Abstract
A bacterium was isolated from the blood culture of a patient with infective endocarditis. The cells were facultative anaerobic, nonsporulating, gram-positive cocci arranged in chains. The bacterium grows on sheep blood agar as alpha-hemolytic, gray colonies of 0.5 to 1 mm in diameter after 24 h of incubation at 37°C in ambient air. Growth also occurs in 10 or 40% bile and on bile esculin agar but not in 6% NaCl. No enhancement of growth is observed in 5% CO2. It is nongroupable with Lancefield groups A, B, C, D, F, or G antisera and is resistant to optochin and bacitracin. The organism is aflagellated and is nonmotile at both 25 and 37°C. It is Voges-Proskauer test positive. It produces leucine arylamidase and β-glucosidase but not catalase, urease, lysine decarboxylase, or ornithine decarboxylase. It hydrolyzes esculin and arginine. It utilizes glucose, lactose, salicin, sucrose, pullulan, trehalose, cellobiose, hemicellulase, mannose, maltose, and starch. 16S rRNA gene sequencing showed that there were 3.6, 3.7, 4.3, 4.7, and 5.9% differences between the 16S rRNA gene sequence of the bacterium and those of Streptococcus gordonii, Streptococcus intermedius, Streptococcus constellatus, Streptococcus sanguis, and Streptococcus anginosus, respectively. The G+C content of it (mean ± standard deviation) was 53.0% ± 2.9%. Based on phylogenetic affiliation, it belongs to the mitis or anginosus group of Streptococcus. For these reasons a new species, Streptococcus sinensis sp. nov., is proposed, for which HKU4 is the type strain. Further studies should be performed to ascertain the potential of this bacterium to become an emerging cause of infective endocarditis.
Since the discovery of PCR and DNA sequencing, comparison of the gene sequences of bacterial species showed that the 16S rRNA gene is highly conserved within a species and among species of the same genus, and hence it can be used as the new gold standard for speciation of bacteria. Using this new standard, phylogenetic trees, based on base differences between species, are constructed, and bacteria are classified and reclassified into new genera (9, 10). Furthermore, noncultivable organisms and organisms with ambiguous biochemical profiles can be classified and identified (11, 12). Recently we have reported the use of this technique for the identification of bacterial strains with ambiguous biochemical profiles (14, 15, 16, 19, 23), species that are rarely encountered clinically (6, 18, 20), and a bacterium that is noncultivable (2); discovery of a novel clinical syndrome (22) and a novel genus and species (24); and characterization of beta-hemolytic Lancefield group G streptococcal bacteremia (21).
In this study, we report the isolation of a bacterial strain from the blood culture of a patient with infective endocarditis. The strain, named HKU4, exhibited phenotypic characteristics that do not fit into patterns of any known species. 16S rRNA gene sequencing showed that there was only 96.4% base identity between the 16S rRNA gene of HKU4 and that of the most closely related species, Streptococcus gordonii. On the basis of these studies we propose a new species, Streptococcus sinensis sp. nov., to describe this bacterium.
MATERIALS AND METHODS
Patient and microbiological methods.
All clinical data were collected prospectively as described in a previous publication by members of our group (7). The BACTEC 9240 blood culture system (Becton Dickinson, Cockeysville, Md.) was used. The bacterium was identified by standard conventional biochemical methods (8). Lancefield serogrouping was performed using Streptex (Murex Biotech Ltd., Dartford, United Kingdom) according to the manufacturer's instructions. In addition, the Vitek System (GPI) (bioMerieux Vitek Hazelwood, Mo.), the API system (20 STREP) (bioMerieux Vitek), and the ATB Expression system (ID32 STREP) (bioMerieux Vitek) were used for the identification of the bacterial isolate in this study. All tests were performed in triplicate with freshly prepared media on separate occasions. Antimicrobial susceptibility was tested by the E-test for penicillin and the Kirby Bauer disk diffusion method for the other antibiotics, and results were interpreted according to the NCCLS criteria for alpha-hemolytic streptococci.
SEM.
