Abstract
Sixty-eight Burkholderia cepacia complex isolates recovered from the sputum of 53 cystic fibrosis patients and 75 isolates collected from the maize rhizosphere were compared to each other to assess their genomovar status as well as some traits related to virulence such as antibiotic susceptibility, proteolytic and hemolytic activities, and transmissibility, in which transmissibility is determined by detection of the esmR and cblA genes. Among the clinical isolates, B. cepacia genomovar III comprised the majority of isolates examined and only a very few isolates were assigned to B. cepacia genomovar I, B. stabilis, and B. pyrrocinia; among the environmental isolates a prevalence of B. cepacia genomovar III and B. ambifaria was observed, whereas few environmental isolates belonging to B. cepacia genomovar I and B. pyrrocinia were found. Antibiotic resistance analysis revealed a certain degree of differentiation between clinical and environmental isolates. Proteolytic activity and onion tissue maceration ability were found to be spread equally among both clinical and environmental isolates, whereas larger percentages of environmental isolates than clinical isolates had hemolytic activity. The esmR gene was found exclusively among isolates belonging to B. cepacia genomovar III, with a marked prevalence in clinical isolates, whereas only one clinical isolate belonging to B. cepacia genomovar III was found to bear the cblA gene. In conclusion, the results of the present study show that the species compositions of the clinical and environmental B. cepacia complex populations examined are quite different and that some of the candidate determinants related to virulence and transmissibility are not confined solely to clinical isolates but are also spread among environmental isolates belonging to different species of the B. cepacia complex.
First described in 1950 by Burkholder (7) as the cause of soft rot in onions, Burkholderia cepacia has been recognized since the late 1970s as an opportunistic human pathogen that causes numerous outbreaks, particularly among cystic fibrosis (CF) patients. B. cepacia is especially problematic in these patients due to the potential severity of respiratory infections, the easy patient-to-patient spread of the organism, and the organism's innate resistance to a wide range of antimicrobial agents (19). While it has emerged as a human pathogen, B. cepacia has attracted increasing interest in agriculture and biotechnology for its potential as a biocontrol and bioremediation agent. The reasons for this interest include the organism's ability to promote plant growth by antagonizing soilborne plant pathogens (5, 6, 21, 30) and to degrade hydrocarbons and thus assist with the bioremediation of contaminated soil and water (16, 23, 25). The mode of acquisition of B. cepacia by CF patients remains unclear, and there is a general consensus that the large-scale use of these organisms would be ill advised until more is known about the presence or absence of the pathogenic potentials of environmental B. cepacia strains.
Recent taxonomic analyses have demonstrated that bacteria identified as B. cepacia actually comprise at least nine distinct species or genomovars, referred to collectively as the B. cepacia complex. These include B. cepacia genomovar I, B. multivorans, B. cepacia genomovar III, B. stabilis, B. vietnamiensis, B. cepacia genomovar VI, B. ambifaria, and B. anthina (10, 11, 17, 37, 38; P. Vandamme et al., unpublished data); in addition, it was recently shown that the species B. pyrrocinia also belongs to the B. cepacia complex (12; P. Vandamme et al., unpublished data).
Although all nine species or genomovars have been recovered from CF patients, B. multivorans and B. cepacia genomovar III account for the majority of isolates from CF patients, especially epidemic clones (26, 29, 35, 37). As far as environmental strains are concerned, in the first extensive study performed with bacterial strains isolated from a single habitat (15), B. cepacia genomovar I, B. cepacia genomovar III, and B. ambifaria have been reported to be the most widespread members of the B. cepacia complex in the rhizosphere of maize; moreover, Balandreau et al. (2) found that a group of B. cepacia-like organisms isolated from the rhizosphere or tissues of maize, wheat, and lupine belonged to B. cepacia genomovar III. However, the risk posed to CF patients by B. cepacia strains present in the environment and the relationship between clinical and environmental isolates remain unclear, with contradictory evidence for phenotypic differences between isolates from CF patients and the environment (19). Previous comparative studies of clinical and environmental isolates which have suggested that environmental and clinical isolates of B. cepacia represent distinct groups, that clinical strains lack the ability to act as phytopathogens, and that environmental isolates are unlikely to be responsible for human infections have been handicapped by the inclusion of only a few environmental isolates (4, 18, 24, 41). Therefore, more extensive studies with a wider collection of isolates from CF patients and environmental isolates are needed to evaluate the role of natural habitats as potential reservoirs of pathogenic strains and the risks associated with the use of strains belonging to the B. cepacia complex for biotechnological purposes.
In the present study, we examined a large collection of clinical and environmental isolates belonging to the B. cepacia complex. Sixty-eight isolates were recovered from 53 Italian CF patients living in 11 regions; 75 isolates were collected from the maize rhizosphere, where the B. cepacia complex represents one of the predominant bacterial groups (13, 14, 20), in three different locations in northern, central, and southern Italy. By investigating the genomovar distributions of clinical and environmental isolates, as well as some traits related to virulence such as proteolytic and hemolytic activities, antibiotic susceptibility, and genetic markers associated with transmissibility, such as the B. cepacia epidemic strain marker (BCESM) encoded by the esmR gene and the cable pilus gene (cblA), we aimed to assess the differences between clinical and environmental isolates of Italian B. cepacia complex bacteria.
