTCAAACCAGGGA | |||
| Reverse | CCAGGGCATCTAGAAACAATCG |
Each PCR mixture contained 2 mM magnesium chloride (MgCl2), 0.1 mM deoxyribonucleotide triphosphate, 0.75 pmol of each primer, 0.1 U of Taq DNA polymerase (Promega), and 0.2 ng of DNA in a total volume of 10 μl. The annealing temperature, the number of cycles, and the expected fragment sizes are presented in Table 1 for each primer pair. Each of the five primer pairs was used individually for PCR for each of the 358 strains. PCR products were run on 1% agarose gel, stained with ethidium bromide, visualized under UV light, photographed using FluoroChem 8800 (Canberra Packard), and scored manually.
Mating type allele determination and analyses.
The serotype- and mating type-specific alleles was determined on the basis of evidence from three sources: (i) crossing experiments; (ii) PCR with serotype- and/or mating type-specific primers at the two gene loci, STE12 and STE20; and (iii) serotype identifications based on earlier published results for these strains (1, 2). In the case of cross amplification by PCR, ambiguities were resolved by comparing directly to crossing experiments and serotype identifications. Specifically, despite extensive searches for highly unique serotype- and mating type-specific primers from the STE20 sequences, cross amplification of serotype A strains was observed occasionally when the serotype D-, MATα-specific primer pair was used in PCR 20; see also Results and Discussion). However, these cross amplifications did not affect our interpretation of mating type alleles in any of the 358 strains (see below).
Comparisons of mating type alleles and self-fertility rates among geographic areas were performed using standard chi-square tests.
Confirmation of a single mating type allele in three serotype AD strains by Southern hybridization.
Previous studies identified that strains of serotype AD were often diploid and contained genetic material from both serotypes A and D (20, 28, 29, 31). Therefore, for serotype AD strains with only one mating type allele as determined by PCR, Southern hybridization was used to confirm that these strains did indeed contain only one MAT allele (MATa or MATα). To perform Southern hybridization, genomic DNAs were first digested with two restriction enzymes XbaI and SacI (MBI Fermentas), separately. Digested samples were electrophoresed in 1% agarose gel and transferred to a nylon membrane (Hybond-N; Amersham). This membrane was separately probed with [32P]dCTP-labeled STE20 gene fragments originally amplified from JEC20 (MATa) and JEC21 (MATα). Under high-stringency conditions (e.g., 65°C washing), the STE20Dα gene fragment can hybridize to Dα and Aα alleles, but not to Aa and Da alleles. Similarly, the STE20Da gene fragment can hybridize to Da and Aa alleles, but not to Dα and Aα alleles (20). Southern hybridization and detection of hybridized bands were performed by standard protocols (27).
RESULTS
Identification of mating types based on crossing experiments on V8 juice agar.
All 358 strains were mixed with tester strains JEC20 and JEC21 on V8 juice medium. Among the 716 combinations, 111 produced hyphae and were scored as compatible with either JEC20 or JEC21. A summary of the crossing experiment is presented in Table 2. Most of the strains (110 of 111) produced hyphae when crossed with JEC20 (MATa), so these 110 strains were scored as MATα. Only strain MAS94-0351 (a serotype AD strain) produced hyphae when crossed with JEC21. The mating success rate did not differ significantly among samples from different geographic areas. However, the rates varied significantly among serotypes (30.2% for serotype A, 66.7% for serotype D, and 26.3% for serotype AD; P < 0.05; Table 2). These mating results excluded the self-fertile strains described below.
TABLE 2.
