Abstract
In Arabidopsis (Arabidopsis thaliana), light-dependent chloroplast movements are induced by blue light. When exposed to low fluence rates of light, chloroplasts accumulate in periclinal layers perpendicular to the direction of light, presumably to optimize light absorption by exposing more chloroplast area to the light. Under high light conditions, chloroplasts become positioned parallel to the incoming light in a response that can reduce exposure to light intensities that may damage the photosynthetic machinery. To identify components of the pathway downstream of the photoreceptors that mediate chloroplast movements (i.e. phototropins), we conducted a mutant screen that has led to the isolation of several Arabidopsis mutants displaying altered chloroplast movements. The plastid movement impaired1 (pmi1) mutant exhibits severely attenuated chloroplast movements under all tested fluence rates of light, suggesting that it is a necessary component for both the low- and high-light-dependant chloroplast movement responses. Analysis of pmi1 leaf cross sections revealed that regardless of the light condition, chloroplasts are more evenly distributed in leaf mesophyll cells than in the wild type. The pmi1-1 mutant was found to contain a single nonsense mutation within the open reading frame of At1g42550. This gene encodes a plant-specific protein of unknown function that appears to be conserved among angiosperms. Sequence analysis of the protein suggests that it may be involved in calcium-mediated signal transduction, possibly through protein–protein interactions.
Plants employ a variety of mechanisms to maximize photosynthetic potential under changing environmental conditions. For example, most plants can biochemically adapt to excessive light by altering the size of light harvesting complexes, dissipating excess energy as heat, scavenging reactive oxygen, and rapidly repairing damaged reaction center proteins (Niyogi, 1999). Developmental changes that occur in response to different light conditions include variations in leaf thickness, density of mesophyll cells, and changes in the orientation of the leaf blade (Boardman, 1977). These responses have been associated with improved photosynthetic potential and/or protection from photodamage. However, their relative importance can be species specific, indicating that some of these responses represent unique adaptive strategies. Another perhaps more ubiquitous plant response to varying light conditions is light-induced chloroplast movement, which has been documented in a large number of species, including alga, moss, ferns, and angiosperms (Zurzycki, 1961; Inoue and Shibata, 1973; Lechowski, 1974; Brugnoli and Björkman, 1992; Dong et al., 1996; Parks et al., 1996; Trojan and Gabrys, 1996; Augustynowicz and Gabrys, 1999; Gorton et al., 1999; Kagawa and Wada, 1999; Kadota et al., 2000). In alga, moss, and ferns, chloroplast migration is induced by both red light (RL) and blue light (BL) (Kagawa and Wada, 1994; Kadota et al., 2000). In angiosperms, chloroplast movements are strictly BL induced and can be separated into two distinct responses (Trojan and Gabrys, 1996; Kagawa and Wada, 2000).
In response to low fluence rates of light, chloroplasts accumulate along periclinal cell walls, perpendicular to the incident light. When exposed to high fluence rates, chloroplasts move to anticlinal walls, parallel to the incident light. Light-induced chloroplast movements are thought to provide an adaptive function (Zurzycki, 1955, 1972; Seitz, 1972; Inoue and Shibata, 1973). Under potentially damaging high light conditions, chloroplasts move to parts of the leaf cell that transmit less light as a protective measure against photodamage (Seitz, 1972). Consistent with this hypothesis, Arabidopsis (Arabidopsis thaliana) mutants that are unable to move their chloroplasts in response to high fluence rates of light were more sensitive to photodamage than the wild type (Kasahara et al., 2002). Similarly, it has been suggested that under light-limiting conditions, the accumulation of chloroplasts perpendicular to the incident light allows for more efficient absorption as demonstrated by increased photosynthesis as chloroplasts move into the light path (Zurzycki, 1955; Lechowski, 1974).
In Arabidopsis, the BL photoreceptors phototropin 1 (phot1) and phot2 mediate light-induced chloroplast movements (Jarillo et al., 2001; Kagawa et al., 2001, 2004; Sakai et al., 2001), with phytochromes playing a minor role in modulating the high-light response (DeBlasio et al., 2003). Interestingly, the putative signal transducers nonphototropic hypocotyl 3 (NPH3) and root phototropism 2 (RPT2), which are required for phototropin signaling during phototropism and stomatal opening (Motchoulski and Liscum, 1999; Sakai et al., 2001), are not involved in BL-induced chloroplast movements (Inada et al., 2004). Although there are several members of the NPH3/RPT2 gene family in Arabidopsis (Motchoulski and Liscum, 1999), none have been implicated in chloroplast movements, and it is possible that phototropin signaling during light-induced chloroplast movements requires a different type of transduction system.
Pharmological studies have revealed that the motility system for light-induced chloroplast movements in angiosperms uses the actin cytoskeleton. For example, addition of cytochalasin and latrunculin disrupt chloroplast movements, whereas colchicine does not, implicating actin and not microtubules to be the candidate cytoskeletal network (Tlalka and Gabrys, 1993). The myosin ATPase inhibitor, N-ethylmaleimide (NEM), also disrupts the responses in Lemna leaves, suggesting that myosin serves as the motor for chloroplast movements (Malec et al., 1996). Furthermore, a subclass of myosin XI has been shown to be associated with plastids in corn (Zea mays; Wang and Pesacreta, 2004). However, because NEM interferes with other types of ATPases (Shimmen and Yakota, 1994; Locoro et al., 1999; Takahashi and Yamaguchi, 1999), it is unclear if its effect on chloroplast movements was a direct result of myosin inactivation. An alternative hypothesis is that actin dynamics may drive chloroplasts movements. It is well known that actin assembly at the rear of Listeria monocytogenes is enough to generate the force required for movement of the bacteria within and between animal cells (Southwick and Purich, 1994; Marchand et al., 1995). Additionally, actin-nucleating proteins such as formin can act as processive motors to drive actin-based propulsion (Golsteyn et al., 1997; Romero et al., 2004). In Arabidopsis cells, chloroplasts can be seen partially or completely encircled by F-actin microfibrils (Kandasamy and Meagher, 1999; Oikawa et al., 2003). In Vallisneria and Adiantum protenemal cells, chloroplast movement was only observed after the thick bundles of microfilaments, found to form around the chloroplasts, fragmented after exposure to light (Kadota and Wada, 1992; Dong et al., 1996). Mutations within Chloroplast Unusual Positioning1 (CHUP1), a Pro-rich, actin-binding protein located on the chloroplast envelope, resulted in loss of chloroplast movement and aggregation of chloroplasts at the bottom of cells (Oikawa et al., 2003). It has been suggested that CHUP1 functions as an anchor, binding chloroplasts to the plasma membrane via the actin cytoskeleton (Wada and Suetsugu, 2004). However, the presence of a putative profilactin-binding motif suggests that CHUP1 may play a more active role in actin assembly (Oikawa et al., 2003).