MATERIALS AND METHODS
B. cepacia complex strains.
Sixty-eight clinical isolates and 75 environmental isolates belonging to the B. cepacia complex were investigated in this study. Clinical isolates were recovered from sputum samples from 53 CF patients who attended the Cystic Fibrosis Centre of Gaslini Hospital (Genoa, Italy) between April 1985 and December 1999. The Cystic Fibrosis Centre of Gaslini Hospital is a large specialized unit to which patients from different regions of Italy have been admitted for many years. Clinical specimens were processed by techniques regularly in use for specimens from CF patients (34). The identities of the isolates were confirmed by standard biochemical procedures (API 20 NE; BioMérieux). The environmental strains of the B. cepacia complex were isolated from the rhizosphere of maize cultivated in three different fields located in northern, central, and southern Italy (5, 13, 14). Representative strains of the current species of the B. cepacia complex were also included (2, 10, 11, 29). All strains were cryopreserved at −80°C in 30% (vol/vol) glycerol.
Species and genomovar identification. (i) Sample preparation prior to PCR amplification
Clinical and environmental strains were grown overnight on nutrient agar (NA; Difco Laboratories) at 37 and 28°C, respectively. Lysis of the cells of clinical strains was performed by the digestion technique described by O'Callaghan et al. (33) in 20 μl of lysis buffer (1× Polymed PCR buffer, 0.5% Tween 20, 200 μg of proteinase K per ml). Lysis of the cells of rhizosphere B. cepacia strains was performed by the procedure described by Di Cello et al. (14).
(ii) Amplification and restriction analysis of 16S rDNA
Amplification analysis of 16S rDNAs were performed with 2 μl of each cell lysate suspension in 20 μl of Polymed Taq buffer with 0.5 U of Taq DNA polymerase (Polymed), as described by Di Cello et al. (14). Preliminary identification was performed on the basis of a previously published PCR-based procedure (15) by digesting each PCR sample in four separate reactions with four of the six enzymes used by Segonds et al. (35) to differentiate Burkholderia species or genomovars; these enzymes, i.e., AluI, CfoI, DdeI, and MspI (New England Biolabs), were selected because of their discriminatory powers.
(iii) recA-based RFLP analyses and genomovar-specific PCR assays.
Genomovar status was determined by restriction fragment length polymorphism (RFLP) analysis of the recA gene, which had previously been amplified with primers selective for bacteria belonging to the B. cepacia complex; the PCR products were digested with the endonucleases HaeIII and MnlI (New England Biolabs) (28). Genomovar assignment was confirmed by PCR of the recA gene with genomovar-specific primers by previously described procedures (28). The identities of B. pyrrocinia isolates were confirmed by DNA-DNA hybridization experiments (P. Vandamme et al., unpublished observations).
Analysis of virulence and transmissibility-related traits. (i) Antibiotic resistance pattern.
Antibiotic susceptibility patterns were determined by the disk diffusion method on Mueller-Hinton medium (Difco Laboratories), according to the criteria of the National Committee for Clinical Laboratory Standards (32). Mueller-Hinton agar plates were inoculated with a bacterial suspension equivalent to a 0.5 McFarland standard, and antibiotic sensitivity disks (Becton Dickinson) were applied. Zones of growth inhibition (in millimeters) were recorded after overnight incubation at 35°C. Pseudomonas aeruginosa ATCC 27853 and Escherichia coli ATCC 35218 were used as control strains. The antibiotics tested were amoxicillin, piperacillin-tazobactam, piperacillin, ticarcillin, ceftazidime, cefoxitin, cefepime, cefotaxime, aztreonam, imipenem, meropenem, nalidixic acid, ciprofloxacin, levofloxacin, tobramycin, amikacin, netilmicin, polymyxin B, minocycline, chloramphenicol, rifampin, and trimethoprim-sulfamethoxazole.
(ii) In vitro maceration of onion tissue.
A culture (100 μl) of each strain of the B. cepacia complex at the log phase of growth (approximately 108 CFU ml−1) was inoculated onto the surface of onion slices prepared as described by Wigley and Burton (40). B. cepacia LMG 1222 and E. coli DH5α were used as positive and negative controls, respectively. The plates were incubated at 28°C, and readings were taken at 48 h of incubation. Maceration was evaluated on a scale of 0 (no maceration of onion tissue) to 4 (100% maceration of onion tissue).
(iii) Proteolytic activity.