Distribution of mating types as determined by crossing experiments
| Geographic location | Crossing | No. of strains (%)
|
||||
|---|---|---|---|---|---|---|
| Serotype
|
Total | |||||
| A | D | AD | B | |||
| San Francisco, California | Mateda | 39 | 3 | 4 | 46 | |
| Self-fertile | 11 | 6 | 1 | 18 (12.77) | ||
| Sterile | 69 | 1 | 7 | 77 | ||
| Total | 119 | 4 | 17 | 1 | 141 | |
| Georgia | Mated | 26 | 4 | 30 | ||
| Self-fertile | 2 | 2 (1.74) | ||||
| Sterile | 78 | 3 | 1 | 1 | 83 | |
| Total | 106 | 7 | 1 | 1 | 115 | |
| Texas | Mated | 22 | 1 | 1 | 24 | |
| Self-fertile | 2 | 2 (2.81) | ||||
| Sterile | 44 | 1 | 45 | |||
| Total | 68 | 1 | 1 | 1 | 71 | |
| Alabama | Mated | 11 | 11 | |||
| Self-fertile | 2 | 2 (6.45) | ||||
| Sterile | 18 | 18 | ||||
| Total | 31 | 31 | ||||
| Total | Mated | 98 (30.2) | 8 (66.7) | 5 (26.3) | 111 (31.0) | |
| Self-fertile | 17 (5.25) | 6 (31.58) | 1 (33.33) | 24 (6.70) | ||
| Sterile | 209 | 4 | 8 | 2 | 223 | |
| Total | 324 | 12 | 19 | 3 | 358 | |
“Mated” means hyphae were formed when strains were crossed to either one of the two tester strains, JEC20 or JEC21. Strains which produced hyphae on their own were not included in the Mated category but in the Self-fertile category.
Twenty-four strains produced hyphae on their own on V8 juice medium. These strains were considered self-fertile. The self-fertility rates varied among serotypes and geographic regions. Specifically, first, strains of serotype AD had a significantly higher self-fertility rate than those of serotypes A and D (31.58% for serotype AD, 5.25% for serotype A, and 0% for serotype D; P < 0.01 by the chi-square test; Table 2). Second, the sample from San Francisco had a higher rate of self-fertility than those from the other three regions (6.45% for Alabama, 1.74% for Georgia, 12.77% for San Francisco, and 2.81% for Texas; P < 0.01 by the chi-square test; Table 2). Excluding the serotype AD strains in the analyses, the sample from San Francisco still had a significantly higher rate of self-fertility than those from the other three areas (P < 0.05)
PCR identification of mating type alleles.
In the initial descriptions of these primers, only strains of serotypes A, D, and AD were tested (20). To confirm the initial observations and to examine whether these primers could be used to determine the mating types of strains of serotypes B and C, two strains each of serotypes B and C with known mating types were screened. Interestingly, the PCR with the STE12α primer pair produced the expected DNA band from MATα strains from both serotypes B and C (Fig. 1). The two MATa strains (E312 and B3184) had no PCR product with the STE12α primer. Furthermore, these two MATa strains had similar-sized DNA bands when a STE20Da primer pair was used during PCR (Fig. 1). No other primer pair produced any band for the four strains with serotype B or C (Fig. 1). Therefore, these primer pairs could be used for determining the mating types of strains of serotypes B and C.
Using these five pairs of primers, the mating type alleles of all 358 strains were determined. In all 324 serotype A strains, the STE20Aα primer pair amplified a 588-bp DNA fragment and the STE12α primer pair amplified a 379-bp DNA fragment. Primer pairs STE20Aa and STE20Da could not amplify any DNA fragment from these serotype A strains. These results indicated that all serotype A strains in this collection had only the MATα allele. Among these 324 Aα strains, 116 also showed a PCR product of 443 bp when the STE20Dα primer pair was used in the PCR amplification. However, STE20Aα primers did not amplify any PCR product from strains of other serotypes.
For the 12 strains of serotype D in this collection, all produced the expected DNA fragment with primers STE20Dα and STE12α. No DNA fragment was produced when primer pairs STE20Da, STE20Aa, and STE20Aα were used. Thus, these 12 strains were determined to be Dα strains.
The primer pair STE12α amplified a DNA fragment from each of the three strains of serotype B in this collection. However, none of the four primer pairs from the STE20 gene amplified any PCR products from these strains. The PCR pattern from these three strains was the same as that of the standard serotype B, MATα strain E275, and unlike the pattern from the serotype B MATa strain E312. Therefore, all three serotype B strains were MATα. The PCR pattern of one of the three serotype B strains (MAS92-0221) is shown in Fig. 1.