Regardless of the exact nature of the motility system involved in BL-induced chloroplasts movements, rearrangements of the actin cytoskeleton as well as myosin motor function can be regulated by calcium (Ca2+) concentrations within the cell (for review, see Wolenski, 1995; Staiger, 2000). Indeed, light-induced chloroplast movements can be affected by altering cytosolic Ca2+ levels (Tlalka and Gabrys, 1993; Tlalka and Fricker, 1999; Sato et al., 2001). Modification of extracellular stores of Ca2+ either by addition of the Ca2+ chelating agent EGTA or the Ca2+ influx inhibitors, lanthanum and gadolinium, had little to no effect on the light-induced chloroplast movements of Lemna trisulca and the fern Adiantum capillus-veneris (Tlalka and Gabrys, 1993; Sato et al., 2001). However, depletion of intracellular stores of Ca2+ by adding the Ca2+ ionophore A 23187 plus EGTA to the external media was sufficient to inhibit both low- and high-light chloroplast migration in Lemna leaves exposed to 30-s pulses of BL (Tlalka and Gabrys, 1993). It has been shown that phot1 and phot2, which are bound to the plasma membrane (Sakamoto and Briggs, 2002), mediate BL-induced increases in cytosolic Ca2+ in Arabidopsis mesophyll cells (Babourina et al., 2002; Harada et al., 2003; Stoelzle et al., 2003). At fluence rates between 0.1 and 50 μmol·m−2·s−1, phot1 induces Ca2+ influx from the apoplast through a channel within the plasma membrane. At fluence rates between 1 and 250 μmol·m−2·s−1, phot2 mediates Ca2+ influx through plasma membrane bound channels as well as release of Ca2+ from internal stores via a phospholipase C-mediated phosphoinositide signaling pathway (Harada et al., 2003). Although these transient increases in cytosolic Ca2+ have been shown to be required for phototropism, inhibition of hypocotyl growth and stomatal regulation (Allen et al., 1999; Shimazaki et al., 1999; Folta et al., 2003; Babourina et al., 2004), it is unclear how or if they are required for blue-light-induced chloroplast movements.
To identify components of the chloroplast movement pathway downstream of the phototropins, we developed a sensitive screening procedure to isolate Arabidopsis mutants with altered chloroplast movements. The screen is based on the proven method of using changes in RL transmittance through intact leaves as a reliable indicator of chloroplast relocation (Trojan and Gabrys, 1996; DeBlasio et al., 2003). In this article, we report on the identification and characterization of plastid movement impaired1 (pmi1), a mutant that exhibits severely attenuated chloroplast movements under both low and high fluence rates of BL. We determined that the pmi1-1 chloroplast movement phenotype is caused by a single nonsense mutation within the gene At1g42550, which encodes a plant-specific protein of unknown function that is conserved between monocots and dicots. Alignment of the deduced amino acid sequence of At1g42550 with a protein sequence found in rice (Oryza sativa) suggests that the PMI1 protein may be involved in phototropin signal transduction to the actin cytoskeleton via interactions with proteins containing C2 Ca2+ binding motifs.
RESULTS
Isolation of Chloroplast Movement Mutants
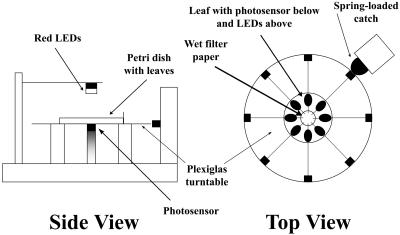
Mutants with altered chloroplast movements were identified using a device that allowed us to monitor the changes in RL transmittance through leaves that occur as a result of BL-induced chloroplast movements (DeBlasio et al., 2003). The apparatus consists of a rotating clear Plexiglas turntable with a red light source (660 nm) mounted above the turntable at the measuring position and a quantum sensor mounted directly below (Fig. 1). Leaves excised from approximately 6000 ethyl methanesulfonate (EMS) mutagenized Arabidopsis plants (M2s) were placed in a modified petri dish with leaf blades located around the perimeter of the dish and their petioles resting on a wet filter paper circle located in the center. After leaves were dark acclimated for 15 to 20 h under saturating humidity, RL transmittance was measured through each leaf using the device in Figure 1. Leaves were then exposed to sequential 1-h treatments of low and high fluence rate BL (1.7 and 70 μmol·m−2·s−1, respectively). RL transmittance was measured after each light treatment and normalized to the dark-acclimated values, and the responses were then compared to the wild type. For all M2 plants that exhibited an abnormal change in RL transmittance in response to the low and/or high fluence rates of BL, leaves from their progeny were subjected to a more sensitive kinetic analysis (DeBlasio et al., 2003).
Figure 1.
Schematic of device used for chloroplast movement mutant screen. Modified petri dishes containing up to eight leaves can be placed on the turntable. The turntable rotates to eight precise positions so that each leaf can be located between the red measuring beam and the photodetector. The design allows for rapid removal and replacement of petri dishes while ensuring that each measurement of light transmittance is made through the same area of each leaf enabling fast and accurate measurements from a large number of leaves. LEDs, light-emitting diodes.
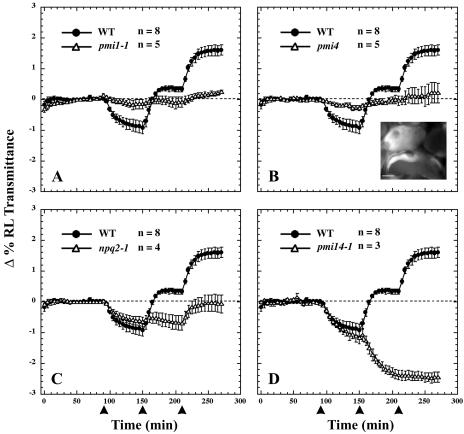
The 15 mutant lines that we have identified so far fall into four phenotypic classes. All previously undescribed mutants have been designated pmi for plastid movement impaired (Fig. 2). Members of the first mutant class display severely attenuated chloroplast movement responses under all fluence rates of BL (Fig. 2A). Another group of mutants also exhibit very little light-directed chloroplast movements, but microscopic analysis revealed defects in chloroplast division since their mesophyll cells on average contained one to three large chloroplasts (Fig. 2B, inset). Other mutants exhibited a normal low light response but an attenuated high light response, including the zeazanthin overproducing mutant npq2-1 (Fig. 2C). Chloroplasts in the mutants of the last phenotypic class (pmi14-1 and pmi14-2) move to the low-light position even when the leaves are exposed to high fluence rates of BL (Fig. 2D). We have so far isolated five pmi mutants with the class I phenotype: three in class II, five in class III, and two in class IV. Complementation analysis showed the two members of class IV to be alleles of the BL photoreceptor gene, PHOT2. The remaining 12 pmi mutants appear to represent unique loci.
Figure 2.
BL-induced chloroplast movements in wild-type and pmi mutants. The plots show the average change (± se) in the percentage of RL transmittance of leaves relative to the average transmittance measured prior to the first BL treatment. RL transmittance was measured in dark-acclimated leaves for 90 min before sequential treatments of 0.3, 20, and 60 μmol·m−2·s−1 of BL (450 nm ± 25 nm) indicated by the arrowheads at 90, 150, and 210 min. Number of leaves (n) is shown for each treatment. Inset is an autofluorescence micrograph of the giant chloroplasts in pmi4 palisade cells. WT, Wild type. Bar = 10 μm.
Chloroplast Distribution in pmi1 Leaf Cells
When exposed to low or high fluence rates of BL, pmi1 leaves exhibited severely attenuated chloroplast movements as indicated by the lack of change in RL transmittance (Fig. 2A). We also analyzed chloroplast movements by time-lapse microscopy in palisade cells of dark-acclimated leaf sections exposed to sequential treatments of low and high fluence rates of white light. As previously shown (Kagawa and Wada, 2000), wild-type chloroplasts migrated to the periclinal regions of the palisade cells upon exposure to low-fluence-rate light (Fig. 3B) and migrated to the anticlinal regions when exposed to high-fluence-rate light (Fig. 3C). Chloroplasts in the pmi1 mutant failed to show any directed movements when exposed to the same light treatments (Fig. 3, E and F).
