Abstract
During senescence, chlorophyll (chl) is metabolized to colorless nonfluorescent chl catabolites (NCCs). A central reaction of the breakdown pathway is the ring cleavage of pheophorbide (pheide) a to a primary fluorescent chl catabolite. Two enzymes catalyze this reaction, pheide a oxygenase (PAO) and red chl catabolite reductase. Five NCCs and three fluorescent chl catabolites (FCCs) accumulated during dark-induced chl breakdown in Arabidopsis (Arabidopsis thaliana). Three of these NCCs and one FCC (primary fluorescent chl catabolite-1) were identical to known catabolites from canola (Brassica napus). The presence in Arabidopsis of two modified FCCs supports the hypothesis that modifications, as present in NCCs, occur at the level of FCC. Chl degradation in Arabidopsis correlated with the accumulation of FCCs and NCCs, as well as with an increase in PAO activity. This increase was due to an up-regulation of Pao gene expression. In contrast, red chl catabolite reductase is not regulated during leaf development and senescence. A pao1 knockout mutant was identified and analyzed. The mutant showed an age- and light-dependent cell death phenotype on leaves and in flowers caused by the accumulation of photoreactive pheide a. In the dark, pao1 exhibited a stay-green phenotype. The key role of PAO in chl breakdown is discussed.
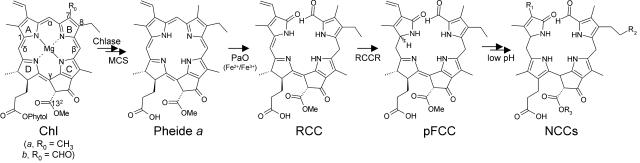
Chlorophyll (chl) degradation is an integral part of leaf senescence and fruit ripening. The fate of chl during senescence has been well established in recent years (for review, see Matile et al., 1999; Hörtensteiner, 1999; Hörtensteiner and Kräutler, 2000; Kräutler, 2003; Eckhardt et al., 2004). Thereby, chl is converted to colorless nonfluorescent chl catabolites (NCCs; Fig. 1) in a pathway that is probably active in all higher plants (Pružinská et al., 2003; Gray et al., 2004).
Figure 1.
The pathway of chl breakdown in higher plants. The chemical constitutions of chl and of chl catabolites are shown. Pyrrole rings (A–D), methine bridges (α–δ), and relevant carbon atoms are labeled. Sites of peripheral modifications as present in different NCCs are indicated (R1–R3). Abbreviations are as indicated in the text. Chlase, chlorophyllase; MCS, Mg-dechelating substance.
Structure elucidation of NCCs from different species has unraveled a common tetrapyrrolic skeleton with an oxygenolytically opened porphyrin macrocycle (Kräutler, 2003). Peripheral modifications at several side chains within different NCCs (Fig. 1, R1–R3) are species specific (Berghold et al., 2002, 2004), and hence have been proposed to occur rather late in the pathway (Hörtensteiner, 1999). Indeed, a primary chl breakdown product (primary fluorescent chl catabolite-1 [pFCC-1]), which exhibits a blue fluorescence, could be identified as a common product of porphyrin ring cleavage (Fig. 1; Mühlecker et al., 1997). Thus, the sequence of reactions is the removal of phytol and magnesium (Mg) by chlorophyllase and Mg-dechelatase, respectively, followed by the conversion of pheophorbide (pheide) a to pFCC-1, which requires the activity of two enzymes, pheide a oxygenase (PAO) and red chl catabolite (RCC) reductase (RCCR; Rodoni et al., 1997; Hörtensteiner, 1999).
PAO is a chloroplast envelope-bound Rieske-type iron-sulfur oxygenase, which is identical to lethal leaf spot 1 (LLS1) from maize (Zea mays) and accelerated cell death 1 (ACD1) from Arabidopsis (Arabidopsis thaliana; Pružinská et al., 2003; Yang et al., 2004). PAO has an intriguing specificity for pheide a, with pheide b inhibiting in a competitive manner (Hörtensteiner et al., 1995; Pružinská et al., 2003). Consequently, all NCCs identified so far from higher plants are derived from chl a (Fig. 1) and chl b is proposed to be reduced to chl a before entering the catabolic pathway through PAO (Hörtensteiner, 1999). In contrast to PAO, RCCR is a soluble protein identical to Arabidopsis ACD2, which mainly localizes to chloroplasts, but in young seedlings has been associated with mitochondria as well (Wüthrich et al., 2000; Mach et al., 2001).
Chl breakdown is a multistep pathway that, in addition to the catabolic enzymes described above, requires transport processes for the export of catabolites to their final destination, the vacuole (Matile et al., 1988). Vacuolar uptake of catabolites has been shown to be mediated by a primary active transport system (Hinder et al., 1996; Tommasini et al., 1998). The complexity of the pathway demands a fine-tuned regulation in order to prevent the accumulation of photodynamic breakdown intermediates, such as pheide a and RCC. Indeed, mutations in or antisense expression of either Pao or Rccr genes have been shown to cause the occurrence of lesion-mimic phenotypes (Greenberg and Ausubel, 1993; Greenberg et al., 1994; Gray et al., 1997; Spassieva and Hille, 2002; Pružinská et al., 2003; Tanaka et al., 2003; Yao et al., 2004), but so far a correlation between the phenotype and an accumulation of the respective intermediates has not been proven unequivocally (Pružinská et al., 2003). Activity of PAO has been demonstrated to be restricted to senescence (Ginsburg et al., 1994), although in a recent investigation limited PAO activity was found in presenescent tissues (Pružinská et al., 2003). In contrast, other chl catabolic enzymes, including RCCR, seem to be constitutively active (Matile et al., 1999). Together, these data indicate an important regulatory role for PAO. Pao/Lls1/Acd1 is considerably expressed before the onset of senescence and also in non-green tissues, but is up-regulated during senescence and upon wounding (Gray et al., 2002; Pružinská et al., 2003; Yang et al., 2004).
The investigation of leaf senescence and of chl breakdown requires a system where senescence can be induced in a controlled manner. In this respect, Arabidopsis is not ideal for several reasons. First, in Arabidopsis, natural senescence coincides with a switch from vegetative to reproductive growth. In addition, rosette leaf senescence is sequential, starting in the older leaves first (Hensel et al., 1993; Miao et al., 2004). Furthermore, the small size of Arabidopsis makes it impossible to use cotyledons for the analysis of chl catabolic enzyme activities or for the identification of chl catabolites, as has been successfully done in canola (Brassica napus; Ginsburg and Matile, 1993; Ginsburg et al., 1994). Nonetheless, we used Arabidopsis as a system to analyze senescence-related chl breakdown. In this article, we identified several Arabidopsis chl catabolites and compared their formation during senescence of detached dark-incubated leaves and individually darkened attached leaves. In particular, we identified two novel modified fluorescent chl catabolites (FCCs), which supports the idea that side chain modifications of chl catabolites occur at the level of FCC. Employing a T-DNA-tagged pao1 mutant, we substantiate the key role of PAO for chl breakdown during senescence. In addition, by a detailed analysis of expression, protein, and activity levels, we show that a major regulation of PAO is at the transcriptional level.