The proteolytic activities of the strains were evaluated by investigating their caseinolytic capacities. All strains were grown on NA supplemented with 4% (wt/vol) skim milk. The plates were incubated at 28 and 37°C and were examined after 48 h of incubation. Clear zones surrounding the bacterial growth indicated casein degradation. B. cepacia LMG 1222 and E. coli DH5α were used as positive and negative controls, respectively.
(iv) Hemolytic activity.
The hemolytic activities of the strains were assessed at 28 and 37°C on NA supplemented with 5% (vol/vol) ram blood. The plates were examined after 48 h, and clear zones surrounding the bacterial growth indicated hemolytic activity. E. coli phy152 and E. coli DH5α were used as positive and negative controls, respectively.
PCR amplification of esmR and cblA genes.
The 1.4-kb esmR sequence encoding BCESM was amplified with specific primers BCESM 1 and BCESM 2 by the procedure described by Mahenthiralingam et al. (27). The 664-bp cblA DNA encoding the cable pilus was amplified with primers CBL1 and CBL2 by the procedure described by Clode et al. (9). All PCRs were performed with lysate cell suspensions and 0.5 U of Taq DNA polymerase (Polymed).
Statistical analysis.
Differences between the antibiotic resistance patterns of clinical and environmental isolates were analyzed by one-way analysis of variance. Antibiotic resistance data were converted to a binary code, and interisolate relationships were measured by the Euclidean metric unweighted pair group average method. All analyses were carried out with StatView 512+ software (BrainPower Inc.).
RESULTS
Species and genomovar distributions of clinical and environmental isolates.
The identification of the different species and genomovars was accomplished by a combination of PCR-based techniques, such as amplification and restriction analysis of 16S rDNA, genomovar-specific PCR tests, and RFLP analyses based on polymorphisms in the recA gene.
As shown in Table 1, not all B. cepacia complex species were found within the two groups of isolates. In fact, B. cepacia genomovar I, B. cepacia genomovar III, B. stabilis, and B. pyrrocinia were found among the clinical isolates, whereas B. cepacia genomovar I, B. cepacia genomovar III, B. ambifaria, and B. pyrrocinia were found among the environmental isolates. It is worth noting that in both the clinical and the environmental groups of isolates, most isolates (86.8 and 53.4%, respectively) belonged to B. cepacia genomovar III. Strains belonging to B. ambifaria were also recovered in large numbers, but only from among the environmental isolates (37.3%).
TABLE 1.
Distribution of genomovars or species among clinical and environmental isolates of the B. cepacia complex
| Genomovar | % of isolates
|
|
|---|---|---|
| ClinicalE | Environmental | |
| B. cepacia genomovar I | 2.9 (2)a | 1.3 (1) |
| B. multivorans | 0.0 | 0.0 |
| B. cepacia genomovar III | 86.8 (59) | 53.4 (40) |
| B. stabilis | 7.4 (5) | 0.0 |
| B. vietnamiensis | 0.0 | 0.0 |
| B. cepacia genomovar VI | 0.0 | 0.0 |
| B. ambifaria | 0.0 | 37.3 (28) |
| B. anthina | 0.0 | 0.0 |
| B. pyrrocinia | 2.9 (2) | 8.0 (6) |
The numbers in parentheses refer to the number of strains belonging to a particular genomovar or species.
Antibiotic resistance.
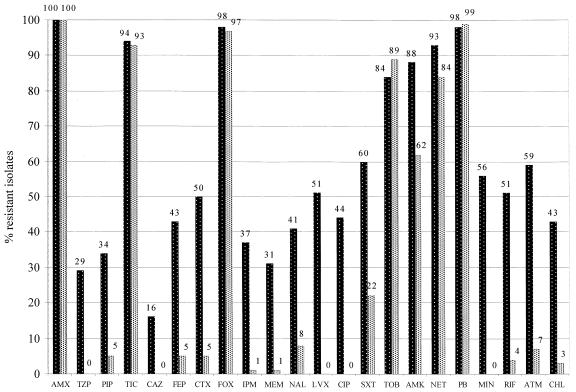
All clinical and environmental isolates were resistant to several antibiotics; the percentages of strains resistant to the various antibiotics varied greatly (Fig. 1). The clinical group of isolates generally had a higher percentage of multidrug-resistant isolates than the environmental group did. In fact, significant differences (P < 0.001) between the two groups of isolates in terms of the number of drugs to which the isolates were resistant (for clinical isolates, mean, 13.00; standard deviation, 4.94; for environmental isolates, mean, 6.81; standard deviation, 1.32) were found. Within the environmental group of isolates, no significant differences in the number of drugs to which the isolates were resistant among the various species were found (P > 0.05), whereas within the clinical group of isolates, most resistant isolates belonged to B. cepacia genomovar III (data not shown). When clinical and environmental isolates of B. cepacia genomovar III were compared to each other, significant differences (P < 0.0001) in the number of drugs to which the isolates were resistant between the two groups of isolates (for clinical isolates, mean, 13.37; standard deviation, 4.99; for environmental isolates, mean, 6.47; standard deviation, 1.81) were found.