Figure 3.
BL-induced chloroplast migration and positioning in wild-type and pmi1 leaf cells. Dark-acclimated wild-type (A–C) and pmi1-1 (D–F) palisade cells were exposed to sequential 1-h treatments of low- and high-fluence-rate white light from below. Micrographs from time points 0 (A and D), 60 (B and E), and 120 (C and F) min are shown. Bar = 10 μm. Cross sections of wild-type and pmi1-1 rosette leaves exposed to 1 h of either darkness (G and J), low BL (H and K), or high BL (I and L) show chloroplast positioning in all cell layers. Bar = 30 μm. Bar graphs depict the percentage (± se) of chloroplasts located along the anticlinal and periclinal cells walls in fixed wild-type and pmi1-1 leaf cross sections and represent averages from 200 to 400 cells per BL treatment.
Analysis of chloroplast position in cross sections (Fig. 3, F and G) showed that in wild-type leaves exposed to low BL (4.5 μmol·m−2·s−1), approximately 80% of the chloroplasts were located along the periclinal walls, whereas under high BL (100 μmol·m−2·s−1), approximately 70% of chloroplasts were along the anticlinal walls (Fig. 3M). In pmi1 leaf sections, chloroplasts were more evenly distributed along both the anticlinal and periclinal cell walls in both light conditions (Fig. 3N). Distribution of chloroplasts within pmi1 cells is different than the chup1 mutant phenotype, in which chloroplasts cluster on the adaxial side of the cells (Oikawa et al., 2003).
No difference was observed between wild type and pmi1 in their phototropic responses to unilaterial low-fluence-rate BL as well as stomatal regulation, indicating that these phototropin-mediated pathways are still functional in the pmi1 mutant (data not shown).
Organization of the pmi1 Actin Cytoskeleton
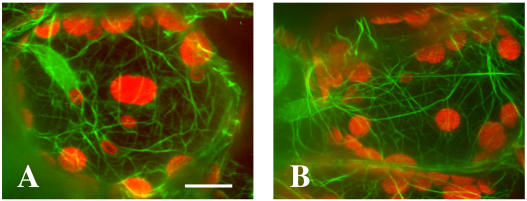
To visualize the actin cytoskeleton in mutant cells, pmi1 plants were crossed to transgenic wild-type plants expressing green fluorescent protein mouse talin (GFP-mtalin) (Kost et al., 1998). Thick actin filaments were distributed over the cell cortex and around chloroplasts in both wild-type and pmi1 palisade cells (Fig. 4). There appeared to be no obvious difference between pmi1 and wild type with respect to the organization of the actin cytoskeleton within these cells.
Figure 4.
Organization of the actin cytoskeleton in wild-type and pmi1 leaf cells. Confocal images of wild-type (A) and pmi1 (B) palisade cells expressing GFP-mtalin. GFP fluorescence is shown in green and chloroplast autofluorescence in red. Images represent projections of 36 optical 0.2-μm sections. Bar = 10 μm.
Identification of the PMI1 Gene
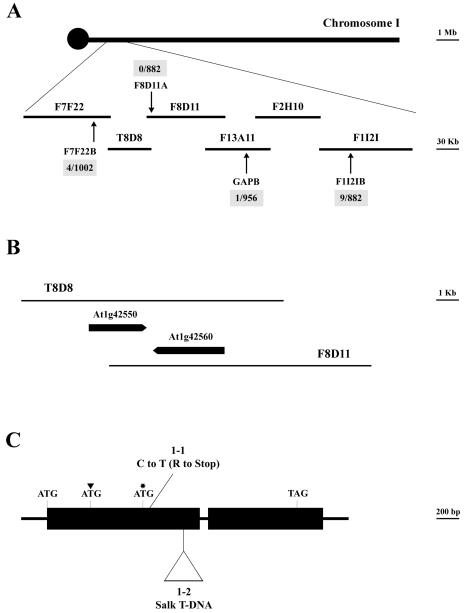
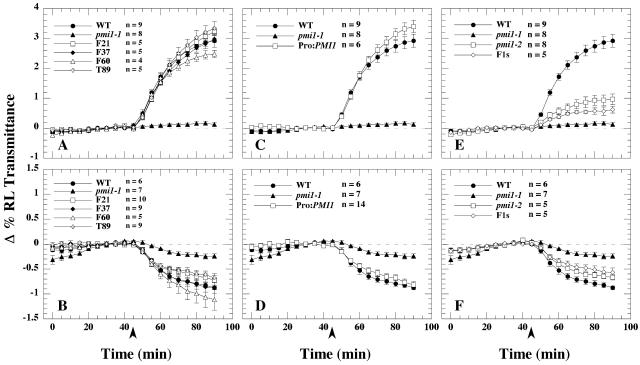
Analysis of F2 progeny obtained from crossing the wild type with pmi1 revealed that the chloroplast movement defect was due to a single, nuclear recessive mutation (data not shown). The pmi1 mutation exhibited 85.1% linkage to the physical marker nga280 on chromosome I. Using a variety of simple sequence length polymorphic (SSLP) and cleaved-amplified polymorphic sequence (CAPS) markers (see “Material and Methods”), we reduced the position of the PMI1 locus to a 217.57-kb region covered by bacterial artificial chromosomes (BACs) T8D8 and F8D11 (Fig. 5A). We were able to rescue the pmi1 chloroplast movement defect with genomic sequence spanning the 8-kb overlap of BAC T8D8 and BAC F8D11 (Fig. 6, A and B). Two genes were located within this region: the putative disease resistance gene Mlo9 (At1g42560) and At1g42550, a gene of unknown function (Fig. 5B). Sequence data for both genes were obtained from several wild-type and pmi1 plants using primers specific for At1g42560 and At1g42550. A single C-to-T nucleotide change was found within the coding region of At1g42550 in the pmi1 mutant (Fig. 5C). The base pair change results in an Arg residue being changed into an early stop codon (Fig. 7). We have designated this mutant allele pmi1-1. Subsequently, we were able to compliment the pmi1-1 chloroplast movement defect by transforming wild-type At1g42550 genomic DNA into mutant plants. The resulting T1 progeny displayed wild-type chloroplast movement responses when exposed to low and high fluence rates of BL (Fig. 6, C and D).
Figure 5.
Mapping of the PMI1 locus and organization of the corresponding PMI1 gene. A, Genetic interval located south of the centromere on chromosome 1 is shown with corresponding SSLP and CAPS markers used to fine map the pmi1-1 EMS mutation. Arrows indicate the positions of the markers and the number of recombinants per number of chromosomes tested. B, The pmi1-1 chloroplast movement defect was rescued by the 8-kb overlapping region of BAC T8D8 and F8D11. The positions of the two genes located in this genetic interval are shown. C, Diagram of the PMI1 gene (T8D8.2). Exons are depicted as thick bars and introns as thin lines. Positions of the initiation and termination codons as well as the pmi1-1 point mutation and pmi1-2 T-DNA insertion are shown. The position of the original annotated and third putative start codons are indicated by an arrowhead and an asterisk, respectively.