RESULTS
Identification and Characterization of Chl Catabolites
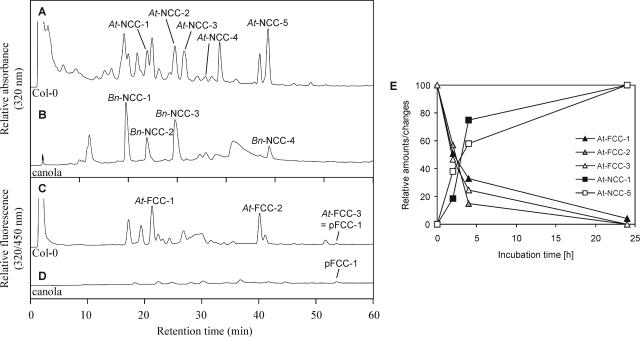
To induce senescence in Arabidopsis, detached leaves of short-day-grown ecotype Columbia (Col-0) plants were incubated in permanent darkness. After extraction, chl catabolites were separated by HPLC (Fig. 2). A total of five different NCCs (Fig. 2A) tentatively numbered At-NCC-1 through At-NCC-5 (for a nomenclature of NCCs, see Ginsburg and Matile, 1993) were identified by their typical UV/Vis spectrum (Kräutler et al., 1991). By comparison, using mass spectroscopy (MS) and HPLC with known NCCs from senescent canola cotyledons (Fig. 2B; Mühlecker and Kräutler, 1996; S. Moser, J. Berghold, T. Müller, S. Hörtensteiner, and B. Kräutler, unpublished data), compounds At-NCC-1, At-NCC-2, and At-NCC-5 were suggested to be identical to canola Bn-NCC-2, Bn-NCC-3, and Bn-NCC-4, respectively. MS analysis (see “Materials and Methods” for MS data) suggested the structures of the remaining NCCs, as summarized in Table I. Thus, three of the NCCs in Arabidopsis are identical to known chl catabolites from canola and are different from each other with respect to modifications at the C82 position. Hence, the ethyl side chain, as present in At-NCC-5, carries a hydroxyl group in At-NCC-2, which is further modified with a Glc moiety in At-NCC-1. In contrast, the major catabolite of canola, Bn-NCC-1, which is malonylated at C82, is absent in Arabidopsis. The remaining NCCs, At-NCC-3 and At-NCC-4, are shown to differ from known NCCs and the elucidation of their chemical structures is underway (S. Moser, T. Müller, S. Hörtensteiner, and B. Kräutler, unpublished data).
Figure 2.
HPLC analysis of chl catabolites in Arabidopsis and canola. Chl catabolites of senescent Arabidopsis (A and C) and canola (B and D) leaves were separated by HPLC as described in “Materials and Methods.” A320 (A and B) and fluorescence (C and D) were recorded. For the identification and characterization of FCCs and NCCs, see Table I. E, Conversion of FCCs to NCCs during incubation of Arabidopsis leaf extracts at pH 5. Formation of NCCs (squares: black, At-NCC-1; white, At-NCC-5) is expressed as difference in peak area at 320 nm relative to the peak area at 0 h (24-h sample set to 100). Decrease in FCCs (triangles: black, At-FCC-1; gray, At-FCC-2; white, At-FCC-3) is expressed as relative difference in fluorescence (320/450 nm; 0-h sample set to 100).
Table I.
Identification of FCCs and NCCs occurring during chl breakdown in Arabidopsis
| Name | R2a | R3a | Identificationb | Identity with | Reference |
|---|---|---|---|---|---|
| At-NCC-1 | O-glucosyl | H | s, c, m | Bn-NCC-2 | Mühlecker et al. (1996); this work |
| At-NCC-2 | OH | H | s, c, m | Bn-NCC-3 | Mühlecker et al. (1996); this work |
| At-NCC-3 | OHc | H | s, m | – | This work |
| At-NCC-4 | O-glucosylc | CH3 | s, m | – | This work |
| At-NCC-5 | H | H | s, c, m | Bn-NCC-4d | This work |
| At-FCC-1 | OHc | H | s, m | – | This work |
| At-FCC-2 | H | H | s, m | – | This work |
| At-FCC-3 | H | CH3 | s, c, m | pFCC-1 | Mühlecker et al. (1997); this work |
R2 and R3 indicate residues at C82 and C132 side positions, respectively, of FCCs or NCCs as shown in Figure 1. R1 is ethyl in all determined cases.
s, Peak identification by UV/Vis spectra (Hörtensteiner, 1999); c, cochromatography with respective standards; m, MS.
In At-NCC-3, the site of hydroxylation is indicated to be C71 (rather than C82); in At-NCC-4 and At-FCC-1, the sites of attachment of the Glc moiety and the OH group, respectively, are not yet defined (S. Moser, T. Müller, S. Hörtensteiner, and B. Kräutler, unpublished data).
S. Moser, J. Berghold, T. Müller, S. Hörtensteiner, and B. Kräutler (unpublished data).
In addition to NCCs, rather large quantities of three different FCCs were identified in Arabidopsis by their typical absorption and fluorescence properties (Fig. 2C; Mühlecker et al., 1997). In contrast, in canola, FCCs occur only in trace amounts (Fig. 2D). MS analysis indicates At-FCC-2 to be the fluorescent isomer of At-NCC-5 (= Bn-NCC-4; Table I), while At-FCC-3 is identical to pFCC-1 (Mühlecker et al., 1997). The structure of At-FCC-1 remains to be established; however, its molecular formula (for MS data, see “Materials and Methods”) indicates that it is an isomer of At-NCC-2 (i.e. of Bn-NCC-3).
It has been shown that in vitro stereoselective nonenzymatic conversion of an FCC to the respective NCC readily occurs at acidic pH (Oberhuber et al., 2003) and it was concluded that in vivo the acidic pH of the vacuole, the final site of chl catabolite accumulation (Matile et al., 1988; Hinder et al., 1996), is responsible for the observed rapid conversion (Oberhuber et al., 2003). To investigate whether the differences in FCC content between Arabidopsis and canola could be due to differences in vacuolar pH, i.e. to a slower FCC-to-NCC conversion after import of FCCs into the vacuoles in Arabidopsis, the pH values of cell saps as representing the vacuolar pH (Barthe and Vaillant, 1993) were determined in leaves. Surprisingly, the pH values of canola (pH 5.82 ± 0.06) and Arabidopsis (5.60 ± 0.13) did not differ significantly. Furthermore, subcellular fractionation studies of senescent Arabidopsis mesophyll cells demonstrated the exclusive localization of FCCs outside the vacuole (A. Pružinská and S. Hörtensteiner, unpublished data). Despite this, incubation of Arabidopsis extracts from senescent leaves at a pH of 5.0 resulted in a fast disappearance of FCC peaks, which was concomitant with an increase in the amount of NCCs (Fig. 2E).
Accumulation of Chl Catabolites Correlates with Progression of Leaf Senescence
Two different methods were used to induce senescence and to investigate the dynamics of the formation of chl catabolites in Arabidopsis: (1) detached leaves incubated in permanent darkness and (2) attached leaves that were covered with aluminum foil (Weaver and Amasino, 2001). Progression of senescence was followed by measuring different senescence parameters, such as loss of chl and protein, and by an increase in the chl a/b ratio (Fig. 3, A and B). Colorless chl catabolites were absent before senescence induction, but the content of the five different At-NCCs increased steadily (Fig. 3C). FCCs and NCCs accumulated simultaneously, since their relative amounts remained constant during the incubation period (Fig. 3D). Similar results as shown for the detached leaf incubations (Fig. 3) were obtained when attached leaves were covered with aluminum foil (data not shown).
Figure 3.
Characterization of protein and chl breakdown during dark-induced senescence of detached Arabidopsis Col-0 leaves. A, Degradation of chl (columns) and changes in the chl a/b ratio (line). B, Degradation of total proteins. C, Accumulation during senescence of five different NCCs (gray, At-NCC-1; black, At-NCC-2; white, At-NCC-3; hatched, At-NCC-4; dotted, At-NCC-5). D, Relative amounts of NCCs (gray) and FCCs (black) remain constant throughout the incubation time. Results of a single representative senescence experiment are shown. Data are means of three replicates. Error bars indicate sd.
Analysis of PAO and RCCR during Leaf Senescence in Arabidopsis
As shown previously (Rodoni et al., 1997), the formation of pFCC-1 from pheide a requires the joint action of membrane-bound PAO and soluble RCCR. Ongoing biochemical analysis aiming at a detailed investigation of this reaction has led to the identification of a new factor that is indispensable for PAO/RCCR activity in vitro (data not shown). This factor most probably is a protein and is required for the formation of RCC; thus, we tentatively name it RCC-forming factor (RFF). RFF is present both in Escherichia coli and in plants, indicating a rather general role. The catalytic properties of RFF will be described in detail elsewhere. Furthermore, work is in progress to identify the molecular nature of RFF.