FIG. 1.
Percentages of clinical (▓) and environmental (░) B. cepacia complex isolates resistant to antibiotics. AMX, amoxicillin; TZP, piperacillin-tazobactam; PIP, piperacillin; TIC, ticarcillin; CAZ, ceftazidime; FEP, cefepime; CTX, cefotaxime; FOX, cefoxitin; IPM, imipenem; MEM, meropenem; NAL, nalidixic acid; LVX, levofloxacin; CIP, ciprofloxacin; SXT, trimethoprim-sulfamethoxazole; TOB, tobramycin; AMK, amikacin; NET, netilmicin; PB, polymyxin B; MIN, minocycline; RIF, rifampin; ATM, aztreonam; CHL, cloramphenicol.
Altogether, 119 different antibiotic resistance patterns were observed. A total of 106 profiles were unique, but 13 profiles were common to two or more isolates. It is worth noting that clinical and environmental isolates did not share common patterns. Moreover, of the 13 profiles mentioned above, only 2 were for clinical isolates; of the 11 profiles to which only environmental isolates belonged, 1 was shared by 8 isolates. The profiles were compared by numerical methods, and 10 different groups or clusters were found on the basis of 60% similarity (Table 2). A highly uneven distribution of clinical and environmental isolates was observed among the 10 clusters. In fact, the largest group, group C, comprised several clinical isolates (n = 13) and most of the environmental (n = 57) isolates. Three groups were heterogeneous and contained isolates of both clinical and environmental origins, and as many as six groups were homogeneous, containing only isolates of clinical origin.
TABLE 2.
Grouping of B. cepacia complex isolates by cluster analysis performed with antibiotic resistance patterns
| Isolate source | No. of isolates in cluster:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | L | |
| CF patients | 4 | 9 | 13 | 4 | 15 | 1 | 5 | 4 | 4 | 8 |
| Environment | 7 | 0 | 57 | 1 | 0 | 0 | 0 | 9 | 0 | 0 |
Pathogenicity tests.
In vitro maceration of onion tissue, casein hydrolysis, and hemolytic activity tests were applied to all isolates to investigate the potential pathogenicities of clinical and environmental isolates for plants and humans.
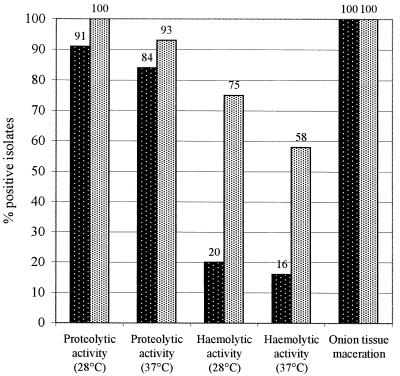
All clinical isolates were able to macerate onion tissue to some degree, as was the case for all environmental isolates (Fig. 2). No significant differences in the maceration abilities were observed either among clinical and environmental isolates (Fig. 2) or among different species (data not shown).
FIG. 2.
Percentages of clinical (▓) and environmental (░) B. cepacia complex isolates capable of proteolysis, hemolysis, and onion tissue maceration.
Most clinical and environmental isolates were able to hydrolyze casein at 28°C (91 and 100%, respectively) and 37°C (84 and 93%, respectively) (Fig. 2). All isolates belonging to B. pyrrocinia and clinical strains belonging to B. cepacia genomovar I and B. stabilis showed marked reductions in their proteolytic abilities at 37°C (Table 3). Among the environmental isolates, B. cepacia genomovar III and B. ambifaria showed only slightly higher percentages of positive strains at 28°C than at 37°C. It is worth noting that no marked differences between clinical and environmental isolates belonging to B. cepacia genomovar III were observed (Table 3).
TABLE 3.
Virulence and transmissibility-related traits in different genomovars or species of the B. cepacia complex
| Test | % of isolates positive by specimen type within each genomovar or speciesa
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
B. cepacia gen. I
|
B. cepacia gen. III
|
B. stabilis
|
B. ambifaria
|
B. pyrrocinia
|
||||||
| Clin (2)b | Env (1) | Clin (59) | Env (40) | Clin (5) | Env (0) | Clin (0) | Env (28) | Clin (2) | Env (6) | |
| Onion maceration | 100 | 100 | 100 | 100 | 100 | — | — | 100 | 100 | 100 |
| Proteolysis | ||||||||||
| 28°C | 100 | 100 | 90 | 100 | 100 | — | — | 100 | 100 | 100 |
| 37°C | 50 | 100 | 93 | 95 | 0 | — | — | 96 | 50 | 67 |
| Hemolysis | ||||||||||
| 28°C | 50 | 0 | 19 | 42 | 0 | — | — | 100 | 100 | 83 |
| 37°C | 0 | 0 | 14 | 35 | 20 | — | — | 96 | 100 | 17 |
| esmR | 0 | 0 | 61 | 15 | 0 | — | — | 0 | 0 | 0 |
| cblA | 0 | 0 | 2 | 0 | 0 | — | — | 0 | 0 | 0 |
Abbreviations and symbols: gen., genomovar; Clin, clinical isolates; Env, environmental isolates; —, isolates were not found.