Figure 6.
BL-induced chloroplast movements of rescued pmi1-1 transgenic and pmi1-2 plants. RL transmittance was measured in dark-acclimated leaves for 45 min before exposure to either 40 (A and C) or 0.3 (B and D) μmol·m−2·s−1 of BL (450 nm ± 25 nm) indicated by the arrowheads. The plots show the average change (± se) in the percentage of RL transmittance of leaves relative to the average transmittance measured for the leaves prior to the BL treatment. Pro:PMI1 is the wild-type PMI1 gene driven by its own promoter. Number of leaves (n) is shown for each treatment.
Figure 7.
Diagram of PMI1 protein. A, The deduced amino acid sequence of PMI1 based on GenBank cDNA sequence and RT-PCR results. The putative cleavage signal is underlined. The raised boxes indicate the coil-coiled sequences and the dotted box the putative bombesin-like domain. The two conserved domains revealed by sequence alignment with orthologous sequences are in boldface. B, Diagram of the PMI1 protein. The positions of the original annotated and third putative start codons are indicated by an arrowhead and an asterisk, respectively. Arrows indicate the position of the pmi1-1 and pmi1-2 mutations. a.a., amino acids.
An Arabidopsis line containing a putative T-DNA insert in At1g42550 (designated Salk_141795) was obtained from the SALK Institute's SIGnAL collection (Alonso et al., 2003). True-breeding homozygous mutant lines exhibited attenuated low and high light chloroplast movement responses, but the defect was less severe than that observed for the pmi1-1 EMS allele (Fig. 6, E and F). DNA sequencing confirmed the presence of a tandem T-DNA insertion between the 468th and 575th codon of At1g42550 within this T-DNA mutant line (data not shown).
F1 progeny obtained from crosses made between pmi1-1 plants with Salk_141795 pollen also exhibited attenuated chloroplast movements when exposed to low and high fluence rates of BL (Fig. 6, E and F), consistent with the mutants being allelic. We have thus designated Salk_141795, pmi1-2. However, when compared to the chloroplast movement responses of pmi1-1 and pmi1-2, F1 leaves were similar to pmi1-2, suggesting that even in a heterozygotic state, the T-DNA mutant allele is able to produce a somewhat functional protein capable of partially inducing both chloroplast movement responses.
Based on the sequence obtained from the pmi1-1 mutant, the results with our transgenic lines, and the identification of a T-DNA insertion allele, we conclude that At1g42550 is the PMI1 locus. According to the annotated cDNA sequence for Accession AY072341, the genomic sequence of At1g42550 is comprised of two exons separated by a 70-bp intron with a 121-bp 5′-untranslated region (UTR) and 217-bp 3′-UTR flanking a 2.124-kb coding region. However, sequence analysis of the 5′ flanking region of At1g42550 revealed an alternative start codon 405 bp upstream of the annotated start site. Using a sense primer starting at this putative start codon, we were able to generate cDNA from wild-type (Columbia [Col]) leaf RNA extract, suggesting that the At1g42550 coding region can be 2.529 kb (Fig. 5C). This extended coding region has been designated T8D8.2 by the National Center for Biotechnology Information (Accession AAG51317). Interestingly, even though only the last 1.926 kb of the At1g42550 open reading frame are located on BAC F8D11, we were still able to rescue the pmi1-1 chloroplast movement defect with clones from this region (Fig. 6, A and B), suggesting that At1g42550 may have a third putative start site 499 bp downstream from the original annotated start site (Fig. 5C). This truncated sequence has been designated F8D11.1 by the National Center for Biotechnology Information (Accession AAG51233).
PMI1 Encodes a Putative Signal Transduction Protein
The predicted amino acid sequence of At1g42550 (Fig. 7A) was analyzed with publicly available programs. SignalP predicted a putative secretory cleavage signal near the N terminus of the protein between residues A15 and Q16 (http://www.cbs.dtu.dk/services/Signalp/). The COILS program predicted the presence of two small coiled-coil regions that could act as sites for protein–protein interactions (Fig. 7). The Myhits motif scan indicated the presence of a bombesin-like domain (Nagalla et al., 1996; Merali et al., 2002) adjacent to the coiled-coil region at the C terminus of the protein, but the putative domain lacks the necessary cleavage signals found in true bombesin proteins (Nagalla et al., 1996).
BLAST searches using the entire amino acid sequence of PMI1 against the National Center for Biotechnology Information database and The Institute for Genomic Research gene indices (http://tigrblast.tigr.org/tgi/) and maize database (http://tigrblast.tigr.org/tgi_maize/index.cgi) identified orthologous protein sequences in rice, corn, soybean (Glycine max), and Medicago trunculata. Orthologous sequences of AtPMI1 were not found in any of the available databases for organisms outside the plant kingdom.
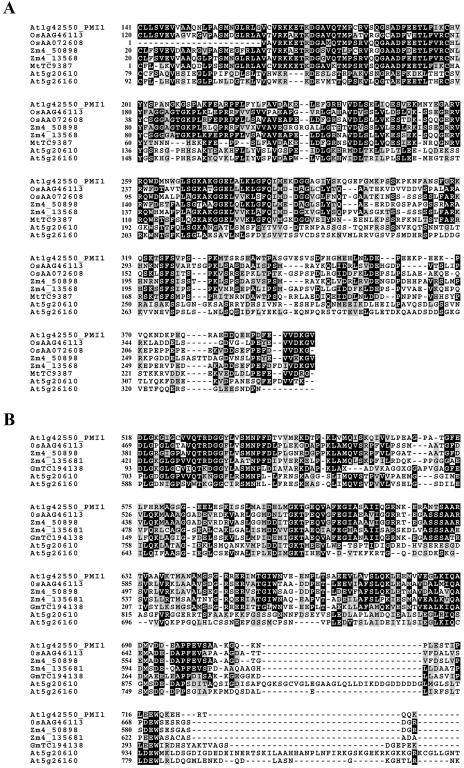
ClustalW alignment of these orthologous amino acid sequences with PMI1 revealed the presence of two highly conserved domains (Figs. 7 and 8). One domain, located just downstream of the putative cleavage signal, shares 48% identity and 64% positive changes to OsAAO72608, a protein identified in a yeast two-hybrid screen as a binding partner of the fungal elicitor protein OsERP3 (Cooper et al., 2003; Figs. 7 and 8A). Although the function of OsAAO72608 is unknown, OsERP3 (Accession AF090698) is a C2 calcium binding protein that is induced in response to stress and fungal pathogen infection (Cooper et al., 2003).
Figure 8.
Alignment of conserved central (A) and C-terminal (B) domains of PMI1 from Arabidopsis with putative orthologous sequences from O. sativa (Os), Z. mays (Zm), G. max (Gm), and M. trunculata (Mt). Orthologous sequences were obtained from a BLAST search of the National Center for Biotechnology Information and The Institute for Genomic Research gene indices and maize database using AtPMI1 as a reference. Sequences were aligned using ClustalW (http://www.ebi.ac.uk/clustalw/) and the output produced by BoxShade (http://www/ch.embnet.org/software/BOX_form.html). Black boxes indicate identity, and gray represents similarity between residues. Alignment of At5g20610 and At5g26160, putative members of the PMI1 gene family, are shown as well as the protein accession numbers for the orthologous sequences.
