Several reports describe the use of Qiagen columns as part of a Legionella PCR (1, 2). However, workers in two laboratories in The Netherlands have found that the Qiagen columns are contaminated with Legionella species.
Nucleic acid sample preparation kits are commercially available from many different suppliers. Some of the most commonly used are the silica-based gel membrane or glass fiber filter columns of Qiagen, which specifically bind DNA while contaminating RNA and proteins are removed. They are widely used to extract DNA from a variety of clinical specimens such as: sputum samples, bronchoalveolar lavage (BAL) samples, aspirates, biopsy specimens, and blood and tissue samples. Recovery of DNA from the columns is highly efficient, and PCR inhibitors are effectively reduced.
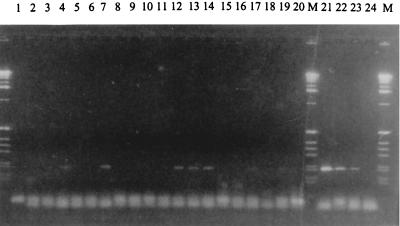
In the Tilburg laboratory, the Qiagen columns (QIAamp DNA mini kit; Qiagen, Hilden, Germany) have been used for the extraction of DNA from sputum samples or BAL samples, to be tested by the Legionella PCR/probe assay. Negative controls, consisting of water (guaranteed DNA and RNA free; Sigma), were processed after every four clinical samples. At one point, it was found that some of the negative controls were positive for Legionella species other than Legionella pneumophila. The occurrence of false positivity coincided with the use of a new batch of QIAamp columns. When water was processed in 20 columns, a Legionella-positive signal was seen after PCR in 4 of 20 columns but never in the unprocessed water (Fig. 1). All the individual components of the Qiagen kit were tested, showing that the source of the contamination was in the columns. The Qiagen distributor in the Netherlands provided the laboratory with 3 other batches of columns. A total 20 columns of each batch were tested. With one batch, all 20 columns tested were negative, but the other 2 batches were contaminated with Legionella DNA.
FIG. 1.
Agarose gel containing the PCR products from DNA- and RNA-free water, processed by QIAmp DNA extraction (lanes 1 to 20), positive controls (lanes 21 to 23), and a negative control (lane 24). Lanes M contain marker DNA.
At the Leiden University Medical Center, a Legionella species-specific real-time PCR assay is being developed, and similar findings were obtained with the Qiagen columns while the specificity of the assay was being determined. Legionella species were extracted with Qiagen columns, with negative extraction controls included after every five samples. When a positive signal was seen in the extraction controls, all potential sources of contamination were investigated and the columns were found to be the source. Other extraction methods have now been employed to continue using the PCR.
The production of columns involves continuous flushing with water (Qiagen distributor [Westburg, Leusden, The Netherlands], personal communication), and during the process Legionella-containing amoeba, DNA, or bacterial cells present in the water may settle on the column material. Pretreatment of columns with DNAase did not eliminate the contamination. In collaboration with the manufacturer, many batches either pretreated with gamma irradiation or from different lots, have been tested. All batches were found to be contaminated; the level of Legionella contamination ranged from 10% to 70%. Our findings emphasize the importance of sufficient negative controls in PCR methods for detection and diagnosis of L. pneumophila and for species other than L. pneumophila, in order to detect the contamination. Although efficiently used for many applications, Qiagen columns at the present time are not suitable for laboratory diagnosis of Legionella infection.
REFERENCES
- 1.Cloud, J. L., K. C., Caroll, P. Pixton, M. Erali, and D. R. Hillyard. 2000. Detection of Legionella species in respiratory specimens using PCR with sequencing confirmation. J. Clin. Microbiol. 38:1709-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jonas, D., A. Rosenbaum, S. Weyrich, and S. Bhakdi. 1995. Enzyme-linked immunoassay for detection of PCR-amplified DNA of Legionellae in bronchoalveolar fluid. J. Clin. Microbiol. 33:1247-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]