Hence, the formation of pFCC-1 from pheide a requires the activity of three enzymes, PAO, RFF, and RCCR. To assess the activities of PAO and RCCR during leaf senescence in Arabidopsis, respective proteins were isolated from detached dark-incubated leaves (Hörtensteiner et al., 1995) and employed in two assays differing in the sources of the respective enzymes used. Thus, for measuring PAO activity, E. coli-expressed His6-AtRCCR was employed together with PAO isolated from the tissues of interest. Conversely, RCCR activity was determined with soluble proteins of the tissues of interest (containing RCCR) incubated together with a partially purified standard PAO preparation (Hörtensteiner et al., 1998). Both assays were supplemented with extracts of E. coli JM109 as a source of RFF and cofactors as described in “Materials and Methods.”
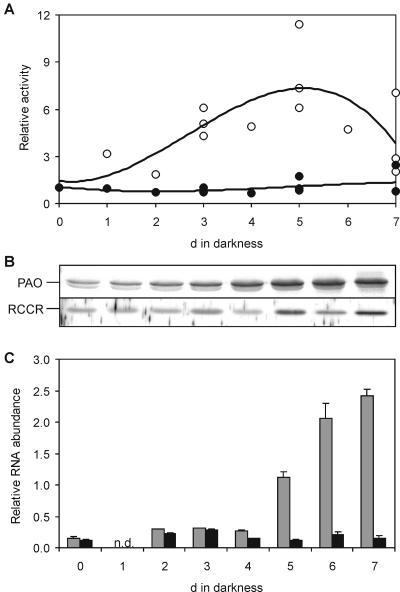
In different independent experiments, PAO activity transiently increased during dark incubation of detached leaves (Fig. 4A). Although the extent of activity induction was variable between experiments, consistently maximal activities were obtained at around day 5. A senescence-related (transient) increase in PAO activity had been observed previously in different species, such as canola and Festuca pratensis (Hörtensteiner et al., 1995; Vicentini et al., 1995), but to date it is not clear whether this is due to an up-regulation of Pao gene expression and/or to a posttranscriptional activation of PAO (Pružinská et al., 2003). To investigate this in more detail, PAO activities were compared with levels of PAO protein as well as with Pao expression. Figure 4, B and C, show the data of a representative experiment, which indicate that PAO protein amounts correlate with Pao gene expression throughout the dark-incubation treatment. Expression and protein abundance are also comparable to PAO activities during early senescence, but in contrast to protein levels, measurable PAO activity dropped toward the end of the senescence period (Fig. 4, A and B). This is explained by the instability of solubilized PAO (Hörtensteiner et al., 1995, 1998), which may become particularly evident when PAO is extracted from tissue at later stages of senescence. Nonetheless, these data indicate that, in Arabidopsis, senescence-related induction of PAO activity is regulated (mainly) at the transcriptional level. For comparison, PAO regulation was analyzed in canola cotyledons. Employing semiquantitative reverse transcription (RT)-PCR and immunoblot analysis, we could again show that, during induction of senescence, an increase of PAO activity (Hörtensteiner et al., 1995) correlates with increased PAO protein levels and Pao gene expression (data not shown).
Figure 4.
Analysis of PAO and RCCR activities (A), protein abundance (B), and gene expression (C) during dark-induced senescence of Arabidopsis leaves. A, For the determination of their activities, PAO and RCCR, respectively, were isolated from the tissues of interest and assessed in RFF-containing assays that were supplemented with purified His6-AtRCCR (determination of PAO activity, white circles) and a standard preparation of PAO (determination of RCCR, black circles), respectively. Values are data from several independent experiments showing the great variability of PAO activity. B, Representative immunoblots of PAO and RCCR extracts used for measuring enzyme activities as shown in A. Gel loadings are based on equal amounts of fresh weight. Blots were labeled and developed as described in “Materials and Methods” using polyclonal antibodies against PAO and RCCR, respectively. C, RNA abundance of Pao (gray) and Rccr (black) was quantified by real-time RT-PCR and normalized to levels of mRNA encoding the actin 2 protein. Values are means of three replicates. Error bars indicate sd. n.d., Not determined.
In contrast to PAO, RCCR activities were almost constant during the course of detached leaf senescence (Fig. 4A) and correlated with RCCR levels and Rccr gene expression (Fig. 4, B and C). This indicates that RCCR is not regulated during leaf senescence.
PAO and RCCR Are Present throughout Arabidopsis Leaf Development
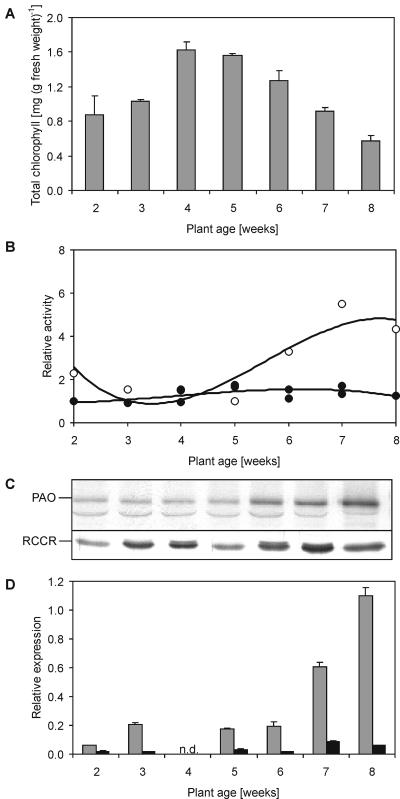
The observation that PAO and RCCR are expressed and active in nonsenescent tissues (see above) allowed us to investigate their activities and expression levels during Arabidopsis rosette leaf development. For this, the entire rosettes of 2- to 8-week-old plants grown under long-day conditions were used. Chl concentrations were at maximum after 4 weeks and then declined (Fig. 5A). The age-dependent decrease in chl was accompanied by the occurrence of colorless chl catabolites starting from week 6, which coincided with yellowing of the oldest rosette leaves (data not shown). PAO activity was present already in young seedlings but started to increase after 5 weeks. Again, PAO activities, PAO protein levels, and Pao expression changed to comparable levels (Fig. 5, B–D). In contrast, expression and activity of RCCR was only weakly regulated.
Figure 5.
Analysis of chl content and characterization of PAO and RCCR during rosette development in Arabidopsis. Entire rosettes of plants grown under long-day conditions were harvested at the indicated times and analyzed for chl content (A), PAO and RCCR activities (B), protein abundance (C), and gene expression (D). A, Values are means of at least three independent samples. Error bars indicate sd. B, For the determination of their activities, PAO and RCCR, respectively, were isolated from the tissues of interest and assessed in RFF-containing assays that were supplemented with purified His6-AtRCCR (determination of PAO activity, white circles) and a standard preparation of PAO (determination of RCCR, black circles), respectively. C, Representative immunoblots of PAO and RCCR extracts used for measuring enzyme activities as shown in B. Gel loadings are based on equal amounts of fresh weight. Blots were labeled and developed as described in “Materials and Methods” using polyclonal antibodies against PAO and RCCR, respectively. D, RNA abundance of Pao (gray) and Rccr (black) was quantified by real-time RT-PCR and normalized to levels of mRNA encoding the actin 2 protein. Values are means of three replicates. Error bars indicate sd. n.d., Not determined.