Another domain conserved between At1g42550 and its orthologs is located at the C terminus of the protein (Figs. 7 and 8B). Unfortunately, we were not able to identify a function for this domain, nor were we able to identify proteins with known function that contained this domain. In the AtPMI1 protein sequence, this domain is separated from the N-terminal domain by a region predicted by the ExPASy ScanProsite program to be enriched with acidic residues Asp (D) and Glu (E; Fig. 7A). Although its biochemical function remains unknown, the C-terminal portion of AtPMI1 appears to be functionally important for chloroplast movement since we were able to complement the pmi1-1 movement defect with constructs containing sequence coding for the last 542 amino acids of the AtPMI1 protein (Fig. 7, start of smaller protein is indicated by the asterisk).
Even though we were unable to identify other Arabidopsis proteins that were homologous to full-length PMI1, we identified two other Arabidopsis proteins (At5g20610 and At5g26160) with similarities to the central and C-terminal domains found in AtPMI1 (Accession NM_122068 and NM_122517). The sequence similarity between At5g20610, At5g26160, and AtPMI1 within these regions is less than that observed for the PMI1-like sequences found in the other plant species, indicating that At5g20610 and At5g26160 are most likely members of the same protein family rather than homologs of AtPMI1 (Table I).
Table I.
Comparison of AtPMI1 domains with putative PMI1s from four other plant species
Values indicate percentage identity (above the diagonal) and similarity (below the diagonal) and were obtained by National Center for Biotechnology Information BLASTP and LALIGN (http://www.ch.embnet.org/software/LALIGN_form.html) with the sequences shown in Figure 7.
| Central Domain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| 1. AtPMI1 | – | 41 | 48 | 40 | 53 | 55 | 32 | 36 |
| 2. OsAA46113 | 58 | – | 46 | 68 | 64 | 60 | 28 | 31 |
| 3. OsAA072608 | 64 | 61 | – | 44 | 77 | 48 | 24 | 34 |
| 4. Zm4_50898 | 58 | 75 | 59 | – | 48a | 38a | 25 | 27 |
| 5. Zm4_13568 | 65 | 48 | 85 | 24a | – | 51a | 27 | 32 |
| 6. MtTC9387 | 72 | 39 | 65 | 29a | 24a | – | 47 | 53 |
| 7. At5g20610 | 57 | 50 | 46 | 44 | 43 | 26 | – | 40 |
| 8. At5g26160 | 59 | 54 | 57 | 52 | 53 | 33 | 57 | – |
| C-Terminal Domain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| 1. AtPMI1 | – | 51 | 51 | 52 | 58 | 32 | 27 | |
| 2. OsAA46113 | 69 | – | 84 | 51 | 46 | 34 | 26 | |
| 3. Zm4_50898 | 71 | 90 | – | 52a | 46a | 33 | 27 | |
| 4. Zm4_13568 | 72 | 70 | 28a | – | 51a | 29 | 24 | |
| 5. GmTC194138 | 72 | 64 | 30a | 26a | – | 56 | 44 | |
| 6. At5g20610 | 53 | 57 | 56 | 54 | 34 | – | 36 | |
| 7. At5g26160 | 46 | 46 | 44 | 43 | 26 | 49 | – |
Expression of PMI1
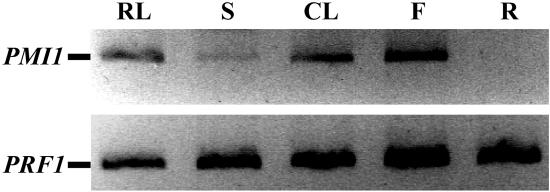
Expression of the AtPMI1 gene (At1g42550) was examined by RT-PCR from total RNA isolated from various organs of wild-type plants (Fig. 9). To avoid contamination from traces of genomic DNA, gene-specific primers flanking the 70-bp intron of At1g42550 were used. We were able to detect AtPMI1 mRNA in leaves, stems, cauline leaves, and whole flowers but not in roots. Since mRNA of the control gene, profilin1, was uniformly present in all these samples, AtPMI1 mRNA is either expressed in roots at extremely low levels not detectable by our RT-PCR conditions or not at all.
Figure 9.
Expression of PMI1 in Arabidopsis tissues. Total RNA was extracted from various wild-type (Col) plant tissues and PMI1 transcript measured by RT-PCR using gene-specific primers spanning the 70-bp intron of At1g42550. The product from the fully spliced mRNA is shown. Primers spanning the two introns of profilin1 were used as a control. RNA was combined from the rosette leaves (RL), stems (S), cauline leaves (CL), flowers (F), and roots (R) of three different 9-week-old plants.
DISCUSSION
In this study, we describe the identification of PMI1, a novel gene required for BL-induced chloroplast movements in Arabidopsis. Mutations within this gene resulted in the attenuation of chloroplast movements under low and high fluence rates of BL, indicating that PMI1 encodes a component involved in both movement responses (Fig. 2A). Cross sections of pmi1-1 leaves revealed that the defect was probably not due to disassociation of the chloroplasts from CHUP1 since chloroplasts in the pmi1-1 mutant were distributed around the periphery of the cells (Fig. 3) and not clustered at the bottom as seen in the previously identified chup1 mutant (Oikawa et al., 2003). The predicted AtPMI1 sequence does not resemble any functionally characterized protein, including proteins known to be associated with actin (Fig. 7). In addition, transgenic pmi1 plants expressing GFP-mtalin revealed no obvious defects in organization of the actin cytoskeleton when compared with the wild type.
Sequence alignment with putative orthlogous sequences found in rice, corn, and various dicots revealed the presence of conserved domains in the central and C-terminal portions of all of these orthologs (Fig. 8). Despite their evolutionary distance, AtPMI1 shares between 40% and 53% of the same residues and 57% to 72% similar residue changes with the corresponding domains in the monocot sequences (Fig. 8). The sequence similarity of the AtPMI1 central domain with that of the small rice protein OsAA072608 (Fig. 8A; Table I) suggests that AtPMI1 may play a role in signal transduction via interaction with Ca2+ binding proteins. OsAA072608 (231 residues) was identified in a yeast two-hybrid screen as a putative binding partner of OsERP3, an elicitor response protein induced by stress and exposure to fungal pathogens (Cooper et al., 2003). In that study, OsERP3 was shown to interact with OsSGT1, a protein first identified in yeast as a component of Skp1-Cullin-F-Box ubiquitin ligases that target proteins for degradation (Kitagawa et al., 1999; Austin et al., 2002). Although the exact nature of OsERP3 function in plant defense is unknown, sequence analysis revealed that the entire OsERP3 protein is homologous to C2-type Ca2+ binding motifs (data not shown). These motifs are found in a variety of signal transduction molecules, including phophastidylinositol-specific phospholipase C, protein kinase C, synaptotagmin, and the defense/stress response copine proteins (Wang et al., 1999; Earles et al., 2001; Jambunathan et al., 2001). Once calcium is bound, these domains often bind to intracellular membranes (Kim et al., 1996). However, there are examples were some proteins have been shown to bind strongly to the Gα subunit of activated G proteins via their C2 domains (Wang et al., 1999).