Bacterial cells were washed twice using milli-Q water. A suspension of the bacterium was settled onto a polycarbonate membrane (Nuclepore) with a pore size of 5 μm for 5 min. The membrane was fixed in 2.5% (wt/vol) glutaraldehyde for 1 h and washed once in 0.1 M sodium cacodylate buffer. Fixed material was dehydrated through a graded ethanol series from 30 to 90% in 20% steps, followed by two changes of absolute ethanol. Each of the stepwise changes was for 15 min. Dehydrated material in absolute ethanol was critical point dried in a BAL-TEC CPD O30 Critical Point Drier using carbon dioxide as the drying agent. Critical dried material was mounted onto an aluminum stub and coated with palladium in the BAL-TEC SCD 005 scanning electron microscopy (SEM) coating system. Coated material was examined in a Leica Cambridge Stereoscan 440 SEM operating at 12 kV, and the specimen stage was tilted at 0°.
Extraction of bacterial DNA for 16S rRNA gene sequencing.
The bacterial DNA extraction method was modified from our previously published protocol (17). Briefly, 80 μl of NaOH (0.05 M) was added to 20 μl of bacterial cells suspended in distilled water, and the mixture was incubated at 60°C for 45 min, followed by addition of 6 μl of Tris-HCl (pH 7.0), achieving a final pH of 8.0. The resultant mixture was diluted 100×, and 5 μl of the diluted extract was used for PCR.
PCR, gel electrophoresis, and 16S rRNA gene sequencing.
PCR amplification and DNA sequencing of the 16S rRNA genes were performed as described in previous publications by members of our group (21, 24). Briefly, DNase I-treated distilled water and PCR master mix (which contains deoxynucleoside triphosphates [dNTPs], PCR buffer, and Taq polymerase) were used in all PCRs by adding 1 U of DNase I (Pharmacia, Uppsala, Sweden) to 40 μl of distilled water or PCR master mix, incubating the mixture at 25°C for 15 min and subsequently at 95°C for 10 min to inactivate the DNase I. The bacterial DNA extracts and the control were amplified with 0.5 μM primers (LPW55 5′-AGTTTGATCCTGGCTCAG-3′ and LPW205 5′-CTTGTTACGACTTCACCC-3′) (Gibco BRL, Rockville, Md.). The PCR mixture (50 μl) contained bacterial DNA, PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 2 mM MgCl2, and 0.01% gelatin), a 200 μM concentration of each dNTP, and 1.0 U of Taq polymerase (Boehringer Mannheim, Mannheim, Germany). The mixtures were amplified in 40 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min, with a final extension at 72°C for 10 min in an automated thermal cycler (Perkin-Elmer Cetus, Gouda, The Netherlands). DNase I-treated distilled water was used as the negative control. Ten microliters of each amplified product was electrophoresed in 1.0% (wt/vol) agarose gel, with a molecular size marker (Lambda DNA AvaII digest; Boehringer Mannheim, Germany) in parallel. Electrophoresis in Tris-borate-EDTA buffer was performed at 100 V for 1.5 h. The gel was stained with ethidium bromide (0.5 μg/ml) for 15 min, rinsed, and photographed under UV light illumination.
The PCR products were gel purified using the QIAquick PCR purification kit (QIAgen, Hilden, Germany). Both strands of the PCR products were sequenced twice with an ABI 377 automated sequencer according to the manufacturer's instructions (Perkin-Elmer, Foster City, Calif.), using the PCR primers (LPW55 and LPW205) and the additional sequencing primers LPW304 5′-CTTACCATGCAGTAAGAT-3′ and LPW306 5′-TGAGATGTTGGGTTAAGT-3′. The sequence of the PCR products were compared with known 16S rRNA gene sequences in the GenBank by multiple sequence alignment using the CLUSTAL W program (13).
Determination of G+C content.