Isolation and Characterization of a Pao T-DNA Insertional Mutant
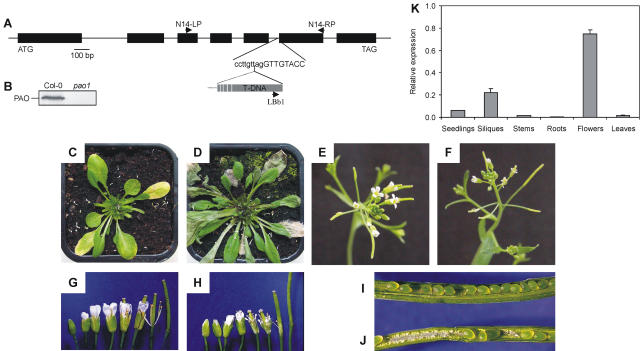
PAO has been described as a key component of chl breakdown in barley (Hordeum vulgare), canola, and maize (Schellenberg et al., 1993; Hörtensteiner et al., 1995; Pružinská et al., 2003). To confirm this role in Arabidopsis, the T-DNA insertion mutant 111333, in which At3g44880 (Pao/Acd1 gene) is tagged, was obtained from the Salk resource (Alonso et al., 2003). T-DNA insertion near the 3′ border of intron 5 of At3g44880 was confirmed by PCR and cloning of the left T-DNA border (Fig. 6A). After identification of homozygote lines, absence of PAO was confirmed by immunoblot analysis (Fig. 6B) and activity measurements (data not shown). This mutant was designated pao1. pao1 developed a lesion-mimic phenotype on the leaves (Fig. 6D), which was similar to the phenotype described for several acd1 mutants (Greenberg and Ausubel, 1993; Yang et al., 2004). The lesions of pao1 occurred in a development-related, light-dependent fashion, coinciding with the initiation of leaf senescence in the wild type (Fig. 6, C and D). Premature cell death was also observed in pao1 flowers (Fig. 6, E–H). In the mutant, petals and sepals started to disintegrate earlier than in the wild type, resulting in flowers that never fully opened. In addition, about 40% of seeds aborted at an early stage of development in pao1 (Fig. 6J; Table II). These data, together with the finding that expression of Pao is rather high in flowers and siliques (Fig. 6K), indicate that, in addition to chl breakdown during senescence, PAO has an important function in flower and/or seed development in Arabidopsis. The data obtained for pao1 were confirmed by constitutive silencing of Pao in Col-0 using an RNA interference strategy (data not shown).
Figure 6.
Characterization of the pao1 mutant and tissue specificity of Pao expression. A, Gene structure of Pao (At3g44880) showing the insertion site of the T-DNA in the pao1 mutant and the location of primers used to identify homozygous knockout lines. B, Immunoblot of protein extracts from senescent Col-0 and pao1 leaves, labeled with anti-PAO monoclonal antibodies. C to J, Phenotype of pao1 (D, F, H, and J) compared to Col-0 (C, E, G, and I). Plants were grown for 4 weeks under long-day conditions. C and D, pao1 (D) exhibits a cell death phenotype, which correlates to senescence in Col-0 (C). Before taking pictures, inflorescences were removed. E and F, In contrast to Col-0 (E), flowers of pao1 (F) do not fully open. G and H, Series of flowers at different stages of development. Sepals and petals senesce earlier in pao1 (H) compared to Col-0 (G). I and J, Early seed abortion occurs in pao1 (J) at a high ratio, but not in the wild type (I). For quantification, see Table II. K, RNA abundance of Pao in different tissues was quantified by real-time RT-PCR and normalized to levels of mRNA encoding the actin 2 protein. Values are means of three replicates. Error bars indicate sd.
Table II.
Seed development in Col-0 and pao1
| Embryos per Silique | Aborted Embryos | |
|---|---|---|
| % | ||
| Col-0 | 31.40 ± 4.90 | 2.29 ± 3.52 |
| pao1 | 28.94 ± 3.78 | 42.25 ± 28.14 |
Pheide a Is Responsible for the Cell Death Phenotype of pao1
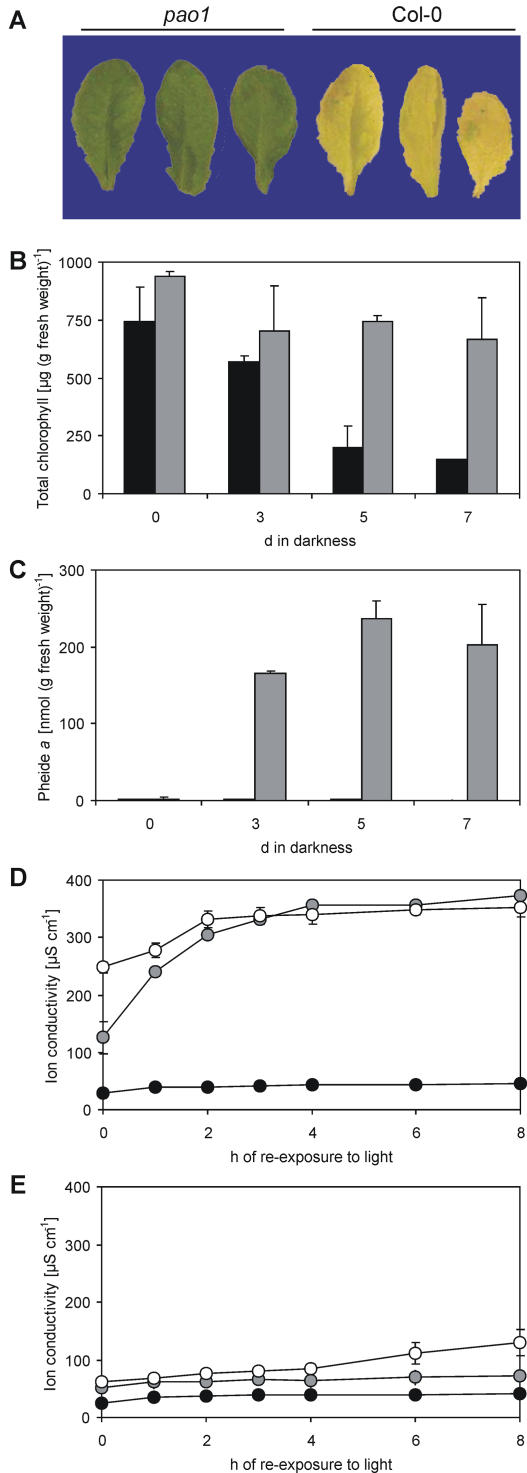
To induce senescence without an interference of light, detached leaves were incubated in permanent darkness. Under these conditions, chl was largely retained in pao1, resulting in a stay-green phenotype (Fig. 7, A and B). The absence of PAO resulted in the accumulation of pheide a in a time-dependent manner (Fig. 7C). Pheide a content positively correlated with the light-dependent death reaction. Thus, cell death, as measured by ion leakage of leaf discs, was faster after longer dark incubation, i.e. at higher pheide a content (Fig. 7D). In contrast, leaf discs stayed intact in the wild type, where pheide a did not accumulate (Fig. 7, C and E). This indicates that pheide a is responsible for the cell death reaction observed in pao1.
Figure 7.
Characterization of chl breakdown in pao1. A, Detached pao1 and Col-0 leaves after 5-d senescence in the dark. B, Degradation of chl in Col-0 (black) and pao1 (gray). C, Accumulation of pheide a in response to dark incubation of Col-0 (black) and pao1 (gray) leaves. D and E, Determination of ion leakage as a measure for cell death in pao1 (D) and Col-0 (E) leaves. Before re-exposure to the light for up to 8 h, leaves were incubated in the dark for 0 (black circles), 3 (gray circles), or 5 (white circles) d. Results of a single representative experiment are shown. Data are means of three replicates. Error bars indicate sd.
DISCUSSION
The Pathway of Chl Breakdown in Arabidopsis
Chl catabolism is an integral process of leaf senescence and fruit ripening. The biochemistry of chl breakdown has been analyzed intensively during recent years, mainly in plant species other than Arabidopsis (Hörtensteiner et al., 1995; Wüthrich et al., 2000; Pružinská et al., 2003). One reason may be that, in Arabidopsis, leaf senescence is induced sequentially in individual leaves within the rosette rather than in the whole plant (Hensel et al., 1993). Thus, developmental senescence is difficult to control and incubation of entire plants in the dark has been shown to inhibit induction of senescence (Weaver and Amasino, 2001). Hence, much senescence-related work in Arabidopsis has been performed with either dark-incubated detached leaves or individually darkened attached leaves. These two systems were employed in this article in order to address chl breakdown in Arabidopsis.