The numbers in parentheses refer to the number of strains belonging to a particular genomovar or species.
In contrast to the in vitro maceration of onion tissue and casein hydrolysis, marked differences in hemolytic activities were found between clinical and environmental isolates (Fig. 2). In fact, surprisingly, a much higher percentage of environmental isolates than clinical isolates showed hemolytic activity at both temperatures used. In particular, more than twice as many environmental than clinical isolates of B. cepacia genomovar III exhibited hemolytic activity, a high percentage of B. ambifaria isolates were positive for hemolytic activity at both temperatures, and B. pyrrocinia isolates showed marked reductions in hemolytic activity at 37°C (Table 3). Furthermore, marked differences were observed when different species were compared to each other, with B. cepacia genomovar I and B. stabilis being the species with the lowest percentage of strains positive for hemolytic activity and B. ambifaria and B. pyrrocinia being the species with the highest percentage of strains positive for hemolytic activity.
PCR detection of esmR and cblA.
The potential pathogenicities of clinical and environmental isolates of B. cepacia complex were also investigated by evaluating the isolates for the presence of two molecular markers related to transmissibility, esmR and cblA, which code for BCESM and the cable pilus, respectively.
All esmR-positive strains belonged exclusively to B. cepacia genomovar III (Table 3). Interestingly, esmR was not confined solely to clinical isolates but was also found among environmental isolates; however, the proportion of clinical isolates positive for esmR (61%) was significantly higher than the proportion of environmental isolates positive for esmR (15%).
Of all the clinical and environmental isolates evaluated for the cblA gene, only one clinical isolate belonging to B. cepacia genomovar III was found to harbor the cblA gene (Table 3).
DISCUSSION
For the first time, to our knowledge, two large sets of clinical and environmental isolates belonging to the B. cepacia complex were compared to each other in the same study to assess their species and genomovar compositions and the presence of traits related to virulence and transmissibility.
The species compositions of the two groups of isolates were found to be quite different. Clinical isolates were assigned almost exclusively to B. cepacia genomovar III, and only a few clinical isolates were assigned to B. cepacia genomovar I, B. stabilis, and B. pyrrocinia. Among the environmental isolates, the predominant species were B. cepacia genomovar III and B. ambifaria, whereas only a few isolates were assigned to B. cepacia genomovar I and B. pyrrocinia. Previous reports (2, 15) have already shown that a significant proportion of isolates from the maize rhizosphere are actually members of B. cepacia genomovar III, the genomovar seemingly best adapted to the lungs of CF patients and the one that includes the major epidemic strains of the B. cepacia complex. Thus, the wide diffusion of B. cepacia genomovar III in both clinical and soil habitats raises serious concern that natural habitats may be potential reservoirs for B. cepacia genomovar III and therefore sources of infection for vulnerable humans. However, the virulence of environmental B. cepacia genomovar III isolates remains to be determined. On the contrary, we found that B. ambifaria, a species that comprises several plant-growth promoting strains (11, 22; L. Chiarini et al., unpublished data), was represented in large numbers among our environmental isolates but was absent from among the clinical isolates. This finding is in agreement with recent results obtained by Fiore et al. (15), who also reported the presence of B. ambifaria strains among environmental strains isolated in Italy, and by Agodi et al. (1), who did not find B. ambifaria within a group of Italian isolates from CF patients. Moreover, LiPuma et al. (26) examined 606 clinical isolates of the B. cepacia complex and found that only 0.7% belonged to B. ambifaria. Assuming that CF patients become infected with B. cepacia complex strains not only by patient-to-patient transmission but also by occasionally picking up strains from the environment, the unequal distribution of B. ambifaria in the clinical and environmental group of isolates suggests that this species may play a minor role as a human pathogen either because its pathogenicity or transmissibility may not be very high or because it may be confined to habitats where the probability of being picked up by humans is very low. In general, the different distributions of species in clinical and natural habitats suggest that there may be differential capacities for pulmonary colonization or interpatient spread, as already pointed out by LiPuma et al. (26).
Such considerations indicate that there is a need for more detailed knowledge about the distributions of each of the species of the B. cepacia complex in different natural habitats and, in particular, for a more precise evaluation of the pathogenicities of environmental and clinical isolates of each species belonging to the B. cepacia complex. Therefore, the distributions of some phenotypic traits and genetic markers related to virulence and transmissibility were investigated in an effort to evaluate the potential pathogenicities of B. cepacia complex isolates in relation to their origins and genomovar status. B. cepacia is endowed with a wide range of potential virulence determinants, but so far none has been proved to play an important role in enhancing disease in humans. Among the candidate virulence determinants, we chose antibiotic susceptibility, proteolytic and hemolytic activities, and two transmissibility markers, esmR and cblA (36). In addition, we investigated the abilities of the isolates to macerate onion tissue, which is a trait related to pathogenicity for plants.