Of the 19 strains of serotype AD, two (MAS92-0022 and MAS92-0793) produced expected DNA fragments from PCR using primer pairs of STE12α, STE20Aα and STE20Da. These two strains were therefore identified as AαDa. For 14 of the 19 strains, the Aa primer pair amplified an 865-bp DNA fragment, the Dα primer pair amplified a 443-bp fragment, and the STE12α primers amplified a 379-bp fragment. Neither the STE20Aα primer nor the STE20Da primer amplified any DNA from these 14 strains, so these 14 strains were classified as AaDα.
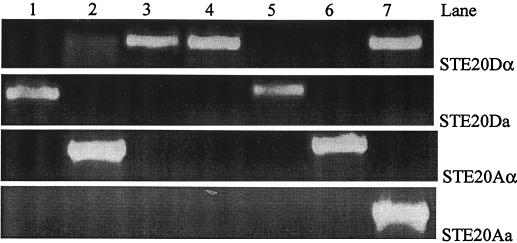
For the remaining three strains of serotype AD, only one mating type allele at the STE20 locus was detected for each strain (Fig. 2). Specifically, strain MAS92-0224 carried only the Dα allele at the STE20 locus but did not contain any other STE20 alleles. However, STE12α did produce the expected DNA fragment for MAS92-0224. Strain MAS92-0855 had the Aα allele at the STE20 locus but no other alleles. Similarly, STE12α amplified an expected DNA fragment for this strain. Strain MAS94-0351 contained the Da allele at the STE20 locus but no other allele (Fig. 2). As expected, the STE12α primer pair failed to amplify any PCR product from strain MAS94-0351.
FIG. 2.
Three strains of serotype AD containing only one STE20 allele were identified by PCR with four serotype- and mating type-specific primers (STE20Aa, STE20Aα, STE20Da, and STE20Dα). Strain MAS94-0351 (lane 1) was serotype AD and contained only the STE20 Da allele but not the STE20 Aα allele or other STE20 alleles. Strain MAS92-0855 (lane 2) was serotype AD and possessed only the STE20 Aα allele but lacked the STE20 Da allele. A weak band was observed with primer pair STE20Dα, likely the result of a cross-reaction (see Results and Discussion). Strain MAS92-0224 (lane 3) was serotype AD and had only the STE20 Dα allele but lacked the STE20 Aa allele. Strains JEC21 (lane 4), JEC20 (lane 5), H99 (lane 6), and MAS92-0153 (lane 7) were used as controls, and their serotypes and mating types were described in the text and the legend of Fig. 1.
Existence of only one mating type allele for the three serotype AD strains was confirmed by Southern hybridization. DNA from MAS92-0224 and MAS92-0855 hybridized to probe STE20Dα but not to probe STE20Da. In contrast, DNA from MAS94-0351 hybridized to probe STE20Da but not to probe STE20Dα (data not show).
Comparison between the two methods for determining mating type alleles.
Overall, the PCR method was more successful in determining mating type alleles than the traditional crossing method. By the PCR method, mating type alleles for all 358 strains were identified (100%). In contrast, the crossing experiment unambiguously identified mating type alleles of only 111 strains (31%). For strains where mating types were unambiguously determined by both methods, the results were completely congruent. All strains which mated with JEC20 also contained alleles from STE12α and either the Aα or Dα allele at the STE20 locus. Of particular notice was strain MAS94-0351. This serotype AD strain possessed only the Da allele at the STE20 locus (Fig. 2), and it mated successfully with the MATα tester strain JEC21.
Geographic distributions of mating type alleles.
Table 3 shows the distribution of mating type alleles in different geographic populations of C. neoformans. In all four populations, Aα was the most common allele (mean, 90.50% [Table 3]). However, the ratio of mating type alleles varied among the four geographic areas. Specifically, first, San Francisco contained a significantly lower frequency of the Aα allele than those of the other three areas (84.39% for San Francisco, 92.17% for Georgia, 95.78% for Texas, and 100% for Alabama; P < 0.01 by the chi-square test). Second, among the 19 serotype AD strains, 17 were collected from San Francisco and 13 (76.47%) contained the Aa allele. Consequently, the frequency of the Aa allele was significantly higher in San Francisco than in the other geographic areas (9.22% for San Francisco,0.87% for Georgia, and 0% for Texas and Alabama; P < 0.01 by the chi-square test). Despite the high incidence of the Aa allele in strains of serotype AD from San Francisco, no serotype A strain with MATa was found in this sample.