Independent of the type of senescence induction, chl was degraded to an identical set of colorless chl catabolites, indicating that there is one common pathway active in breaking down chl during senescence in Arabidopsis. In addition, the amounts of accumulating colorless catabolites roughly matched the amounts of degraded chl, indicating that, in Arabidopsis, as has been shown for canola (Ginsburg and Matile, 1993), degradation of chl exclusively occurs through the formation of FCCs (and subsequently NCCs) catalyzed by PAO/RCCR. A comparison of the structures of NCCs from canola and Arabidopsis uncovered similarities, but also major differences. Thus, structures of three of the five At-NCCs are identical to canola NCCs, but a malonylated NCC, the major catabolite in canola, is absent from Arabidopsis. Furthermore, two additional (new) NCCs are found in Arabidopsis, but are absent from canola. These findings indicate that the two closely related species contain different sets of chl catabolic enzymes that are responsible for the species-specific modifications of side chains present in NCCs.
In other plant species analyzed so far, NCCs, but not FCCs, have been identified as final chl catabolites (Curty and Engel, 1996; Mühlecker and Kräutler, 1996; Kräutler, 2003; Berghold et al., 2004). Thus, it was surprising to find rather large quantities of FCCs during chl breakdown in Arabidopsis. The conversion of FCCs to NCCs occurs after import into the vacuole due to the acidic vacuolar milieu (Oberhuber et al., 2003). A high FCC-to-NCC ratio (Fig. 3D) may be explained by a rather high vacuolar pH in Arabidopsis. Our data show that vacuolar pH values are similar in canola and Arabidopsis, thus excluding this possibility. Alternatively, catabolite transport at the tonoplast could be slower in Arabidopsis than in other species. Such transport has been shown to be a primary active process in barley (Hinder et al., 1996) and most probably involves members of the multidrug resistance-associated protein subfamily of ATP-binding cassette transporters (Lu et al., 1998; Tommasini et al., 1998). Vacuolar uptake experiments were performed with NCCs, but inhibition studies, together with data of FCC-to-NCC conversion at low pH, suggest that the true substrates for vacuolar import are FCCs (Hörtensteiner, 1999). Consequently, modifications that are present at different side groups of the tetrapyrrolic skeleton in Arabidopsis NCCs (Table I) most likely occur at the level of FCCs outside the vacuole. This hypothesis is supported by the identification in this study of two modified FCCs, At-FCC-1 and At-FCC-2 (Fig. 2; Table I). Hence, Arabidopsis proves to be a suitable system to study the biochemical processes that are involved in the late steps of chl catabolism and are responsible for peripheral modifications of FCCs.
PAO Has a Key Role in Chl Breakdown
In contrast to chlorophyllase, where two genes have been identified in Arabidopsis (Tsuchiya et al., 1999), PAO and RCCR are single-copy genes. We have analyzed properties of these two proteins and their respective genes during leaf senescence. Our data indicate an important regulatory role for PAO, but not for RCCR. Thus, PAO activity, protein abundance, and Pao gene expression positively correlate with rates of chl breakdown (Figs. 4 and 5). In contrast, RCCR protein and RNA levels do not change significantly during development and senescence (Figs. 4 and 5; Mach et al., 2001). Regulation of PAO at the posttranscriptional level had been suggested from phosphatase treatments that inhibited PAO activity (M. Roca and S. Hörtensteiner, unpublished data; Pružinská et al., 2003). The results presented here show that such regulation would not necessarily be required to explain the observed senescence-related increase in PAO activity. In addition, we were unable to repeat the original phosphatase experiments in this work (data not shown). Thus, we conclude that regulation of PAO occurs at the transcriptional level.
The analysis of pao1, a knockout mutant in the Pao gene, further substantiates a key role for PAO during chl catabolism. In contrast to mutants in RCCR, which are still able to produce FCCs and NCCs (A. Pružinská and S. Hörtensteiner, unpublished data), absence of PAO completely inhibits chl breakdown at the level of pheide a. Hence, like in the stay-green mutant Bf 993 of F. pratensis and in an introgression mutant of Lolium temulentum (Roca et al., 2004), most pigment is retained as chl during dark-induced senescence in pao1 (Fig. 7). In maize lls1, which is affected in the ortholog of Arabidopsis PAO, chl retention is accompanied by the retention of chl-binding proteins. This phenomenon has been observed in several instances of stay-green mutants that are affected in PAO activity (Hilditch et al., 1989; Bachmann et al., 1994; Pružinská et al., 2003). Together, these data indicate the existence of a feedback mechanism, which limits metabolism of chl in mutants that are unable to degrade chl beyond pheide a. Whether such a proposed mechanism would act on chlorophyllase, the first enzyme of chl breakdown, or on so-far unknown proteases that would be involved in the degradation of chl-binding proteins, remains to be shown. The strategy of plants to avoid extensive chl breakdown if the degradation pathway is blocked has its explanation in the phenotype observed in the pao1 mutant. pao1 shows a cell death phenotype in leaves and flowers, which is age and light dependent (Figs. 6 and 7). A lesion-mimic phenotype has also been described for other PAO mutants, such as acd1 and lls1 (Greenberg and Ausubel, 1993; Gray et al., 1997). Analysis of maize double mutants of lls1 with oil-yellow or iojap, which have drastically reduced chl content, indicated that a chl-derived compound mediates cell death (Gray et al., 2002). Here we show that the cell death-promoting substance in pao1 is pheide a (Fig. 7), which has been demonstrated to be a potent phototoxin (Jonker et al., 2002). Light absorption by pheide a is believed to cause the production of singlet oxygen, which in turn induces cell death. Such a singlet oxygen-triggered cell death reaction has been described in the flu mutant, where the disruption of feedback inhibition of chl biosynthesis causes the accumulation of photodynamic protochlorophyllide in the dark (Meskauskiene et al., 2001). In this system, the EXECUTER protein has been shown to be a (early) component of a cell death-signaling pathway (Wagner et al., 2004). In order to analyze whether cell death in pao1 proceeds by the same pathway, we started to produce and analyze pao1/ex1 double mutants.
MATERIALS AND METHODS
Plant Material and Senescence Induction
The Columbia (Col-0) ecotype of Arabidopsis (Arabidopsis thaliana) was used as the wild type. Seeds of the T-DNA insertion mutant SALK_111333 were obtained from the Nottingham Arabidopsis Stock Centre (Nottingham, UK). Presence of the T-DNA insertion within the Pao gene (At3g44880) in SALK_111333 was confirmed by sequencing the PCR product generated from genomic DNA with the gene-specific primer N14-RP (5′-GGCTCACCTGACGCTTGGTTA-3′) and the T-DNA left border primer, LBb1 (5′-GCGTGGACCGCTTGCTGCAAT-3′). Homozygous lines from the segregating population of SALK_111333 seeds were identified by PCR using primers LBb1, N14-RP, and N14-LP (5′-CGACGGTGACAATTCAAAGGG-3′). One of the homozygous lines was renamed pao1 and used for further analysis.
Plants were grown on soil either in short-day (8/16 h) or long-day (16/8 h) growth rooms under fluorescent light of 60 to 120 μmol photons m−2 s−1 at 22°C. For senescence induction, leaves from 3- to 4-week-old (long-day) or 8-week-old (short-day) plants were excised and incubated in permanent darkness on wet filter paper for up to 7 d at ambient temperature. Alternatively, individual attached leaves were wrapped with aluminum foil.
Canola (Brassica napus) was grown on soil and senescence of cotyledons was induced by dark incubation of 10-d-old seedlings in the dark (Hörtensteiner et al., 1995).