Genomic DNA was prepared according to a published protocol (1), and the G+C content was determined by thermal denaturation (4). Briefly, the temperature of the genomic DNA in SSC (0.15 M NaCl with 0.015 M sodium citrate) buffer (25 μg/ml) was increased slowly (0.5°C/min) from 25°C, and the absorbance of the solution at 260 nm was monitored continuously against a blank containing SSC buffer only. The Tm of the DNA is defined as the temperature at 50% hyperchromicity. The G+C content of the genomic DNA was calculated by the following formula: percent (G+C) = 2.44Tm − 169.
Phylogenetic characterization.
The phylogenetic relationships of strain HKU4 to the other Streptococcus species was determined using the PileUp method with GrowTree (Genetics Computer Group, Inc.). A total of 1,373 nucleotide positions were included in the analysis.
Nucleotide sequence accession number.
The 16S rRNA gene sequence of HKU4 has been lodged within the GenBank sequence database under accession no. AF432856.
RESULTS
Patient.
A 42-year-old Chinese woman was admitted to hospital in November 2000 because of fever, chills, and rigor for 1 week. She had mitral regurgitation as a result of chronic rheumatic heart disease. Examination showed an oral temperature of 38.5°C, a grade 3/6 pansystolic murmur over the cardiac apex radiating to the left axilla (compatible with her mitral regurgitation), and a 3-cm erythematous nodule over the left palm (compatible with Janeway lesion due to septic embolism). The hemoglobin level was 11.5 g/dl, total white cell count, 7.5 × 109/liter, with a neutrophil count of 6.0 × 109/liter, lymphocyte count of 1.0 × 109/liter, and monocyte count of 0.4 × 109/liter, and the platelet count was 243 × 109/liter. The serum albumin, globulin, creatinine, urea, bilirubin, alkaline phosphatase, alanine aminotransferase, and aspartate aminotransferase were within normal limits. The erythrocyte sedimentation rate was 37 mm/h. A presumptive diagnosis of infective endocarditis was made. Three sets of blood cultures were obtained, and therapy with empirical intravenous ampicillin and gentamicin was commenced. Both transthoracic and transesophageal echocardiograms did not reveal any vegetations. On day 1 postincubation, all three pairs of blood cultures turned positive with the same gram-positive coccus (strain HKU4). The patient responded to a total of 4 weeks of ampicillin and 2 weeks of gentamicin.
Phenotypic characteristics.
Strain HKU4 is a gram-positive, non-spore-forming coccus arranged in chains. It grows on sheep blood agar as alpha-hemolytic, gray colonies of 0.5 to 1 mm in diameter after 24 h of incubation at 37°C in ambient air. No enhancement of growth is observed in 5% CO2. It also grows in a microaerophilic or anaerobic environment, in 10 or 40% bile, and on bile esculin agar but does not grow in 6% NaCl. It is nongroupable with Lancefield groups A, B, C, D, F, or G antisera, is resistant to optochin and bacitracin, and is nonmotile at both 25 and 37°C. The biochemical profile of strain HKU4 is shown in Table 1. It is Voges-Proskauer test positive. It produces leucine arylamidase and β-glucosidase but not catalase, urease, lysine decarboxylase, or ornithine decarboxylase. It hydrolyzes esculin and arginine. It utilizes glucose, lactose, salicin, sucrose, pullulan, trehalose, cellobiose, hemicellulase, mannose, maltose, and starch. The Vitek system (GPI) showed that it was 99% Streptococcus intermedius; the API system (20 STREP) showed that it was 51.3% Streptococcus intermedius, 25.8% Lactococcus lactis, and 21.5% Streptococcus anginosus; and the ATB Expression system (ID32 STREP) showed that it was 99.9% Streptococcus anginosus. It is sensitive to penicillin (MIC = 0.064 μg/ml), ceftriaxone, cefepime, clindamycin, erythromycin, ofloxacin, tetracycline, and vancomycin.
TABLE 1.