Our results suggest that clinical and environmental B. cepacia strains might be functionally equivalent in terms of several traits related to virulence for humans and plants. In fact, proteolytic activity was found to be equally spread within both groups of isolates, as was onion tissue maceration ability. The latter property, which is related to pathogenicity for plants, was previously thought to be characteristic only of environmental strains (42), but at present most clinical isolates including epidemic strains readily cause soft rot of onions (8). On the contrary, the antibiotic resistance profiles of the B. cepacia complex isolates were associated with their origins; in fact, clinical isolates generally exhibited resistance to larger numbers of antibiotics and a higher degree of resistance to the single antibiotics tested than the environmental isolates did. In particular, clinical isolates of B. cepacia genomovar III were far more resistant than environmental isolates belonging to the same genomovar. In a recent study, Berg et al. (3) analyzed the antibiotic susceptibilities of Stenotrophomonas maltophilia isolates from CF patients and the environment and found no significant differences between the two groups of isolates, suggesting that S. maltophilia isolates do not acquire antibiotic resistance during antibiotic therapy in the clinical or hospital environment. In the case of B. cepacia complex isolates, our results suggest that antibiotic therapy may play a fundamental role in the acquisition of antibiotic resistance.
When looking for the hemolytic activity of B. cepacia complex isolates, a characteristic considered to be typical of virulent strains, we found that it was partially associated with their origin; i.e., more environmental isolates than clinical isolates showed hemolytic activity. In particular, the percentage of hemolytic environmental B. cepacia genomovar III isolates was markedly higher than the percentage of clinical ones. Furthermore, it is worth noting that almost all isolates belonging to B. ambifaria were hemolytic. The low percentage of clinical isolates positive for hemolytic activity correlates well with earlier findings of Nakazava et al. (31) and Vasil et al. (39), whereas, to our knowledge, this is the first extensive survey of environmental isolates for hemolytic activity. The reason for such marked differences between clinical and environmental isolates is unclear. Vasil et al. (39) observed that the loss of hemolytic activity in clinical isolates belonging to the B. cepacia complex is associated with genetic rearrangements. Therefore, we hypothesize that hemolytic activity may not be a primary virulence factor in patients with CF.
The esmR marker, related to strain transmissibility (27), was found not only in a large proportion (61%) of clinical isolates belonging to B. cepacia genomovar III but also in a large proportion of environmental isolates (15%) of the same genomovar. The presence of BCESM DNA among environmental B. cepacia genomovar III isolates reinforces the hypothesis expressed above that the environment may serve as a reservoir of pathogenic strains belonging to this genomovar. In the case of the cable pilin subunit gene (cblA), only one isolate from a CF patient contained the cblA gene. The low frequency of the cblA gene casts further doubts on its reliability as a marker for virulence or transmissibility, as already pointed out by LiPuma et al. (26).
In summary, our results suggest that there may be differences in the pathogenic potentials of the various species of the B. cepacia complex. Moreover, we observed that some of the candidate determinants related to virulence and transmissibility are not confined solely to clinical isolates but are spread also among environmental isolates belonging to different species of the B. cepacia complex. A fuller appreciation of the species-related differences in pathogenicity and of the risks posed by strains likely to be encountered in the natural environment implies a more in-depth comprehension not only of the biological mechanisms involved in pathogenicity but also of the ecophysiology of each species belonging to the B. cepacia complex.
Acknowledgments
We are grateful to G. Gaslini Children's Hospital for financial support. This work was partially supported by a grant (to S.P.) from the Ligurian Cystic Fibrosis Foundation.
We thank Verena Peggion and Alessia Fiore for many helpful discussions and Gabriella Seri for technical assistance.