In plants, induction of the phosphoinostide signaling pathway often results in release of Ca2+ from intracellular stores (Drøbak and Ferguson, 1985; Read et al., 1993), and there is evidence that components in the pathway can interact with actin-binding proteins such as profilin (Aderem, 1992; Xu et al., 1992). It has already been established that BL-induced chloroplast movements require both the actin cytoskeleton and release of Ca2+ from intracellular stores (Tlalka and Gabrys, 1993; Malec et al., 1996; Tlalka and Fricker, 1999; Sato et al., 2000, 2001). Because of its high sequence similarity to OsAAO72608, it is possible that the central domain of AtPMI1 may also function as a binding partner to C2 calcium binding proteins.
Complementation of the pmi1-1 movement defect with clones that only contained sequence coding for the last 542 amino acids of AtPMI1 (Fig. 7, start site is indicated by the asterisk) suggests that this portion of the protein may be all that is required for BL-induced chloroplast movements (Fig. 6, A and B). This region of the protein includes the last 96 residues of the putative C2 interacting domain, the coil-coil, and the conserved C-terminal domain of unknown function. However, we cannot dismiss the possibility that these transgenic pmi1-1 plants could still be producing a truncated protein corresponding to the first 316 amino acids and that the presence of both fragments lead to the rescue of the movement defect.
In other BL-induced responses, such as phototropism and stomatal regulation, phototropin signaling requires the putative signal transducers NPH3 and RPT2 (Motchoulski and Liscum, 1999; Sakai et al., 2000). However, NPH3 and RPT2 do not appear to be necessary for phototropin-induced chloroplast movements (Inada et al., 2004). The identification of PMI1 as an essential component of both the chloroplast accumulation and avoidance responses suggests that signaling during chloroplast movements may involve a mechanism that is different from other phototropin-dependent responses or that it works in another NPH3-like signaling branch specific to chloroplast movements. That PMI1 may be a Ca2+ responsive molecule and Ca2+ is an important regulator of both actin dynamics and chloroplast movement suggests that PMI1 functions in some aspect of phototropin signaling or in the motility mechanism itself.
Although the exact mechanism of action of PMI1 is not yet known, it appears to represent part of a plant-specific mechanism since we were unable to identify orthologs of AtPMI1 in nonplant organisms. Considering that chloroplasts within pmi1 cells do not cluster like they do in chup1, PMI1 appears to represent another type of gene involved in BL-induced chloroplast movements and may represent a new protein family as indicated by the presence of two other Arabidopsis genes that share similar domains (At5g20610 and At5g26160). Identification of proteins that interact with PMI1 along with determination of its cellular localization will be important for understanding PMI1 function in BL-induced chloroplasts movements and will lead to a better knowledge of how environmental stimuli modulate activities of the actin cytoskeleton.
MATERIALS AND METHODS
Growth Conditions
Seeds were sown in water-soaked Scott's Plug or Metro mix and incubated at 4°C in darkness for 3 to 4 d. Plants used for the mutant screen were grown at 23°C in a Percival growth chamber with a 12-h photoperiod using white light (80–100 μmol·m−2·s−1) provided by a mixture of cool-white fluorescent and incandescent bulbs. Plants for all other experiments were grown with a 12-h photoperiod in a temperature-controlled room (average 23°C) under white light (60–80 μmol·m−2·s−1) provided by cool-white fluorescent bulbs. Seedlings were fertilized with K-grow all-purpose plant food (Kmart, Troy, MI) every 2 weeks.
Mutants were isolated from Arabidopsis (Arabidopsis thaliana; Col) M2 seeds mutagenized with 0.3% EMS (Sigma-Aldrich, St. Louis). Salk_141795 T3 seeds were obtained from the Arabidopsis Stock Center (Ohio State University, Columbus, OH), and npq2-1 mutant seeds (Col ecotype) were provided by Kris Niyogi (University of California, Berkeley, CA).
Mutant Screen
Rosette leaves were excised from 6-week-old EMS-mutagenized M2 plants and dark acclimated for 17 to 24 h sandwiched between an inverted petri dish bottom and lid. Eight leaves were arranged in each petri dish so that their petioles pointed toward the center and were positioned on moistened Whatman filter paper (42.5 mm diameter; Clifton, NJ). RL transmittance was measured through each leaf using a custom device (Fig. 1) that consists of a clear Plexiglas turntable with a LI-190SA quantum sensor (connected to a LI-COR LI-189 quantum radiometer photometer; LI-COR, Lincoln, NE) mounted below the turntable. Three red light-emitting diodes positioned directly above the quantum sensor provided the RL used for light transmittance measurements. The turntable was built to hold an inverted petri dish in a specified orientation, so when the turntable is rotated to eight precise positions, each of the eight leaves can be located in turn between the red light-emitting diodes and the quantum sensor for light transmittance measurements. The device design ensures that each transmittance measurement is made through the same 5-mm-diameter area of each leaf, even after a petri dish is removed and later returned to the turntable. RL transmittance was measured through leaves before and after exposure to sequential 1-h treatments of low (5 μmol·m−2·s−1) and high (70 μmol·m−2·s−1) broadband BL (480 nm ± 50 nm) given from above. For each leaf, the change in the percentage of RL transmittance was calculated as ((It/Io)*100)/I), where Io and It are the incident and transmitted RL fluence rates, respectively, and I is the percentage of RL transmittance value measured in the dark-acclimated leaves prior to the first BL treatment.
The lower fluence rate of BL was provided by filtering light from cool-white fluorescent light bulbs through blue Plexiglas. Light from halogen flood lamps (150 W Quartzline; General Electric, Fairfield, CT) was filtered through 7 cm of 1.5% (w/v) CuSO4·7H2O (Sigma-Aldrich) and blue Plexiglas to obtain the high-fluence-rate BL. Both BL sources provided similar spectral outputs peaking near 480 with approximately 100-nm half-bandwidth (measured with a LI-COR 1800 spectroradiometer).
Initially, one leaf per M2 plant was tested using the screening device described above. If a leaf failed to show wild-type changes in RL transmittance when exposed to either of the fluence rates of BL, two to three more leaves were tested from the corresponding M2. Plants that failed the second and third rounds of screening were allowed to set seed and their progeny subjected to the more sensitive sequential BL-induced RL transmittance measurements previously described by DeBlasio et al. (2003). Approximately 6,000 M2 plants have been screened.
Microscopy
Excised rosette leaves from 5-week-old wild-type and pmi1-1 plants were dark acclimated for 18 h in a humid environment. For analysis of chloroplast movements in live palisade cells, leaves were then cut into 1-mm sections under a very low fluence rate of BL and mounted under a coverslip on a microscope slide with distilled water. The slide was placed on a Nikon E800 microscope stage (Melville, NY) and the leaf sections exposed to sequential 1-h treatments of low- and high-fluence-rate white light from below. Images of the palisade cell layer at 600× magnification were captured every 1.5 min using a Hamamatsu ORCA-ER charge-coupled device camera (Hamamtsu City, Japan) and MetaMorph software (Universal Imaging, Downingtown, PA).
For cross sections, RL transmittance through each dark-acclimated leaf was measured before being placed in darkness, low-fluence-rate (4.5 μmol·m−2·s−1), or high-fluence-rate-broadband BL (100 μmol·m−2·s−1) for 1 h. RL transmittance was measured again after each light treatment to document the chloroplast movement response that had occurred. Leaves were then cut into 1-mm strips and vacuum infiltrated with 5% gluteraldahyde and 4% formaldahyde. Leaf sections were subjected to an acetone dehydration series and then embedded into soft Spur's resin. Sections (1 μm) were obtained using an automated ultramicrotome Sorvall MT-2, and stained with 1% toluidine blue and Borax solution. Bright-field images of cross sections from each light treatment were captured at 20× magnification using the same camera system described above.