TABLE 3.
Distribution of mating type alleles as determined by PCR with serotype- and mating type allele-specific primers in C. neoformans
| Locationa | No. of strains (%) with serotype- and mating type-specific allele
|
Total no. of strains | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Aα | Dα | AaDα | AαDa | ADα | AαD | ADa | Bα | ||
| SF | 119 (84.39) | 4 (2.84) | 13 (9.22) | 2 (1.42) | 1 (0.71) | 1 (0.71) | 1 (0.71) | 141 | |
| GA | 106 (92.17) | 7 (6.09) | 1 (0.87) | 1 (0.87) | 115 | ||||
| TX | 68 (95.78) | 1 (1.41) | 1 (1.41) | 1 (1.41) | 71 | ||||
| AL | 31 (100) | 31 | |||||||
| Total | 324 (90.50) | 12 (3.35) | 14 (3.91) | 2 (0.56) | 1 (0.28) | 1 (0.28) | 1 (0.28) | 3 (0.84) | 358 |
Abbreviations: SF, San Francisco; GA, Georgia; TX, Texas; AL, Alabama.
DISCUSSION
This study identified mating type allele distribution for an epidemiologically comprehensive collection of the human-pathogenic fungus C. neoformans (1, 2). Two methods were used in our identification: (i) crossing on V8 juice agar medium; and (ii) PCR with serotype- and mating type-specific primers. Overall, these two methods produced congruent results. Similar to previous analyses of other samples, this collection of C. neoformans had a significantly skewed distribution of mating type alleles in favor of MATα. All MATa alleles were found in strains of serotype AD. Interestingly, most of the MATa alleles in serotype AD strains were Aa, and most of these were from San Francisco.
Mating type allele identification methods.
Similar to previous studies (12, 15, 17), the traditional crossing method was able to identify mating types of only a portion of the collection. In contrast, the PCR method determined the mating types of all 358 strains. The PCR method was highly efficient and less susceptible to environmental variables and was essential for determining strains containing Aa (20). In addition, using serotype- and mating type-specific primers at the STE20 locus, we were able to distinguish AaDα from AαDa for strains within the serotype AD group.
Despite these advantages, the PCR method has its drawbacks as well. First, DNA fragments of about 30% of Aα strains were amplified by the serotype D-, MATα-specific primer pair at the STE20 locus. Nucleotide identity between Aα and Dα alleles at the STE20 locus was 93 to 95% (22). Therefore, it was difficult to design primers that differed by more than a couple of bases within the STE20 locus (20). Similar results were obtained in genomic regions unrelated to the mating type locus (32). Primer pairs from all four genes used in the study by Xu et al. (32) amplified genes from strains in all five serotypes, serotypes A, B, C, D, and AD. Second, because primers were designed based on only a few sequences from common laboratory strains, gene sequence variation at these primer sites among clinical or natural strains was generally unknown. Therefore, if mutations existed at these primer sites, standard PCR conditions could result in failure to detect the potential allele. Under these circumstances, changes of PCR conditions were needed to identify mating types. In our work, the primer pair STE20Aα failed to amplify the expected product from 70 Aα strains when the annealing temperature was set at 55°C as calculated from the primer sequences (data not shown). A touch-down protocol (annealing temperatures, 55 to 47°C) was used to alleviate this problem. At present, the extent of sequence variation among strains within the same serotype group at the mating-related gene loci remains to be determined. However, these problems do highlight the necessity of a combination of methods with various conditions involving both traditional (e.g., crossing experiment and serotype identification) and molecular methods in epidemiological studies of C. neoformans.
Different self-fertility rate among serotypes.