REFERENCES
- 1.Agodi, A., E. Mahenthiralingam, M. Barchitta, V. Gianninò, A. Sciacca, and S. Stefani. 2001. Burkholderia cepacia complex infection in Italian patients with cystic fibrosis: prevalence, epidemiology, and genomovar status. J. Clin. Microbiol. 39:2891-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balandreau, J., V. Viallard, B. Cournoyer, T. Coenye, S. Laevens, and P. Vandamme. 2001. Burkholderia cepacia genomovar III is a common plant-associated bacterium. Appl. Environ. Microbiol. 67:982-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg, G., N. Roskot, and K. Smalla. 1999. Genotypic and phenotypic relationships between clinical and environmental isolates of Stenotrophomonas maltophilia. J. Clin. Microbiol. 37:3594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bevivino, A., S. Tabacchioni, L. Chiarini, M. V. Carusi, M. Del Gallo, and P. Visca. 1994. Phenotypic comparison between rhizosphere and clinical isolates of Burkholderia cepacia. Microbiology 140:1069-1077. [DOI] [PubMed] [Google Scholar]
- 5.Bevivino, A., S. Sarrocco, C. Dalmastri, S. Tabacchioni, C. Cantale, and L. Chiarini. 1998. Characterization of a free-living maize-rhizosphere population of Burkholderia cepacia: effect of seed treatment on disease suppression and growth promotion of maize. FEMS Microbiol. Ecol. 27:225-237. [Google Scholar]
- 6.Bevivino, A., C. Dalmastri, S. Tabacchioni, and L. Chiarini. 2000. Efficacy of Burkholderia cepacia MCI 7 in disease suppression and growth promotion of maize. Biol. Fertil. Soils 31:225-231. [Google Scholar]
- 7.Burkholder, W. H. 1950. Sour skin, a bacterial rot of onion bulbs. Phytopathology 40:115-117. [Google Scholar]
- 8.Butler, S. L., C. J. Doherty, J. E. Hughes, J. W. Nelson, and J. R. W. Govan. 1995. Burkholderia cepacia and cystic fibrosis: do natural environments present a potential hazard? J. Clin. Microbiol. 33:1001-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clode, F. E., M. E. Kaufmann, H. Malnick, and T. L. Pitt. 2000. Distribution of genes encoding putative transmissibility factors among epidemic and nonepidemic strains of Burkholderia cepacia from cystic fibrosis patients in the United Kingdom. J. Clin. Microbiol. 38:1763-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coenye, T., J. J. LiPuma, D. Henry, B. Hoste, K. Vandemeulebroucke, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia cepacia genomovar VI, a new member of the Burkholderia cepacia complex isolated from cystic fibrosis patients. Int. J. Syst. Evol. Microbiol. 51:271-279. [DOI] [PubMed] [Google Scholar]
- 11.Coenye, T., E. Mahenthiralingam, D. Henry, J. J. LiPuma, S. Laevens, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex comprising biocontrol and cystic-fibrosis related isolates. Int. J. Syst. Evol. Microbiol. 51:1481-1490. [DOI] [PubMed] [Google Scholar]
- 12.Coenye, T., P. Vandamme, J. R. W. Govan, and J. J. LiPuma. 2001. Taxonomy and identification of the Burkholderia cepacia complex. J. Clin. Microbiol. 39:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalmastri, C., L. Chiarini, C. Cantale, A. Bevivino, and S. Tabacchioni. 1999. Soil type and maize cultivar affect the genetic diversity of maize root-associated Burkholderia cepacia populations. Microb. Ecol. 38:274-283. [DOI] [PubMed] [Google Scholar]
- 14.Di Cello, F., A. Bevivino, L. Chiarini, R. Fani, D. Paffetti, S. Tabacchioni, and C. Dalmastri. 1997. Biodiversity of a Burkholderia cepacia population isolated from maize rhizosphere at different plant growth stages. Appl. Environ. Microbiol. 63:4485-4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiore, A., S. Laevens, A. Bevivino, C. Dalmastri, S. Tabacchioni, P. Vandamme, and L. Chiarini. 2001. Burkholderia cepacia complex: distribution of genomovars among isolates from the maize rhizosphere in Italy. Environ. Microbiol. 3:137-143. [DOI] [PubMed] [Google Scholar]
- 16.Folsom, B. R., P. J. Chapman, and P. H. Pritchard. 1990. Phenol and trichloroethylene degradation by Pseudomonas cepacia G4: kinetics and interactions between substrates. Appl. Environ. Microbiol. 56:1279-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillis, M., T. V. Van, R. Bardin, M. Goor, P. Hebbar, A. Willems, P. Segers, K. Kersters, T. Heulin, and M. P. Fernandez. 1995. Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int. J. Syst. Bacteriol. 45:274-289. [Google Scholar]
- 18.Gonzalez, C. F., and A. K. Vidaver. 1979. Bacteriocin, plasmid and pectolytic diversity in Pseudomonas cepacia of clinical and plant origin. J. Gen. Microbiol. 110:161-170. [DOI] [PubMed] [Google Scholar]
- 19.Govan, J. R. W., J. E. Hughes, and P. Vandamme. 1996. Burkholderia cepacia: medical, taxonomic and ecological issues. J. Med. Microbiol. 45:395-407. [DOI] [PubMed] [Google Scholar]
- 20.Hebbar, K. P., M. H. Martel, and T. Heulin. 1994. Burkholderia cepacia, a plant growth promoting rhizobacterial associate of maize, p. 201-203. In M. H. Ryder, P. M. Stephens, and G. D. Bowen (ed.), Improving plant productivity with rhizosphere bacteria. Division of Soils, Commonwealth Scientific and Industrial Research Organisation, Adelaide, Australia.