For analysis of the pmi1 actin cytoskeleton, wild-type (Landsberg erecta) plants expressing GFP-mtalin were used to cross-pollinate pmi1-1. F2 progeny were screened for the presence of the pmi1-1 homozygous genotype by DNA sequencing. Pmi1-1 mutants with high levels of GFP-mtalin expression were chosen for further analysis. Leaves were cut into 1-mm sections and mounted under a coverslip on a microscope slide with distilled water. Actin cytoskeleton was visualized using a Spinning Disk Confocal microscope (Nikon) mounted with a fluorescein isothiocyanate excitation and emission filter (480 nm). Chlorophyll autofluorescence was visualized using a long-pass emission filter (660 nm). Images represent the projection of 36 optical 0.2-μm sections taken at 1000× magnification. Wild-type GFP-mtalin seeds were graciously donated to us by Zhenbiao Yang (University of California, Riverside, CA).
Genetic Mapping of PMI1
Recombinant plants were generated by crossing pmi1-1 homozygous mutant plants (Col) with wild-type Landsberg erecta plants. F2 progeny were screened for the pmi1-1 chloroplast movement defect using the device described above. Movement-defective F2 progeny were then tested for Landsberg-associated SSLP markers nga280 (Bell and Ecker, 1994), F1I2IB (forward, 5′-GTTGTTGTCTTTTCTTCCA-3′; reverse, 5′-AAGTGTGAAAGATTAGTT-3′), F8D11A (forward, 5′-TATTTGTATTATCGTCTTTA-3′; reverse, 5′-TTCCATGCTTGTAGGTTTTC-3′), and F7F22B (forward, 5′-TAAGTCATCCTCGTCCATCA-3′; reverse 5′-CATGGTTATCGGGCACACAA-3′). The CAPS marker GAPB was also used (forward, 5′-TCTGATCAGTTGCAGCTATG-3′; reverse 5′-GGCACTATGTTCAGTGCTG-3′). All original SSLP markers were made with the help of the Cereon database.
Complementation of the pmi1-1 Mutation
For rescue of pmi1-1, pBeloBAC plasmids containing Arabidopsis BAC F8D11 and T8D8, respectively (Arabidopsis Stock Center, Ohio State University), were partially digested with Sau3AI (New England Biolabs, Beverly, MA) to produce an enrichment of 12-kb fragments. These fragments were then ligated to BamHI-digested binary vector pCLD04541 (Arabidopsis Stock Center, Ohio State University) at 13°C overnight. Plasmid DNA was then introduced into Escherichia coli (DH5α) using a Gigapack III XL cosmid packaging kit (Stratagene, Cedar Creek, TX).
A 3.8-kb fragment of At1g42550 genomic DNA, carrying the At1g42550 coding region, 3′ untranslated sequence, and the 5′ flanking region, including the At1g42550 promoter, was amplified by PCR from wild-type (Col) DNA using the PAGE purified primers T8D8.2Pro-F (5′- GGATCCCGGTAAATCCATTAGACTGAAACTGTAG-3′) and F8D11.1 3′-UTR-R (5′-TCACATTGCCCCTTTACGCTC-3′) (Integrated DNA Technologies, Coralville, IA). The At1g42550 genomic fragment was cloned into pGEM-T vector using a TA cloning kit (Invitrogen, Calsblad, CA) and transferred to the pCLD04541 vector. Plasmid DNA was then introduced into E. coli (DH10B) by electroporation.
Colonies with BAC and wild-type At1g42550 inserts were selected for by growth on Luria-Bertani medium supplemented with tetracycline (50 μg/mL; Sigma-Aldrich), 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (BioVectra, Prince Edward Island, Canada), and isopropyl β-d-1-thiogalactopyranoside (Sigma-Aldrich). Insert of DNA fragments was confirmed by PCR using BAC- and At1g42550-specific primers. Clones containing inserts were mated to Agrobacterium tumefaciens strain GV3101 using the helper strain pRK2013. Transformed Agrobactrium were selected for by growth on Luria-Bertani medium containing 50 μg/mL gentamycin (Sigma-Aldrich), 25 μg/mL kanamycin, and 2.5 μg/mL tetracycline. Cloned DNA was introduced into pmi1-1 mutant plants via floral dip mediated transformation (Clough and Bent, 1998). Transformants were selected for by growing seeds on half-strength Murashige and Skoog basal salt mix (Gibco/Life Technologies, Grand Island, NY) supplemented with 0.8% Bacto-agar and 50 μg/mL kanamycin (Sigma-Aldrich). Kanamycin-resistant plants were transferred to soil and screened for wild-type chloroplast movements. The chloroplast movement phenotype was also confirmed in the T2 generation.
Sequence Analysis
Sequencing of At1g42550 and At1g42560 from wild-type (Col) and pmi1-1 mutant DNA was preformed at the Indiana University Molecular Institute (Bloomington, IN) using capillary 3100 Genetic Analyzers (Applied Biosystems, Foster City, CA) and ABI Prism BigDye Terminator Cycle Sequencing v3.0 Ready Reaction with AmpliTaq DNA Polymerase (Applied Biosystems). DNA for sequencing was isolated from M4 and isogenic pmi1-1 plants. Sequences from the At1g42550-specific primers F8D11.1B-F (5′-CGGCGAACGGTAAAGGTAGTCCAG-3′), F8D11.1 F8D11.1A-F (5′-GGGATGAAACCGAGTAGTAAACCT-3′), and F8D11-1-R (5′-ATCCCGTGAAAATCCGAAACTGAC-3′) were used to identify the pmi1-1 point mutation.
Sequence analysis was performed using the following programs: COILS (http://www.ch.embnet.org/software/COILS_form.html), SignalP (http://www.cbs.dtu.dk/services/Signalp/), Myhits Motif_scan (http://myhits.isb-sib.ch/cgi-bin/motif_scan), and ExPASy Scanprosite (http://us.expasy.org/cgi-bin/scanprosite). Sequence homology searches were performed using the National Center for Biotechnology Information and The Institute for Genomic Research BLAST servers. Sequence alignments were carried out by the ClustalW method (Thompson et al., 1994) using the EMBL server (http://www.ebi.ac.uk/clustalw/). The output was produced by BoxShade (http://www.ch.embnet.org/software/BOX_form.html).
Percentage identity and similarity between the sequences shown in Figure 8 were determined by protein–protein BLAST at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/blast/Blast) or LALIGN (http://www.ch.embnet.org/software/LALIGN_form.html). BLOSUM62 scoring matrix was used for both programs. Gap costs for LALIGN were −14 opening gap penalty and −4 extending gap penalty. For National Center for Biotechnology Information gap penalties were 11 existence and 1 extension. Low complexity filter was not used.