When nitrogen starved, haploid MATα strains can produce hyphae on their own. This phenomenon was termed haploid fruiting. In this analysis, 17 strains of Aα and one strain of Bα underwent haploid fruiting on V8 juice agar medium. None of the 12 Dα strains underwent haploid fruiting on this medium. This rate of haploid fruiting (∼5%) was significantly lower than a previous observation based on a small set of laboratory strains (30).
Six of 19 serotype AD strains (31.6%) also produced hyphae on their own. All six self-fertile strains contained both a MATα allele and a MATa allele in their genome. Previous studies demonstrated that strains of serotype AD were generally diploid or aneuploid (20, 29) and that these strains are the results of recent hybridization between strains of serotypes A and D (31, 32). Because most of the serotype AD strains analyzed here contained both mating type alleles (MATa and MATα), they were expected to be self-fertile. Though strains of serotype AD had a significantly higher self-fertility than those of serotypes A and D, not all strains of serotype AD were self-fertile.
Skewed ratio of mating type alleles.
Among the 358 isolates, 357 contained a MATα allele. Only the serotype AD strain MAS94-0351 lacked the MATα allele. This strain contained the MATa allele from serotype D (Fig. 2). No strain with only a MATa allele was found in strains of serotypes A, B, and D in this sample. Such an imbalance was also reported in other studies of clinical and natural isolates (12, 17, 23). Several attributes of strains with MATα alleles could have contributed to its prevalence in this sample. First, a laboratory study identified that a MATα strain was more virulent than its congenic MATa strain in a mouse model. Therefore, MATα strains might more readily infect and be maintained in humans (16). All the strains analyzed here were from human hosts, not the environment. Second, unlike MATa strains, MATα strains could undergo haploid fruiting (30). The ability to undergo haploid fruiting and the production of basidiospores could aid the dispersal and survival of MATα strains in the environment. Because of their small size (diameter of 1.8 to 2.0 μm versus 5 to 10 μm for vegetative cells), basidiospores have been traditionally regarded as the infecting agent for humans (3).
Clonal population structure.
The skewed ratio of mating type alleles could also contribute to the clonal population structure of this pathogen. In a previous study, Brandt et al. (2) revealed a significant clonal structure in this set of strains. They found that a few multilocus enzyme electrophoresis genotypes dominated all four geographic populations. Wide distributions of one or few genotypes were consistent with clonal population structure (35). While asexual reproduction seemed to be the predominant mode of reproduction of C. neoformans in the United States (2), sexual reproduction could also play a role in the generation of genetic variation in this species. Indeed, the isolation of both AaDα and AαDa strains from human hosts suggested that populations from certain geographic areas, e.g., San Francisco, might contain significant evidence for sexual mating and recombination.
Several studies have demonstrated evidence of recombination in C. neoformans. For example, parsimony analysis of the DNA sequence data for the URA5 gene returned 1,276 most parsimonious trees, which was unexpected for a strictly clonal organism (9). In addition, using multiple gene genealogy analysis, Xu et al. identified genealogical incongruences among different genes for a group of 34 strains collected from different parts of the world (32). More recently, using a gene genealogical analysis, Xu et al. (31) identified that strains of serotype AD were the products of recent hybridizations between strains of serotypes A and D. At least three independent hybridization events were inferred among the 14 strains of serotype AD that they screened (31). The coexistence of sexual and asexual reproduction could contribute to the long-term survival and success of C. neoformans.
High incidence of AaDα from San Francisco.
Geographical differences in mating type allele distribution have been observed in environmental populations of serotype B strains from Australia (12). Unlike that study, the majority of the allelic differences observed here were for the Aa allele found only in strains of serotype AD and mostly from San Francisco. These results suggested that serotype A, MATa strains must exist in North America, and San Francisco should be a good candidate area for further investigations.
