Over the past decade, PCR-based methodologies have been introduced to complement or even replace histopathologic study of biopsy specimens for the diagnosis of Whipple's disease (12). However, positive PCR results have been reported on testing small-bowel and saliva specimens from asymptomatic patients (3, 4, 11). Although these results have not been independently confirmed (8), they have nonetheless led to a poor predictive diagnostic value for these house-made PCR assays that lack of controls. In response to this concern, we evaluated a quantitative real-time PCR (LightCycler; Roche, Mannheim, Germany) combining rapid cycling with fluorescence-based identification of PCR products in glass capillaries (13, 14) for the diagnosis of Whipple's disease with two pairs of primers targeting different genes. We determined a diagnostic cutoff value based on evaluating the detection level by a titration of cells infected with Tropheyma whipplei, the agent of the disease (6, 10) and negative controls (uninfected cells). A positive PCR result was defined by a fluorescent signal equivalent to that derived from at least 10 copies of standard control DNA (Fig. 1). The assay was evaluated on seven frozen duodenal biopsies, one frozen lymph node biopsy and one frozen cardiac valve obtained from nine patients with histologically proven Whipple's disease (9) and a control group composed of 150 duodenal biopsy specimens, 20 lymph node biopsy samples, and 100 saliva specimens from people with no suspicion of Whipple's disease.
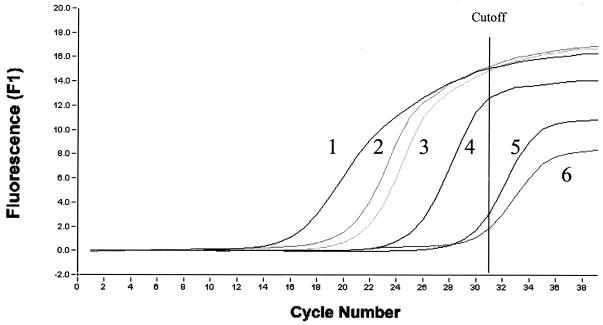
FIG. 1.
Detection of T. whipplei DNA using the LightCycler technique performed with 16S-23S rDNA(ITS)-derived primers. The amplification of copies of the DNA of 104 copies from DNA standard T. whipplei (1), positive duodenal biopsy from patient 2 (2), positive duodenal biopsy from patient 6 (3), 10 copies from DNA standard T. whipplei (4), a negative duodenal biopsy from control group (5), uninfected cells using as negative control (6).
We used primers tws3f and tws4r to target a 489-bp fragment of the 16S-23S ribosomal DNA intergenic spacer (ITS) (5) and primers TWRPOB.F and TWRPOB.R to target a 650-bp fragment of the β-subunit of the RNA polymerase gene (rpoB) (1). Either 25 mg of tissue or 1 ml of aspirate from each sample was used for DNA extraction, as previously described (3). Mixes were prepared by following the manufacturer's instructions (FastStart DNA Master SYBR Green; Roche). The LightCycler PCR result, with both primer pairs, was positive for each of the nine patients with Whipple's disease, with each yielding similar quantitative results (Table 1). All the 150 duodenal biopsy samples, 20 lymph node biopsy samples, and 100 saliva samples from the control group failed to yielded a significant PCR product signal.
TABLE 1.
Number of DNA copies detected for nine patients with Whipple's disease
| Patient/sex/agea | Sample | Number of copies detected
|
|
|---|---|---|---|
| ITS | rpoB | ||
| 1/M/48 | Lymph node tissue | 150,000 | 170,000 |
| 2/M/62 | Duodenal biopsy specimen | 30,000 | 58,000 |
| 3/M/46 | Duodenal biopsy specimen | 300 | 120 |
| 4/F/33 | Duodenal biopsy specimen | 100 | 100 |
| 5/M/57 | Duodenal biopsy specimen | 20,000 | 15,000 |
| 6/F/71 | Duodenal biopsy specimen | 10,000 | 6,500 |
| 7/M/61 | Duodenal biopsy specimen | 500 | 500 |
| 8/M/66 | Duodenal biopsy specimen | 900 | 1,000 |
| 9/M/63 | Cardiac valve specimen | 900 | 1,200 |
M, male; F, female. Ages are given in years.