Pao RNA Interference Plants
A full-length cDNA clone of Pao (pda07874) was obtained from the RIKEN Tsukuba Institute BioResourceCenter (Seki et al., 2002). Primer combinations R4XN (5′-GACCTCGAGCCATGGAATGTGCCAACCCCGTTC-3′)/R4K (5′-CCGGTACCTTGTGATGAGCAAAATC-3′) and R4B (5′-GATTTGGATCCGAATGTGCC-3′)/R4C (5′-CCATCGATGAGCAAAATCTATATGG-3′) were used to amplify by PCR two 480-bp fragments of Pao. Appropriate restriction sites linked to the primers enabled a two-step cloning of the PCR products in opposite directions into pHannibal (Wesley et al., 2001). After digestion with NotI, the RNA interference construct was introduced into NotI-restricted pGreen0029 (Hellens et al., 2000) to produce pGpao-c. Arabidopsis (Col-0) plants were transformed by the floral-dip method (Sidler et al., 1998). From the original 10 kanamycin-resistant T1 plants that were analyzed for the accumulation of pheide a after senescence induction, two lines (IIIa and IIIb) were selected for further analysis.
Analysis of Chl and Chl Catabolites
Chl and Green Catabolites
Chl was isolated from leaf tissue by homogenization in liquid nitrogen and subsequent 3-fold extraction into 80% (v/v) acetone containing 1 μm KOH. After centrifugation (2 min, 16,000g), supernatants were combined and chl concentrations were determined spectrophotometrically (Strain et al., 1971).
For extraction of green polar chl catabolites (pheide and chlorophyllide), leaf material (four to eight 1-cm leaf discs) was homogenized in liquid nitrogen and resuspended in 100 μL 0.5 m Tris-HCl, pH 8.0. After the addition of 400 μL acetone and centrifugation (2 min, 16,000g), the supernatant was analyzed by HPLC essentially as described (Langmeier et al., 1993). Pigments were identified by their absorption spectra and quantified using authentic standards (Ginsburg and Matile, 1993; Hörtensteiner et al., 1995).
Colorless Chl Catabolites
Colorless chl catabolites were extracted as described (Pružinská et al., 2003) and either directly or after concentration on a C18-SepPak cartridge (Waters; Mühlecker et al., 1997) analyzed by HPLC. The reversed-phase system consisted of a C18 Hypersil ODS column (250 × 4.6 mm; 5 μm; MZ-Analysentechnik), which was developed with a gradient (flow rate 0.5 mL min−1) of solvent B (20% [v/v] 25 mm potassium phosphate buffer, pH 7.0, and 80% methanol) in solvent A (50 mm potassium phosphate, pH 7.0) as follows: 25% to 75% over 60 min, 75% to 100% over 5 min, and 100% solvent B for 5 min. Peak detection was with sequential monitoring using a System Gold 168 photodiode array detector (200–600 nm; Beckman Coulter) and a RF-10AXL fluorescence detector (excitation at 320 nm, emission at 450 nm; Shimadzu Corporation). Analysis of peaks was performed with a 32K workstation (Beckman Coulter). Chl catabolites were identified by their absorption (FCCs and NCCs; Hörtensteiner, 1999) and fluorescence (FCCs) properties. Peak areas of NCCs were quantified with a standard Cj-NCC-1 (Oberhuber et al., 2001) at defined concentrations (log ɛ315 nm = 4.2; Kräutler et al., 1991).
MS
A Finnigan MAT 95-S mass spectrometer (Thermo Electron Corporation) in the positive-ion mode was employed. Settings for electrospray ionization (ESI)-MS are as follows: flow rate, 600 μL min−1; spray voltage, 3.2 kV; solvent, methanol:water 1:1 (v/v); values are m/z (% relative intensity). For high-resolution fast atom bombardment (HR FAB)-MS, a cesium ion gun at 20 keV was employed; matrix, glycerin; values are m/z (measured versus calculated values).
At-NCC-1: ESI-MS, 815.3 (100, [M + Na + H]+), 831.4 (90, [M + K + H]+); HR FAB-MS, 793.328 ([M + H]+; calc. 793.3291). At-NCC-2: ESI-MS, 669.2 (95, [M + K + H]+), 707.2 (100, [M + 2K + H]+). At-NCC-3: ESI-MS, 669.2 (100, [M + K + H]+), 707.2 (30, [M + 2K + H]+); HR FAB-MS, 631.280 ([M + H]+; calc. 631.2762). At-NCC-4: ESI-MS, 845.4 (100, [M + K + H]+), 883.1 (15, [M + 2K + H]+). At-NCC-5: ESI-MS, 615.2 (20, [M + H]+), 653.2 (100, [M + K + H]+); HR FAB-MS, 615.283 ([M + H]+; calc. 615.2813). At-FCC-1: ESI-MS, 625.2 (20, [M + K + H-CO2]+), 663.2 (100, [M + 2K + H-CO2]+). At-FCC-2: ESI-MS, 609.2 (100, [M + K + H-CO2]+), 615.3 (25, [M + H]+), 653.3 (25, [M + K + H]+). At-FCC-3: ESI-MS, 629.3 (50, [M + H]+), 667.3 (100, [M + K + H]+).
FCC-to-NCC Conversion
A sample of colorless chl catabolites extracted from senescent Col-0 leaves and concentrated on a C18-SepPak cartridge (see above) was acidified to pH 5.0 by the addition of 0.25 volumes of 0.2 m citrate buffer. After degassing, the mixture was incubated at ambient temperature in the dark. Samples were withdrawn and analyzed by HPLC as indicated in Figure 2E.
pH Determination
To measure the pH values of leaf extracts, leaves were homogenized in liquid nitrogen and defrosted. After filtration through miracloth (Calbiochem), extracts were diluted 4-fold and 10-fold, respectively, with MilliQ-water (Millipore) and the pH values were determined using a Knick pH meter (Escolab) equipped with an Orion 8102 Ross electrode (Cambridge Scientific Products).
cDNA Cloning of His6-AtRCCR
Primers AtFOR1 (5′-GGGATCCATGGAAGACCACGACG-3′) and AtRCCR5 (5′-TTCTGCAGAGAACACCGAAAGCT-3′), with pGEM-AtRCCR as template (Wüthrich et al., 2000), were used to amplify a truncated fragment of Arabidopsis Rccr covering codons from Ser-40, the predicted cleavage site of the chloroplast transit peptide of AtRCCR, to the end of the protein. BamHI and PstI sites located within the primers were employed for in-frame ligation of this fragment to the His6-tag of pQE30 (pQE-1.1). After confirming the fidelity of pQE-1.1 by sequencing, His6-AtRCCR was expressed in Escherichia coli JM109 (Wüthrich et al., 2000).
RNA Isolation and Real-Time PCR
RNA was prepared using the Plant RNeasy kit (Qiagen). After DNA digestion with RQ1 RNase-free DNase (Promega), 1 μg of RNA was reverse transcribed using the RETROscript kit (Ambion). Quantitative PCR was performed in a LightCycler (Roche Diagnostics) using the QuantiTect SYBR Green PCR kit (Qiagen). Ten- to 100-fold dilutions of first-strand cDNA reaction mixes (corresponding to 0.3–3 ng of RNA) were employed in 20-μL reactions and were used to calculate the real-time PCR efficiency of each sample. The relative expression ratios of target genes (Pao, Rccr) were calculated in comparison to a reference gene (Act2; Kürsteiner et al., 2003). The following specific primers were used: Act2 (forward, 5′-TGGAATCCACGAGACAACCTA-3′ and reverse, 5′-TTCTGTGAACGATTCCTGGAC-3′); Pao (Acd1; forward, 5′-ACGGCATGGTAAGAGTCAGC-3′ and reverse, 5′-AAACCAGCAAGAACCAGTCG-3′); and Rccr (Acd2; forward, 5′-ATCGCCTCCAATCACAACTC-3′ and reverse, 5′-TTAGCACAAGCGACTTGGAA-3′).
Protein Extraction and Immunoblot Analysis
His6-AtRCCR and Total E. coli Proteins
For the isolation of proteins from E. coli, JM109 or JM109 expressing His6-AtRCCR from pQE-1.1 were used. Soluble proteins were extracted and His6-AtRCCR purified on a HiTrap chelating column (General Electric Healthcare) according to published procedures (Wüthrich et al., 2000).