Biochemical profile of strain HKU4 by conventional biochemical tests and Vitek GPI, API 20 STREP, and ATB ID32 STREP systems
| Biochemical reaction or enzyme | Result by testing method
|
|||
|---|---|---|---|---|
| Conventional | Vitek GPI | API 20 STREP | ATB ID32 STREP | |
| Catalase | − | − | ||
| Resistance to bacitracin | + | + | ||
| Resistance to optochin | + | + | ||
| Growth in 6% NaCl | − | − | ||
| Growth in 10% bile | + | |||
| Growth in 40% bile | + | + | ||
| Esculin hydrolysis | + | + | + | |
| Hippurate hydrolysis | − | − | ||
| Arginine hydrolysis | + | + | + | + |
| Lysine decarboxylase | − | |||
| Ornithine decarboxylase | − | |||
| Urease | − | − | − | |
| Voges-Proskauer test | + | + | + | |
| Tetrazolium reduction | + | |||
| Resistance to novobiocin | + | + | ||
| Resistance to polymyxin B | + | |||
| Utilization of: | ||||
| Hemicellulase | + | |||
| Dextrose | + | |||
| Lactose | + | + | + | + |
| Mannitol | − | − | − | − |
| Raffinose | − | − | − | − |
| Salicin | + | + | ||
| Sorbitol | − | − | − | − |
| Sucrose | + | + | + | |
| Trehalose | + | + | + | + |
| Arabinose | − | − | − | − |
| Pyruvate | − | |||
| Pullulan | + | + | ||
| Inulin | − | − | − | |
| Melibiose | − | − | − | |
| Melezitose | − | − | ||
| Cellobiose | + | + | ||
| Ribose | − | − | − | − |
| Xylose | − | − | ||
| D-glucose | + | |||
| D-mannose | + | |||
| Maltose | + | |||
| Starch | + | |||
| Glycogen | − | − | ||
| Arabitol | − | |||
| Methyl-B-d-glucopyranoside | + | |||
| Tagatose | − | |||
| Cyclodextrin | − | |||
| Pyrrolidonylarylamidase | − | − | ||
| α-galactosidase | − | − | ||
| β-glucuronidase | − | − | ||
| β-galactosidase | − | − | ||
| Leucine arylamidase | + | |||
| β-glucosidase | + | |||
| α-galactosidase | − | |||
| Alanine-phenylalanine-proline arylamidase | − | |||
| Acetyl-β-glucosaminidase | − | |||
| Glycyl-tryptophane arylamidase | − | |||
| β-mannosidase | − | |||
| Nitrate reduction | − | |||
| Alkaline phosphatase | − | − | − | |
| Identification | 99% S. intermedius | 51.3% S. intermedius, 25.8% L. lactis, 21.5% S. anginosus | 99.9% S. anginosus | |
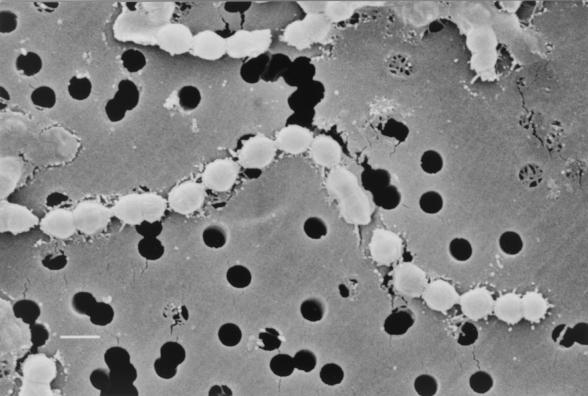
SEM.
A scanning electron micrograph of Streptococcus sinensis is shown in Fig. 1. Bacterial cells were cocci arranged in chains.
FIG. 1.
Scanning electron micrograph of S. sinensis. The bacterium is arranged in chains and is aflagellated. Cells vary in diameter from 0.82 to 0.98 μm. Bar, 1 μm.
Molecular characterization by 16S rRNA gene sequencing, determination of G+C content, and phylogenetic characterization.