Compared to the other three regions, San Francisco is cooler in the summer than the southern part of the United States but is warmer in the winter than Alabama and Georgia. Eucalyptus trees are common in the San Francisco Bay area. These trees were identified as potential hosts for strains of serotypes B and C in both Southern California and Australia (3). Both mating type alleles, MATa and MATα, were found in strains obtained from eucalyptus trees (12). Whether these conditions were responsible for the high incidence of AaDα in San Francisco is not known. Indeed, the potential genetic diversity of environmental strains of C. neoformans is still largely unexplored. Aside from eucalyptus trees, there are many other potential reservoirs for C. neoformans in the environment, including pigeon droppings (3), bat guano (18), wasp nests (11), slime flux of mesquite (7), and hollows formed by decaying wood in living trees (19).
Origin and stability of strains of serotype AD.
Several recent studies indicated that strains of serotype AD were generally diploid or aneuploid (6, 20) and typically contained genetic material from strains of both serotypes A and D (20, 31). If serotype AD strains were the results of hybridization between strains of serotypes A and D, these strains were expected to contain one mating type allele from each partner. However, in the present study, three serotype AD strains contained only a single STE20 allele (Fig. 2). The existence of a single STE20 allele in these strains was likely the result of deletion and/or chromosome loss of one mating type allele after the hybridization. In C. neoformans, genetic changes have been reported for strains which underwent prolonged asexual propagation and improper storage in the laboratory (8). Lengeler et al. also observed allelic losses for many of the loci examined in a small collection of serotype AD strains (20).
It should be noted, however, that an alternative hypothesis regarding the relationship between strains of serotype AD and those of serotypes A and D has not been critically examined and refuted. In this alternative hypothesis, strains of serotype AD could give rise to serotype A or D. Such a process could occur through the loss of alleles either sexually through meiosis or asexually through mitosis. Indeed, laboratory crosses between strains of serotypes A and D have produced progenies with serotype A, D, or AD (20; J. Xu, unpublished data). Unfortunately, our present data are not sufficient to unambiguously infer the frequency or the directions of these relationships among the 358 strains. Multiple gene genealogies are needed to critically examine this issue (32, 35).
Conclusion.
Using both traditional and molecular approaches, we identified the mating types of 358 strains of C. neoformans collected from four areas in the United States. Among the four serotypes (A, B, D, and AD) identified, three (A, B, and D) contained only MATα strains. In the fourth serotype group AD, 14 of the 19 strains had the MATa allele from serotype A and the MATα allele from serotype D. Of the 14 AaDα strains, one was from Georgia and 13 were from San Francisco. Our results suggested that, as in Africa (20) and Europe (29) where serotype A and MATa strains were recently found, these strains, once thought to be extinct, likely also exist in North America.
Acknowledgments
We thank Mary Brandt, Dee Carter, Joe Heitman, and John Perfect for generously contributing strains of C. neoformans for this study. We thank Heather Yoell for helpful comments on the manuscript and Christopher Somers and Vicky Kjoss for technical assistance.
We are indebted to Thomas G. Mitchell and Rytas Vilgalys for their financial help in the initial collection and transfer of strains in this study. This research was supported in part by grants to J. Xu from McMaster University, the Natural Sciences and Engineering Research Council (NSERC) of Canada, the Canadian Foundation for Innovation (CFI), and the Ontario Innovation Trust (OIT).
REFERENCES
- 1.Brandt, M. E., L. C. Hutwagner, R. J. Kuykendall, R. W. Pinner, and The Cryptococcal Disease Active Surveillance Group. 1995. Comparison of multilocus enzyme electrophoresis and random amplified polymorphic DNA analysis for molecular subtyping of Cryptococcus neoformans. J. Clin. Microbiol. 33:1890-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandt, M. E., L. C. Hutwagner, L. A. Klug, W. S. Baughman, D. Rimland, E. A. Graviss, R. J. Hamill, C. Thomas, P. G. Pappas, A. L. Reingold, R. W. Pinner, and The Cryptococcal Disease Active Surveillance Group. 1996. Molecular subtype distribution of Cryptococcus neoformans in four areas of the United States. J. Clin. Microbiol. 34:912-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casadevall, A., and J. R. Perfect. 1998. Cryptococcus neoformans. American Society for Microbiology, Washington, D.C.