Extraction of Soluble Plant Proteins
Proteins were extracted from Arabidopsis leaves and precipitated with ammonium sulfate according to published procedures (Rodoni et al., 1997; Wüthrich et al., 2000; Pružinská et al., 2003).
Extraction of PAO
PAO was isolated from canola or Arabidopsis chloroplast membranes by Triton X-100 solubilization according to a standard procedure (Hörtensteiner et al., 1995). Canola PAO was partially purified on EAH Sepharose 4B (General Electric Healthcare; Hörtensteiner et al., 1998). In contrast to canola, where typically at least 50 g of leaf tissue were used, extraction of PAO from Arabidopsis required downscaling of the above method and a minimum of 2 g of leaves was used for each extraction.
Immunoblot Analysis
After separation by SDS-PAGE, proteins were transferred to nitrocellulose membranes according to standard procedures. Proteins were labeled with antibodies against Arabidopsis RCCR (1:1,000; Wüthrich et al., 2000) or PAO (monoclonal, 1:500; polyclonal, 1:2,000; Gray et al., 2004), and thereafter with alkaline phosphatase-conjugated second antibodies, and visualized using bromochloroindolyl phosphate/nitroblue tetrazolium as substrate.
Protein Quantification
Protein content was measured according to Bradford (1976) using bovine serum albumin as standard.
Enzyme Assays
PAO and RCCR activities were assessed in different assays according to published procedures (Hörtensteiner et al., 1995; Wüthrich et al., 2000). Briefly, assays (total volume of 50 μL) contained different combinations of PAO (equivalent to 0.5 g of tissue), E. coli (50 μg) protein extracts as a source of RFF, and purified His6-AtRCCR (2.9 μg) or ammonium sulfate-precipitated proteins extracted from Col-0 (equivalent to 13.3 mg of leaf tissue) as a source of RCCR. The assays were supplemented with 0.5 mm pheide a (Hörtensteiner et al., 1995), 10 μg ferredoxin (Fd), and a Fd-reducing system consisting of 2 mm Glc-6-P, 1 mm NADPH, 50 milliunits of Glc-6-P dehydrogenase, and 5 milliunits of Fd-NADP+ oxidoreductase. After 1-h incubation at 25°C, reactions were terminated by the addition of 80 μL methanol. Formation of pFCC-1 was followed by reversed-phase HPLC with 36% (v/v) 50 mm potassium phosphate buffer, pH 7.0, in methanol as solvent. Activities are determined as integrated fluorescence units (320/450 nm) of pFCC-1 (FUpFCC). Identity of pFCC-1 was confirmed with authentic standards (Kräutler et al., 1997; Mühlecker et al., 1997, 2000).
Ion Leakage
For ion conductivity analysis, senescence was induced in detached leaves that were incubated in the dark for up to 7 d. Eight leaf discs (1-cm diameter) were excised and transferred to 6-well cell culture plates (Sarstedt) containing 5 mL water. After reexposure to light (150 μmol photons m−2 s−1) for up to 8 h, ion leakage from the leaf discs as a measure of cellular damage was determined by measuring the conductivity of the solution with a CDH-42 conductivity meter (Omega Engineering).
Acknowledgments
We thank John Gray, University of Toledo, for the generous gift of antibodies against PAO/ACD1, and Esther Tapernoux-Lüthi, University of Zurich, for her help in sequencing.
This work was supported by the Swiss National Science Foundation (grant nos. 3100–063628 and 3100A0–105389), by the National Centre of Competence in Research (NCCR), Plant Survival, research program of the Swiss National Science Foundation, and the Austrian National Science Foundation (FWF; P–16097).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.065870.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmermann J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bachmann A, Fernández-López J, Ginsburg S, Thomas H, Bouwcamp JC, Solomos T, Matile P (1994) Stay-green genotypes of Phaseolus vulgaris L.: chloroplast proteins and chlorophyll catabolites during foliar senescence. New Phytol 126: 593–600 [Google Scholar]
- Barthe P, Vaillant V (1993) Changes in the buffering capacity of cell sap in senescing rose petals. Sci Hortic 54: 165–174 [Google Scholar]
- Berghold J, Breuker K, Oberhuber M, Hörtensteiner S, Kräutler B (2002) Chlorophyll breakdown in spinach: on the structure of five nonfluorescent chlorophyll catabolites. Photosynth Res 74: 109–119 [DOI] [PubMed] [Google Scholar]
- Berghold J, Eichmüller C, Hörtensteiner S, Kräutler B (2004) Chlorophyll breakdown in tobacco: on the structure of two nonfluorescent chlorophyll catabolites. Chem Biodivers 1: 657–668 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Curty C, Engel N (1996) Detection, isolation and structure elucidation of a chlorophyll a catabolite from autumnal senescent leaves of Cercidiphyllum japonicum. Phytochemistry 42: 1531–1536 [Google Scholar]
- Eckhardt U, Grimm B, Hörtensteiner S (2004) Recent advances in chlorophyll biosynthesis and breakdown in higher plants. Plant Mol Biol 56: 1–14 [DOI] [PubMed] [Google Scholar]
- Ginsburg S, Matile P (1993) Identification of catabolites of chlorophyll porphyrin in senescent rape cotyledons. Plant Physiol 102: 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg S, Schellenberg M, Matile P (1994) Cleavage of chlorophyll-porphyrin. Requirement for reduced ferredoxin and oxygen. Plant Physiol 105: 545–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J, Close PS, Briggs SP, Johal GS (1997) A novel suppressor of cell death in plants encoded by the Lls1 gene of maize. Cell 89: 25–31 [DOI] [PubMed] [Google Scholar]
- Gray J, Janick-Bruckner D, Bruckner B, Close PS, Johal GS (2002) Light-dependent death of maize lls1 cells is mediated by mature chloroplasts. Plant Physiol 130: 1894–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J, Wardzala E, Yang M, Reinbothe S, Haller S, Pauli F (2004) A small family of LLS1-related non-heme oxygenases in plants with an origin amongst oxygenic photosynthesizers. Plant Mol Biol 54: 39–54 [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Ausubel FM (1993) Arabidopsis mutants compromised for the control of cellular damage during pathogenesis and aging. Plant J 4: 327–341 [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Guo A, Klessig DF, Ausubel FM (1994) Programmed cell death in plants: a pathogen-triggered response activated coordinately with multiple defense functions. Cell 77: 551–563 [DOI] [PubMed] [Google Scholar]
- Hellens R, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hensel LL, Grbic V, Baumgarten DA, Bleecker AB (1993) Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in Arabidopsis. Plant Cell 5: 553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilditch PI, Thomas H, Thomas BJ, Rogers LJ (1989) Leaf senescence in a non-yellowing mutant of Festuca pratensis: proteins of photosystem II. Planta 177: 265–272 [DOI] [PubMed] [Google Scholar]
- Hinder B, Schellenberg M, Rodoni S, Ginsburg S, Vogt E, Martinoia E, Matile P, Hörtensteiner S (1996) How plants dispose of chlorophyll catabolites. Directly energized uptake of tetrapyrrolic breakdown products into isolated vacuoles. J Biol Chem 271: 27233–27236 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S (1999) Chlorophyll breakdown in higher plants and algae. Cell Mol Life Sci 56: 330–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtensteiner S, Kräutler B (2000) Chlorophyll breakdown in oilseed rape. Photosynth Res 64: 137–146 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S, Vicentini F, Matile P (1995) Chlorophyll breakdown in senescent cotyledons of rape, Brassica napus L.: enzymatic cleavage of phaeophorbide a in vitro. New Phytol 129: 237–246 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S, Wüthrich KL, Matile P, Ongania K-H, Kräutler B (1998) The key step in chlorophyll breakdown in higher plants. Cleavage of pheophorbide a macrocycle by a monooxygenase. J Biol Chem 273: 15335–15339 [DOI] [PubMed] [Google Scholar]