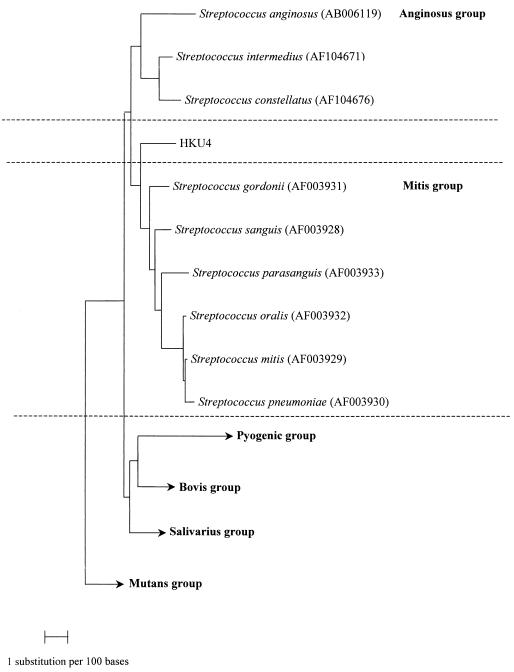
PCR of the 16S rRNA gene of strain HKU4 showed a band at 1,512 bp. There was 0.2% difference between the 16S rRNA gene sequence of strain HKU4 and that of a 16S rRNA gene sequence amplified from the aortic valve of a patient with infective endocarditis, recently deposited in GenBank (GenBank accession no. AY049738); 3.6% difference between the 16S rRNA gene sequence of strain HKU4 and that of S. gordonii (GenBank accession no. AF003931); 3.7% difference between the 16S rRNA gene sequence of strain HKU4 and that of S. intermedius (GenBank accession no. AF104671); 4.3% difference beween the 16S rRNA gene sequence of strain HKU4 and that of S. constellatus (GenBank accession no. AF104676); 4.7% difference between the 16S rRNA gene sequence of strain HKU4 and that of Streptococcus sanguis (GenBank accession no. AF003928); and 5.9% difference between the 16S rRNA gene sequence of strain HKU4 and that of S. anginosus (GenBank accession no. AB006119). The G+C content of strain HKU4 (mean ± standard deviation) was 53.0% ± 2.9%. Based on phylogenetic affiliation, it belongs to the mitis or anginosus group of Streptococcus (Fig. 2).
FIG. 2.
Phylogenetic tree showing the relationship of S. sinensis sp. nov. to the related Streptococcus species. A total of 1,373 nucleotide positions in each 16S rRNA gene were included in the analysis. The scale bar indicates the estimated number of substitutions per 100 bases using the Jukes-Cantor correction. Names and accession numbers are given as cited in the GenBank database.
DISCUSSION
We report the isolation of HKU4 from the blood cultures of a Chinese patient with chronic rheumatic heart disease. The clinical significance of the bacterium was made evident by its isolation from multiple blood cultures in a patient with infective endocarditis, fulfilling one major (isolation of the bacterium from the patient's blood culture) and three minor (fever, preexisting chronic rheumatic heart disease, and thromboembolic phenomenon) criteria (3). The 16S rRNA gene of HKU4 exhibited less than 97% nucleotide identity with the 16S rRNA gene of all previously described bacterial strains of the Streptococcus genus. The most closely related species is S. gordonii, a viridans Streptococcus that belongs to the mitis group of the Streptococcus genus (5).