- 4.Chaturvedi, S., B. Rodeghier, J. Fan, C. M. McClelland, B. L. Wickes, and V. Chaturvedi. 2000. Direct PCR of Cryptococcus neoformans MATα and MATa pheromones to determine mating type, ploidy, and variety: a tool for epidemiological and molecular pathogenesis studies. J. Clin. Microbiol. 38:2007-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke, D. L., G. L. Woodlee, C. M. McClelland, T. S. Seymour, and B. L. Wickes. 2001. The Cryptococcus neoformans STE11α gene is similar to other fungal mitogen-activated protein kinase kinase kinase (MAPKKK) genes but is mating type specific. Mol. Microbiol. 40:200-213. [DOI] [PubMed] [Google Scholar]
- 6.Cogliati, M., M. C. Esposto, D. L. Clarke, B. L. Wickes, and M. A. Viviani. 2001. Origin of Cryptococcus neoformans var. neoformans diploid strains. J. Clin. Microbiol. 39:3889-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evenson, A., and J. W. Lamb. 1964. Slime flux of mesquite as a new saprophytic source of Cryptococcus neoformans. J. Bacteriol. 88:542.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franzot, S. P., J. Mukherjee, R. Cherniak, L. C. Chen, J. S. Hamdam, and A. Casadevall. 1998. Microevolution of a standard strain of Cryptococcus neoformans resulting in differences in virulence and other phenotypes. Infect. Immun. 66:89-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franzot, S. P., J. S. Hamdam, B. P. Currie, and A. Casadevall. 1997. Molecular epidemiology of Cryptococcus neoformans in Brazil and the United States: evidence for both local genetic differences and a global clonal population structure. J. Clin. Microbiol. 35:2243-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fry, W. E., and S. B. Goodwin. 1997. Re-emergence of potato and tomato late blight in the United States. Plant Dis. 81:1349-1357. [DOI] [PubMed] [Google Scholar]
- 11.Gezuele, E., L. Calegari, D. Sanabria, G. Davel, and E. Civila. 1993. Isolation in Uruguay of Cryptococcus neoformans var. gatti from a nest of the wasp Polybia occidentalis. Rev. Iber. Micol. 10:5-6. [Google Scholar]
- 12.Halliday, C. L., T. Bui, M. Krockenberger, R. Malik, D. H. Ellis, and D. A. Carter. 1999. Presence of α and a mating types in environmental and clinical collections of Cryptococcus neoformans var. gattii strains from Australia. J. Clin. Microbiol. 37:2920-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heitman, J., B. Allen, J. A. Alspaugh, and K. J. Kwon-Chung. 1999. On the origins of congenic MATα and MATa strains of the pathogenic yeast Cryptococcus neoformans. Fung. Genet. Biol. 28:1-5. [DOI] [PubMed] [Google Scholar]
- 14.Kabasawa, K., H. Itagaki, R. Ikeda, T. Shinoda, K. Kagaya, and Y. Fukazawa. 1991. Evaluation of a new method for identification of Cryptococcus neoformans which uses serologic tests aided by selected biological tests. J. Clin. Microbiol. 29:2873-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon-Chung, K. J., B. L. Wickes, L. Stockman, G. D. Roberts, D. Ellis, and D. H. Howard. 1992. Virulence, serotype, and molecular characteristics of environmental strains of Cryptococcus neoformans. Infect. Immun. 60:602-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon-Chung, K. J., J. C. Edman, and B. L. Wickes. 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60:1869-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwon-Chung, K. J., and J. E. Bennett. 1978. Distribution of α and a mating types of Cryptococcus neoformans among natural and clinical isolates. Am. J. Epidemiol. 108:337-340. [DOI] [PubMed] [Google Scholar]
- 18.Lazera, M. S., B. Wanke, and M. M. Nishikawa. 1993. Isolation of both varieties of Cryptococcus neoformans from saprophytic sources in the city of Rio de Janeiro, Brazil. J. Med. Vet. Mycol. 31:449-454. [Google Scholar]