The epidemiology of Whipple's disease remains unclear. The bacterium may well be present in the environment (7); thus humans may be regularly exposed to, or colonized by, but not necessarily infected with, T. whipplei. In such circumstances, the potential ability of quantitative PCRs to differentiate between environmental contamination or low-level colonization and the higher concentration of bacteria associated with clinical manifestation becomes apparent. Thus, introduction of an evaluated and commercially available LightCycler PCR procedure seems well adapted to the diagnosis of Whipple's disease.
Acknowledgments
We thank Richard Birtles for reviewing the manuscript.
REFERENCES
- 1.Drancourt, M., A. Carlioz, and D. Raoult. 2001. rpoB sequence analysis of cultured Tropheryma whippelii. J. Clin. Microbiol. 39:2425-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dutly, F., and M. Altwegg. 2001. Whipple's disease and “Tropheryma whippelii.” Clin. Microbiol. Rev. 14:561-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dutly, F, P. Hinrickson, T. Seidel, S. Morgenegg, M. Altwegg, and P. Bauerfeind. 2000. Tropheryma whippelii DNA in saliva of patients without Whipple's disease. Infection 28:219-222. [DOI] [PubMed] [Google Scholar]
- 4.Ehrbar, H. U., P. Bauerfeind, F. Dutly, H. R. Koelz, and M. Altwegg. 1999. PCR-positive tests for Tropheryma whippelii in patients without Whipple's disease. Lancet 353:2214.. [DOI] [PubMed] [Google Scholar]
- 5.Hinrickson, H. P., F. Dutly, and M. Altwegg. 1999. Homogeneity of 16S-23S ribosomal intergenic spacer regions of Tropheryma whippelli in Swiss patients with Whipple's disease. J. Clin. Microbiol. 37:152-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.La Scola, B., F. Fenollar, P. E. Fournier, M. Altwegg, M. N. Mallet, and D. Raoult. 2001. Description of Tropheryma whipplei gen. nov., sp. nov., the Whipple's disease bacillus. Int. J. Syst. Evol. Microbiol. 51:1471-1479. [DOI] [PubMed] [Google Scholar]
- 7.Maiwald, M., F. Schuhmacher, H. J. Ditton, and A. von Herbay. 1998. Environmental occurrence of the Whipple's disease bacterium (Tropheryma whippelii). Appl. Environ. Microbiol. 64:760-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maiwald, M., A. von Herbay, D. H. Persing, P. Schawn Mitchell, M. F. Abdelmalek, J. N. Thorvilson, D. N. Fredricks, and D. A. Relman, 2001. Tropheryma whippelii DNA is rare in the intestinal mucosa of patients without other evidence of Whipple's disease. Ann. Intern. Med. 134:115-119. [DOI] [PubMed] [Google Scholar]
- 9.Pron, B., C. Poyart, T. Abachin, C. Fest, C. Belanger, P. Bonnet, J. F. Capelle, A. Bretagne, L. Fabianek, H. Girard, H. Hagège, and P. Berche. 1999. Diagnosis and follow-up of Whipple's disease by amplification of the 16S rRNA gene of Tropheryma whippelii. Eur. J. Clin. Microbiol. Infect. Dis. 18:62-65. [DOI] [PubMed] [Google Scholar]
- 10.Raoult, D., M. L. Birg, B. La Scola, P. E. Fournier, M. Enea, H. Lepidi, V. Roux, J. C. Piette, F. Vandenesch, D. Vital-Durand, and T. J. Marrie. 2000. Cultivation of the bacillus of Whipple's disease. N. Engl. J. Med. 34:620-625. [DOI] [PubMed] [Google Scholar]
- 11.Street, S., H. D. Donoghue, and G. H. Neild. 1999. Tropheryma whippelii DNA in saliva of healthy people. Lancet 354:1178-1179. [DOI] [PubMed] [Google Scholar]
- 12.Wilson, K. H., R. Blitchington, R. Frothingham, and J. A. Wilson. 1991. Phylogeny of the Whipple's disease-associated bacterium. Lancet 338:474-475. [DOI] [PubMed] [Google Scholar]
- 13.Wittwer, C. T., M. G. Hermann, A. A. Moss, and R. P. Rasmussen. 1997. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques 22:134-138. [DOI] [PubMed] [Google Scholar]
- 14.Wittwer, C. T., K. M. Ririe, R. V. Andrew, D. A. David, R. A. Gundry, and U. J. Balis. 1997. The LightCycler: a microvolume multisample fluorimeter with rapid temperature control. BioTechniques 22:176-181. [DOI] [PubMed] [Google Scholar]