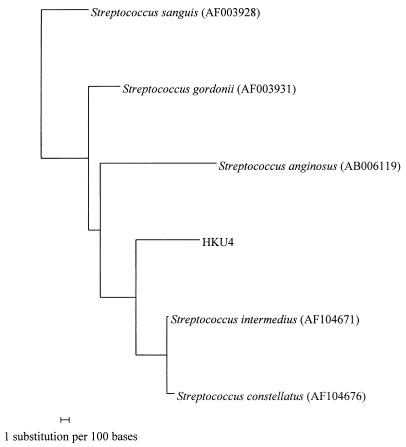
Further phylogenetic studies would be necessary for the determination of the exact phylogenetic position of HKU4. Although the most closely related species of HKU4 based on 16S rRNA gene sequences is S. gordonii, there is evidence showing that it may not in fact be the closest relative. First, there are only very small differences between the 16S rRNA gene sequences of HKU4 and those of S. gordonii (3.6%), S. intermedius (3.7%), S. constellatus (4.3%), S. sanguis (4.7%), and S. anginosus (5.9%). Second, when the phylogenetic relationship of the hypervariable region of the 16S rRNA gene of HKU4 (bases 55 to 231) is determined (Fig. 3), the most closely related species is S. intermedius, whereas S. gordonii is more distantly related. Third, phenotypically the species most closely resembling HKU4 is S. intermedius (a member of the anginosus group) (Table 2). The only difference between S. intermedius and HKU4 is that most S. intermedius isolates, but not HKU4, produce alkaline phosphatase. In fact, HKU4 was identified as S. intermedius at 99 and 57.3% confidence by the Vitek (GPI) and API (20 STREP) systems, whereas none of the three commercial systems identified HKU4 as S. gordonii. Further experiments, such as comparison of the sequences of other rRNA genes, as well as other essential genes, among these Streptococcus species and HKU4 would be necessary in order to delineate the exact phylogenetic relationships of HKU4 with the other streptococci.
FIG. 3.
Phylogenetic tree showing the relationship of S. sinensis sp. nov. to the five closely related Streptococcus species using the sequence of the hypervariable region in the 16S rRNA genes. One hundred seventy-seven nucleotide positions in each 16S rRNA gene were included in the analysis. The scale bar indicates the estimated number of substitutions per 100 bases using the Jukes-Cantor correction. Names and accession numbers are given as cited in the GenBank database.
TABLE 2.
Comparison of the major phenotypic characteristics among S. intermedius, S. anginosus, S. constellatus, S. gordonii, S. sanguis, and HKU4
| Phenotypic characteristic | Result for Streptococcus species
|
|||||
|---|---|---|---|---|---|---|
| S. intermedius | S. anginosus | S. constellatus | S. gordonii | S. sanguis | HKU4 | |
| Hemolysis | α, β | α, β | α, β | α | α | α |
| Lancefield serogroup | −, F, G | −, A, C, F, G | −, A, G | − | − | − |
| Hydrolysis of: | ||||||
| Esculin | + | + | + | + | Va | + |
| Hippurate | − | − | − | − | − | − |
| Arginine | + | + | + | + | V | + |
| Voges-Proskauer test | + | + | + | −b | − | + |
| Pyrrolidonylaryl-amidase | − | − | − | − | − | − |
| α-galactosidase | − | + | − | V | V | − |
| β-glucuronidase | − | − | − | − | − | − |
| β-galactosidase | − | − | − | + | − | − |
| Alkaline phosphatase | + | + | + | + | V | − |
| Utilization of: | ||||||
| Ribose | − | − | − | − | − | − |
| Mannitol | − | + | − | − | − | − |
| Sorbitol | − | − | − | − | − | − |
| Lactose | + | + | − | + | + | + |
| Trehalose | + | + | + | + | + | + |
| Inulin | − | − | − | + | V | − |
| Raffinose | − | + | − | − | V | − |
V, variable.
The biochemical test results of S. intermedius, S. anginosus, S. constellatus, S. gordonii, and S. sanguis that are different from those of HKU4 are shown in bold.
S. sinensis may be an emerging cause of infective endocarditis. First, besides isolating HKU4 in our patient with infective endocarditis, 16S rRNA gene sequence with only three base differences from HKU4 have been detected in the aortic valve of a patient with infective endocarditis (GenBank accession no. AY049738). This shows that S. sinensis is probably not one of those bacteria that are isolated only once in humans. Furthermore, its presence in both Asia and Europe implies that S. sinensis may be a bacterium of global importance. Second, as HKU4 was identified as S. intermedius or S. anginosus by commercial kits, some of the S. intermedius or S. anginosus strains may actually be S. sinensis. Amplification of the 16S rRNA genes of these isolates using S. sinensis-specific primers would determine the proportion of S. intermedius and S. anginosus strains that are actually S. sinensis.