- 19.Lazera, M. S., F. D. A. Pires, L. Camillo-Coura, M. M. Nishikawa, C. C. F. Bezerra, L. Trilles, and B. Wanke. 1996. Natural habitat of Cryptococcus neoformans var. neoformans in decaying wood forming hollows in living trees. J. Med. Vet. Mycol. 34:127-131. [PubMed] [Google Scholar]
- 20.Lengeler, K. B., G. M. Cox, and J. Heitman. 2001. Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and are heterozygous at the mating-type locus. Infect. Immun. 69:115-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lengeler, K. B., R. B. Davison, C. D'Souza, T. Harashima, W.-C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lengeler, K. B., P. Wang, G. M. Cox, J. R. Perfect, and J. Heitman. 2000. Identification of the MATa mating-type locus of Cryptococcus neoformans reveals a serotype A MATa strain thought to have been extinct. Proc. Natl. Acad. Sci. USA 97:14455-14460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madrenys, N., C. De Vroey, C. R. Wuytack, and J. M. Torres-Rodriguez. 1993. Identification of the perfect state of Cryptococcus neoformans from 195 clinical isolates including 84 from AIDS patients. Mycopathologia 123:65-68. [DOI] [PubMed] [Google Scholar]
- 24.Moore, T. D., and J. C. Edman. 1993. The alpha-mating type locus of Cryptococcus neoformans contains a peptide pheromone gene. Mol. Cell. Biol. 13:1962-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paugam, A., J. Dupouy-Camet, P. Blanche, J. P. Gangneaux, C. Tourte-Schaefer, and D. Sicard. 1993. Increased fluconazole resistance of Cryptococcus neoformans isolated from a patient with AIDS and recurrent meningitis. Clin. Infect. Dis. 17:431-436. [DOI] [PubMed] [Google Scholar]
- 26.Pfaller, M., J. Zhang, S. Messere, M. Tumbeerland, E. Mbidde, C. Jessup, and M. Ghannoum. 1998. Molecular epidemiology and antifungal susceptibility of Cryptococcus neoformans isolates from Uganda AIDS patients. Diagn. Microbiol. Infect. Dis. 32:191-199. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, E., F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 28.Takeo, K., R. Tanaka, H. Taguchi, and K. Nishimura. 1993. Analysis of ploidy and sexual characteristics of natural isolates of Cryptococcus neoformans. Can. J. Microbiol. 39:958-963. [DOI] [PubMed] [Google Scholar]
- 29.Viviani, M. A., M. C. Esposto, M. Cogloati, M. T. Montagna, and B. L. Wickes. 2001. Isolation of a Cryptococcus neoformans serotype A MATa strain from the Italian environment. Med. Mycol. 39:383-386. [DOI] [PubMed] [Google Scholar]
- 30.Wickes, B. L., M. E. Mayorga, U. Edman, and J. C. Edman. 1996. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the α-mating type. Proc. Natl. Acad. Sci. USA 93:7327-7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu, J., G. Luo, R. J. Vilgalys, M. E. Brandt, and T. G. Mitchell. 2002. Multiple origins of hybrid strains of Cryptococcus neoformans with serotype AD. Microbiology 148:203-212. [DOI] [PubMed] [Google Scholar]
- 32.Xu, J., R. J. Vilgalys, and T. G. Mitchell. 2000. Multiple gene genealogies reveal recent dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Mol. Ecol. 9:1471-1481. [DOI] [PubMed] [Google Scholar]
- 33.Xu, J., A. R. Ramos, R. J. Vilgalys, and T. G. Mitchell. 2000. Clonal and spontaneous origins of fluconazole resistance in Candida albicans. J. Clin. Microbiol. 38:1214-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu, J., C. Onyewu, H. J. Yoell, R. Y. Ali, R.J. Vilgalys, and T. G. Mitchell. 2001. Dynamic and heterogeneous mutations to fluconazole resistance in Cryptococcus neoformans. Antimicrob. Agents Chemother. 45:420-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu, J., and T. G. Mitchell. 2002. Strain variation and clonality in species of Candida and Cryptococcus neoformans, p. 739-750. In R. A. Calderone and R. L. Cihlar (ed.), Fungal pathogenesis: principles and clinical applications. Marcel Dekker, Inc., New York, N.Y.