- 21.Hebbar, K. P., M. H. Martel, and T. Heulin. 1998. Suppression of pre- and postemergence damping-off in corn by Burkholderia cepacia. Eur. J. Plant Pathol. 104:29-36. [Google Scholar]
- 22.Heungens, K., and J. L. Parke. 2000. Zoospore homing and infections events: effects of the biocontrol bacterium Burkholderia cepacia AMMDR1 on two oomycete pathogens of pea (Pisum sativum L.). Appl. Environ. Microbiol. 66:519-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmes, A., J. Govan, and R. Goldstein. 1998. Agricultural use of Burkholderia (Pseudomonas) cepacia: a threat to human health? Emerg. Infect. Dis. 4:221-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, W. M., S. D. Tyler, and K. R. Rozee. 1994. Linkage analysis of geographic and clinical clusters in Pseudomonas cepacia infections by multilocus enzyme electrophoresis and ribotyping. J. Clin. Microbiol. 32:924-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kilbane, J. J., D. K. Chatterjee, and A. M. Chakrabarty. 1983. Detoxification of 2,4,5-trichlorophenoxyacetic acid from contaminated soil by Pseudomonas cepacia. Appl. Environ. Microbiol. 45:1697-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LiPuma, J. J., T. Spilker, L. H. Gill, P. W. Campbell III, L. Liu, and E. Mahenthiralingam. 2001. Disproportionate distribution of Burkholderia cepacia complex species and transmissibility markers in cystic fibrosis. Am. J. Respir. Crit. Care Med. 164:92-96. [DOI] [PubMed] [Google Scholar]
- 27.Mahenthiralingam, E., D. A. Simpson, and D. P. Speert. 1997. Identification and characterization of a novel DNA marker associated with epidemic Burkholderia cepacia strains recovered from patients with cystic fibrosis. J. Clin. Microbiol. 35:808-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahenthiralingam, E., J. Bischof, S. K. Byrne, C. Radomski, J. E. Davies, Y. Av-Gay, and P. Vandamme. 2000. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J. Clin. Microbiol. 38:3165-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. W. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38:910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLoughlin, T. J., J. P. Quinn, A. Bettermann, and R. Bookland. 1992. Pseudomonas cepacia suppression of sunflower wilt fungus and role of antifungal compounds in controlling the disease. Appl. Environ. Microbiol. 58:1760-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakazava, T., Y. Yamada, and M. Ishibashi. 1987. Characterization of hemolysin in extracellular products of Pseudomonas cepacia. J. Clin. Microbiol. 25:195-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial susceptibility testing, ninth informational supplement. Document M100-S9. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 33.O'Callaghan, E. M., M. S. Tanner, and G. J. Boulnois. 1994. Development of a PCR probe test for identifying Pseudomonas aeruginosa and Pseudomonas (Burkholderia) cepacia. J. Clin. Pathol. 47:222-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saiman, L., D. Schidlow, and A. Smith (ed.). 1994. Concepts in care: microbiology and infectious diseases in cystic fibrosis, vol. V, Suppl. 1. Cystic Fibrosis Foundation, Bethesda, Md.
- 35.Segonds, C., T. Heulin, N. Marty, and G. Chabanon. 1999. Differentiation of Burkholderia species by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene and application to cystic fibrosis isolates. J. Clin. Microbiol. 37:2201-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Speert, D. P. 2001. Understanding Burkholderia cepacia: epidemiology, genomovars, and virulence. Infect. Med. 18:49-56. [Google Scholar]
- 37.Vandamme, P., B. Holmes, M. Vancanneyet, T. Coenye, B. Hoste, R. Coopman, H. Reverts, S. Lauwers, M. Gillis, K. Kersters, and J. R. W. Govan. 1997. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 47:1188-1200. [DOI] [PubMed] [Google Scholar]
- 38.Vandamme, P., E. Mahenthiralingam, B. Holmes, T. Coenye, B. Hoste, P. De Vos, D. Henry, and D. P. Speert. 2000. Identification and population structure of Burkholderia stabilis sp. nov. (formerly Burkholderia cepacia genomovar IV). J. Clin. Microbiol. 38:1042-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vasil, M. L., D. P. Krieg, J. S. Kuhns, J. W. Ogle, V. D. Shortridge, R. M. Ostroff, and A. I. Vasil. 1990. Molecular analysis of hemolytic and phospholipase C activities of Pseudomonas cepacia. Infect. Immun. 58:4020-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wigley, P., and N. F. Burton. 1999. Genotypic and phenotypic relationships in Burkholderia cepacia isolated from cystic fibrosis patients and the environment. J. Appl. Microbiol. 86:460-468. [DOI] [PubMed] [Google Scholar]
- 41.Yohalem, D. S., and J. W. Lorbeer. 1994. Multilocus isoenzyme diversity among strains of Pseudomonas cepacia isolated from decayed onions, soils, and clinical sources. Syst. Appl. Microbiol. 17:116-124. [Google Scholar]
- 42.Yohalem, D. S., and J.W. Lorbeer. 1997. Distribution of Burkholderia cepacia phenotypes by niche, method of isolation and pathogenicity to onion. Ann. Appl. Biol. 130:467-479. [Google Scholar]