Description of S. sinensis, sp. nov.
Streptococcus means chain-forming coccus; sinensis, in honor of China, indicates the place where the bacterium was discovered.
Its cells are facultative anaerobic, nonsporulating, gram-positive cocci arranged in chains. It grows on sheep blood agar as alpha-hemolytic, gray colonies of 0.5 to 1 mm in diameter after 24 h of incubation at 37°C in ambient air. Growth also occurs in 10 or 40% bile and on bile esculin agar but not in 6% NaCl. No enhancement of growth is observed in 5% CO2. It is nongroupable with Lancefield groups A, B, C, D, F, or G antisera and is resistant to optochin and bacitracin. The organism is aflagellated and is nonmotile at both 25 and 37°C. It is Voges-Proskauer test positive. It produces leucine arylamidase and β-glucosidase but not catalase, urease, lysine decarboxylase, and ornithine decarboxylase. It hydrolyzes esculin and arginine. It utilizes glucose, lactose, salicin, sucrose, pullulan, trehalose, cellobiose, hemicellulase, mannose, maltose, and starch. (Table 1). The moles percent G+C content of the DNA of the strain is 53.0% ± 2.9%. The organism was isolated from the blood culture of a patient with infective endocarditis. The type strain of S. sinensis is strain HKU4.
Acknowledgments
This work was partly supported by the University Development Fund, University Research Grant Council, and the Committee for Research and Conference Grant, The University of Hong Kong.
REFERENCES
- 1.Ausubel, F. M., R. Brent, and R. E. Kingston (ed.). 1998. Preparation of genomic DNA from bacteria, p. 2.4.1-2.4.2. In Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Cheuk, W., P. C. Y. Woo, K. Y. Yuen, P. H. Yu, and J. K. C. Chan. 2001. Intestinal inflammatory pseudotumor with regional lymph node involvement: identification of a new bacterium as the etiologic agent. J. Pathol. 192:289-292. [DOI] [PubMed] [Google Scholar]
- 3.Durack, D. T., A. S. Lukes, and D. K. Bright. 1994. New criteria for diagnosis of infective endocarditis: utilization of specific echocardiographic findings. Duke Endocarditis Service. Am. J. Med. 96:200-209. [DOI] [PubMed] [Google Scholar]
- 4.Goodfellow, M. 1985. Chemical methods in bacterial systematics, p. 67-93. Academic Press, London, United Kingdom.
- 5.Kawamura, Y., X. G. Hou, F. Sultana, H. Miura, and T. Ezaki. 1995. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int. J. Syst. Bacteriol. 45:406-408. [DOI] [PubMed] [Google Scholar]
- 6.Lau, S. K. P., P. C. Y. Woo, J. L. L. Teng, K. W. Leung, and K. Y. Yuen. Identification by 16S ribosomal RNA gene sequencing of Arcobacter butzleri bacteraemia in a patient with acute gangrenous appendicitis. Mol. Pathol., in press. [DOI] [PMC free article] [PubMed]
- 7.Luk, W. K., S. S. Wong, K. Y. Yuen, P. L. Ho, P. C. Y. Woo, R. A. Lee, and P. Y. Chau. 1998. Inpatient emergencies encountered by an infectious disease consultative service. Clin. Infect. Dis. 26:695-701. [DOI] [PubMed] [Google Scholar]
- 8.Murray, P. R., E. J. Baro, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.). 1999. Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 9.Olsen, G. J., R. Overbeek, N. Larsen, T. L. Marsh, M. J. McCaughey, M. A. Maciukenas, W. M. Kuan, T. J. Macke, Y. Xing, and C. R. Woese. 1992. The ribosomal database project. Nucleic Acids Res. 20(Suppl.):2199-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olsen, G. J., and C. R. Woese. 1993. Ribosomal RNA: a key to phylogeny. FASEB J. 7:113-123. [DOI] [PubMed] [Google Scholar]