- Jonker JW, Buitelaar M, Wagenaar E, van der Valk MA, Scheffer GL, Scheper RJ, Plösch T, Kuipers F, Oude Elferink RPJ, Rosing H, et al (2002) The breast cancer resistance protein protects against a major chlorophyll-derived dietary phototoxin and protoporphyria. Proc Natl Acad Sci USA 99: 15649–15654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kräutler B (2003) Chlorophyll breakdown and chlorophyll catabolites. In KM Kadish, KM Smith, R Guilard, eds, The Porphyrin Handbook, Vol 13. Elsevier Science Publishing, New York, pp 183–209
- Kräutler B, Jaun B, Bortlik K-H, Schellenberg M, Matile P (1991) On the enigma of chlorophyll degradation: the constitution of a secoporphinoid catabolite. Angew Chem Int Ed Engl 30: 1315–1318 [Google Scholar]
- Kräutler B, Mühlecker W, Anderl M, Gerlach B (1997) Breakdown of chlorophyll: partial synthesis of a putative intermediary catabolite. Helv Chim Acta 80: 1355–1362 [Google Scholar]
- Kürsteiner O, Dupuis I, Kuhlemeier C (2003) The pyruvate decarboxylase1 gene of Arabidopsis is required during anoxia but not other environmental stresses. Plant Physiol 132: 968–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmeier M, Ginsburg S, Matile P (1993) Chlorophyll breakdown in senescent leaves: demonstration of Mg-dechelatase activity. Physiol Plant 89: 347–353 [Google Scholar]
- Lu Y-P, Li Z-S, Drozdowicz Y-M, Hörtensteiner S, Martinoia E, Rea PA (1998) AtMRP2, an Arabidopsis ATP binding cassette transporter able to transport glutathione S-conjugates and chlorophyll catabolites: functional comparisons with AtMRP1. Plant Cell 10: 267–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach JM, Castillo AR, Hoogstraten R, Greenberg JT (2001) The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc Natl Acad Sci USA 98: 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matile P, Ginsburg S, Schellenberg M, Thomas H (1988) Catabolites of chlorophyll in senescing barley leaves are localized in the vacuoles of mesophyll cells. Proc Natl Acad Sci USA 85: 9529–9532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matile P, Hörtensteiner S, Thomas H (1999) Chlorophyll degradation. Annu Rev Plant Physiol Plant Mol Biol 50: 67–95 [DOI] [PubMed] [Google Scholar]
- Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K (2001) FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 98: 12826–12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Laun T, Zimmermann P, Zentgraf U (2004) Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol 55: 853–867 [DOI] [PubMed] [Google Scholar]
- Mühlecker W, Kräutler B (1996) Breakdown of chlorophyll: constitution of nonfluorescing chlorophyll-catabolites from senescent cotyledons of the dicot rape. Plant Physiol Biochem 34: 61–75 [Google Scholar]
- Mühlecker W, Kräutler B, Moser D, Matile P, Hörtensteiner S (2000) Breakdown of chlorophyll: a fluorescent chlorophyll catabolite from sweet pepper (Capsicum annuum). Helv Chim Acta 83: 278–286 [Google Scholar]
- Mühlecker W, Ongania K-H, Kräutler B, Matile P, Hörtensteiner S (1997) Tracking down chlorophyll breakdown in plants: elucidation of the constitution of a “fluorescent” chlorophyll catabolite. Angew Chem Int Ed Engl 36: 401–404 [Google Scholar]
- Oberhuber M, Berghold J, Breuker K, Hörtensteiner S, Kräutler B (2003) Breakdown of chlorophyll: a nonenzymatic reaction accounts for the formation of the colorless “nonfluorescent” chlorophyll catabolites. Proc Natl Acad Sci USA 100: 6910–6915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhuber M, Berghold J, Mühlecker W, Hörtensteiner S, Kräutler B (2001) Chlorophyll breakdown - on a nonfluorescent chlorophyll catabolite from spinach. Helv Chim Acta 84: 2615–2627 [Google Scholar]
- Pružinská A, Anders I, Tanner G, Roca M, Hörtensteiner S (2003) Chlorophyll breakdown: pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc Natl Acad Sci USA 100: 15259–15264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca M, James J, Pružinská A, Hörtensteiner S, Thomas H, Ougham H (2004) Analysis of the chlorophyll catabolism pathway in leaves of an introgression senescence mutant of Lolium temulentum. Phytochemistry 65: 1231–1238 [DOI] [PubMed] [Google Scholar]
- Rodoni S, Mühlecker W, Anderl M, Kräutler B, Moser D, Thomas H, Matile P, Hörtensteiner S (1997) Chlorophyll breakdown in senescent chloroplasts. Cleavage of pheophorbide a in two enzymic steps. Plant Physiol 115: 669–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg M, Matile P, Thomas H (1993) Production of a presumptive chlorophyll catabolite in vitro: requirement for reduced ferredoxin. Planta 191: 417–420 [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y, et al (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145 [DOI] [PubMed] [Google Scholar]
- Sidler M, Hassa P, Hasan S, Ringli C, Dudler R (1998) Involvement of an ABC transporter in a developmental pathway regulating hypocotyl cell elongation in the light. Plant Cell 10: 1623–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassieva S, Hille J (2002) A lesion mimic phenotype in tomato obtained by isolating and silencing an Lls1 homologue. Plant Sci 162: 543–549 [Google Scholar]
- Strain HH, Cope BT, Svec WA (1971) Analytical procedures for the isolation, identification, estimation and investigation of the chlorophylls. Methods Enzymol 23: 452–476 [Google Scholar]
- Tanaka R, Hirashima M, Satoh S, Tanaka A (2003) The Arabidopsis-accelerated cell death gene ACD1 is involved in oxygenation of pheophorbide a: inhibition of pheophorbide a oxygenase activity does not lead to the “stay-green” phenotype in Arabidopsis. Plant Cell Physiol 44: 1266–1274 [DOI] [PubMed] [Google Scholar]
- Tommasini R, Vogt E, Fromenteau M, Hörtensteiner S, Matile P, Amrhein N, Martinoia E (1998) An ABC transporter of Arabidopsis thaliana has both glutathione-conjugate and chlorophyll catabolite transport activity. Plant J 13: 773–780 [DOI] [PubMed] [Google Scholar]
- Tsuchiya T, Ohta H, Okawa K, Iwamatsu A, Shimada H, Masuda T, Takamiya K (1999) Cloning of chlorophyllase, the key enzyme in chlorophyll degradation: finding of a lipase motif and the induction by methyl jasmonate. Proc Natl Acad Sci USA 96: 15362–15367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicentini F, Hörtensteiner S, Schellenberg M, Thomas H, Matile P (1995) Chlorophyll breakdown in senescent leaves: identification of the biochemical lesion in a stay-green genotype of Festuca pratensis Huds. New Phytol 129: 247–252 [DOI] [PubMed] [Google Scholar]
- Wagner D, Przybyla D, op den Camp R, Kim C, Landgraf F, Lee KP, Würsch M, Laloi C, Nater M, Hideg E, et al (2004) The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306: 1183–1185 [DOI] [PubMed] [Google Scholar]
- Weaver LM, Amasino RM (2001) Senescence is induced in individually darkened Arabidopsis leaves, but inhibited in whole darkened plants. Plant Physiol 127: 876–886 [PMC free article] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang M, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbot D, Stoutjesdijk PA, et al (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Wüthrich KL, Bovet L, Hunziker PE, Donnison IS, Hörtensteiner S (2000) Molecular cloning, functional expression and characterisation of RCC reductase involved in chlorophyll catabolism. Plant J 21: 189–198 [DOI] [PubMed] [Google Scholar]
- Yang M, Wardzala E, Johal GS, Gray J (2004) The wound-inducible Lls1 gene from maize is an orthologue of the Arabidopsis Acd1 gene, and the LLS1 protein is present in non-photosynthstic tissues. Plant Mol Biol 54: 175–191 [DOI] [PubMed] [Google Scholar]
- Yao N, Eisfelder BJ, Marvin J, Greenberg JT (2004) The mitochondrion—an organelle commonly involved in programmed cell death in Arabidopsis thaliana. Plant J 40: 596–610 [DOI] [PubMed] [Google Scholar]