- 11.Relman, D. A., J. S. Loutit, T. M. Schmidt, S. Falkow, and L. S. Tompkins. 1990. The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N. Engl. J. Med. 323:1573-1580. [DOI] [PubMed] [Google Scholar]
- 12.Relman, D. A., T. M. Schmidt, R. P. MacDermott, and S. Falkow. 1992. Identification of the uncultured bacillus of Whipple's disease. N. Engl. J. Med. 327:293-301. [DOI] [PubMed] [Google Scholar]
- 13.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woo, P. C. Y., E. Y. L. Cheung, K. W. Leung, and K. Y. Yuen. 2001. Identification by 16S ribosomal RNA gene sequencing of an Enterobacteriaceae species with ambiguous biochemical profile from a renal transplant recipient. Diagn. Microbiol. Infect. Dis. 39:85-93. [DOI] [PubMed] [Google Scholar]
- 15.Woo, P. C. Y., A. M. Y. Fung, S. S. Y. Wong, H. W. Tsoi, and K. Y. Yuen. 2001. Isolation and characterization of a Salmonella enterica serotype typhi variant and its clinical and public health implications. J. Clin. Microbiol. 39:1190-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo, P. C. Y., P. K. L. Leung, K. W. Leung, and K. Y. Yuen. 2000. Identification by 16S ribosomal RNA gene sequencing of an Enterobacteriaceae species from a bone marrow transplant recipient. Mol. Pathol. 53:211-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woo, P. C. Y., C. Y. Lo, S. K. Lo, H. Siau, J. S. M. Peiris, S. S. Y. Wong, W. K. Luk, T. M. Chan, W. W. Lim, and K. Y. Yuen. 1997. Distinct genotypic distributions of cytomegalovirus (CMV) envelope glycoprotein in bone marrow and renal transplant recipients with CMV disease. Clin. Diagn. Lab. Immunol. 4:515-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woo, P. C. Y., H. W. Tsoi, K. W. Leung, P. N. L. Lum, A. S. P. Leung, C. H. Ma, K. M. Kam, and K. Y. Yuen. 2000. Identification of Mycobacterium neoaurum isolated from a neutropenic patient with catheter-related bacteremia by 16S rRNA sequencing. J. Clin. Microbiol. 38:3515-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woo, P. C. Y., A. S. P. Leung, K. W. Leung, and K. Y. Yuen. 2001. Identification of slide-coagulase positive, tube-coagulase negative Staphylococcus aureus by 16S ribosomal RNA gene sequencing. Mol. Pathol. 54:244-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woo, P. C. Y., K. T. K. Chong, K. W. Leung, T. L. Que, and K. Y. Yuen. 2001. Identification of Arcobacter cryaerophilus isolated from a traffic accident victim with bacteraemia by 16S ribosomal RNA gene sequencing. Diagn. Microbiol. Infect. Dis. 40:125-127. [DOI] [PubMed] [Google Scholar]
- 21.Woo, P. C. Y., A. M. Y. Fung, S. K. P. Lau, S. S. Y. Wong, and K. Y. Yuen. 2001. Group G beta-hemolytic streptococcal bacteremia characterized by 16S ribosomal RNA gene sequencing. J. Clin. Microbiol. 39:3147-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woo, P. C. Y., J. H. C. Li, W. M. Tang, and K. Y. Yuen. 2001. Acupuncture mycobacteriosis. N. Engl. J. Med. 345:842-843. [DOI] [PubMed] [Google Scholar]
- 23.Woo, P. C. Y., A. M. Y. Fung, S. K. P. Lau, and K. Y. Yuen. 2002. Identification by 16S ribosomal RNA gene sequencing of Lactobacillus salivarius bacteremic cholecystitis. J. Clin. Microbiol. 40:265-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuen, K. Y., P. C. Y. Woo, J. L. L. Teng, K. W. Leung, M. K. M. Wong, and S. K. P. Lau. 2001. Laribacter hongkongensis gen. nov., sp. nov., a novel gram-negative bacterium isolated from a cirrhotic patient with bacteremia and empyema. J. Clin. Microbiol. 39:4227-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]