PCR Analysis of the T-DNA Mutant
PCR amplification and DNA sequencing T-DNA left border specific primer (Lba-1 R, 5′-TGGTTCACGTAGTGGGCCATCG-3′) with two gene-specific primers (F8D11.1A-F, 5′-GGGATGAAACCGAGTAGTAAACCT-3′; F8D11.1A-R, 5′-AATACCTCCAAGATCAACTAATGC-3′) confirmed the presence of the T-DNA insert in the Salk_141795 true-breeding and F1 lines. Genomic DNA was extracted from 6-week-old wild-type (Col) and Salk-141795 T4 plants that exhibited chloroplast movement defects by first pulverizing leaf tissue frozen in liquid N2 and incubating at 65°C in 2% cetyl-trimethyl-ammonium bromide (Sigma-Aldrich). After 16 h, DNA was isolated by chloroform extraction followed by ethanol precipitation. PCR reactions were conducted for 40 cycles (94°C for 15 s, annealing at 55°C for 30 s, and a 1-min extension at 72°C).
RT-PCR Analysis of AtPMI1 Length and Expression
Total RNA was isolated using the RNeasy plant mini kit (Qiagen, Valencia, CA) from 100 to 150 μg of rosette leaves, stems, cauline leaves, whole flowers, and roots, combined from three 9-week-old wild-type plants (Col). RT-PCR was conducted using the SuperScript III OneStep RT-PCR kit with Plantinum taq (Invitrogen). One microgram of total RNA for each tissue was used per reaction and a 30-min incubation at 55°C followed by 2 min at 94°C and 40 cycles of 94°C for 15 s, 55°C for 30 s, and 68°C for 1 min were used for amplification as instructed by the kit protocol. Forty microliters of a 50-μL reaction were subjected to electrophoresis in a 3% (w/v) Metaphore agarose gel (Cambrex Bio Science Rockland, Rockland, ME). To determine the length of the AtPMI1 coding region, primers specific to the beginning and end of the annotated 2.6-kb open reading frame (forward 5′-TTCTTCTACATGGCAGGAGAATA-3′; reverse 5′-GGATCCATGCAATTTCACATCAGGGA-3′) were used to generate cDNA from wild-type rosette tissue. The resulting RT-PCR product was sequenced to confirm generation of cDNA. Primers flanking the 70-bp intron of At1g42550 (forward, 5′-CTCGGATCTCGGTAAAGGCATTGG-3′; reverse, 5′-TTGCTCCCCTGTTTTCCCCATTAG-3′) were used to detect the presence of the PMI1 transcript in the various plant tissues. Primers flanking both introns of profilin1 (forward, 5′-TGTCGAAGGCAACCATCTCACC-3′; reverse, 5′-ATCGTAGAAGCCAAAGACCAAAGC-3′) were used as a control.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY072341.
This work was supported by a grant from the National Science Foundation (IBN–0080783) and a U.S. Department of Agriculture National Needs Fellowship (98–38420–584).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.061887.
References
- Aderem A (1992) Signal transduction and the actin cytoskeleton: the roles of MARCKS and profilin. Trends Biochem Sci 17: 438–443 [DOI] [PubMed] [Google Scholar]
- Allen GJ, Kwak JM, Chu SP, Llopis J, Tsien RY, Harber JF, Schroeder JI (1999) Cameleon calcium indicator reports cytoplasmic calcium dynamics in Arabidopsis guard cells. Plant J 19: 735–747 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen HM, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Augustynowicz J, Gabrys H (1999) Chloroplast movements in fern leaves: correlation of movement dynamics and environmental flexibility of the species. Plant Cell Environ 22: 1239–1248 [Google Scholar]
- Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JD, Parker JE (2002) Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295: 2077–2080 [DOI] [PubMed] [Google Scholar]
- Babourina O, Godfrey L, Voltchamskii K (2004) Changes in ion fluxes during phototropic bending in etiolated oat coleoptiles. Ann Bot (Lond) 94: 187–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babourina O, Newman I, Shabala S (2002) Blue light-induced kinetics of H+ and Ca2+ fluxes in etiolated wild-type and phototropin-mutant Arabidopsis seedlings. Proc Natl Acad Sci USA 99: 2433–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR (1994) Assignment of 30 microsatellite loci to the linkage map of Arabidposis. Genomics 19: 137–144 [DOI] [PubMed] [Google Scholar]
- Boardman NK (1977) Comparative photosynthesis of sun and shade plants. Annu Rev Plant Physiol 28: 355–377 [Google Scholar]
- Brugnoli E, Björkman O (1992) Chloroplast movements in leaves: influence on chlorophyll fluorescence and measurements of light-induced absorbance changes related to change in pH and zeaxanthin formation. Photosynth Res 32: 23–35 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cooper B, Clarke JD, Budworth P, Kreps J, Hutchison D, Park S, Guimil S, Dunn M, Luginbühl P, Ellero C, Goff SA, Glazebrook J (2003) A network of rice genes associated with stress response and seed development. Proc Natl Acad Sci USA 100: 4945–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBlasio SL, Mullen JL, Luesse DL, Hangarter RP (2003) Phytochrome modulation of blue-light-induced chloroplast movements in Arabidopsis. Plant Physiol 133: 1471–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong XJ, Ryu JH, Takagi S, Nagai R (1996) Dynamic changes in the organization of microfilaments associated with the photocontrolled motility of chloroplasts in the epidermal cells of Vallisneria. Protoplasma 195: 18–24 [Google Scholar]
- Drøbak BK, Ferguson IB (1985) Release of Ca2+ from plant hypocotyl microsomes by inositol 1,4,5-triphosphate. Biochem Biophys Res Commun 130: 1241–1246 [DOI] [PubMed] [Google Scholar]
- Earles CA, Bai J, Wang P, Chapman ER (2001) The tandem C2 domains of synaptotagmin contain redundant Ca2+ binding sites that cooperate to engage t-SNAREs and trigger exocytosis. J Cell Biol 154: 1117–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folta KM, Lieg EJ, Durham T, Spalding EP (2003) Primary inhibition of hypocotyl growth and phototropism depend differentially on phototropin-mediated increases in cytoplasmic calcium induced by blue light. Plant Physiol 133: 1464–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golsteyn RM, Louvard D, Friederich E (1997) The role of actin binding proteins in epithelial morphogenesis: models based upon Listeria movement. Biophys Chem 68: 73–82 [DOI] [PubMed] [Google Scholar]
- Gorton HL, Williams WE, Vogelmann TC (1999) Chloroplast movement in Alocasia macrorrhiza. Physiol Plant 106: 421–428 [Google Scholar]
- Harada A, Sakai T, Okada K (2003) phot1 and phot2 mediate blue light-induced transient increases in cytosolic Ca2+ differently in Arabidopsis leaves. Proc Natl Acad Sci USA 100: 8583–8588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, Shibata K (1973) Light-induced chloroplast rearrangements and their action spectra as measured by absorption spectrophotometry. Planta 114: 341–358 [DOI] [PubMed] [Google Scholar]
- Inada S, Ohgishi M, Mayama T, Okada K, Sakai T (2004) RPT2 is a signal transducer involved in phototropic response and stomatal opening by association with phototropin 1 in Arabidopsis thaliana. Plant Cell 16: 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambunathan N, Siani JM, McNellis TW (2001) A humidity-sensitive Arabidopsis copine mutant exhibits precocious cell death and increased disease resistance. Plant Cell 13: 2225–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo JA, Gabrys H, Capel J, Alonso JM, Ecker JR, Cashmore AR (2001) Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature 410: 952–954 [DOI] [PubMed] [Google Scholar]
- Kadota A, Sato Y, Wada M (2000) Intracellular chloroplast photorelocation in the moss Physcomitrella patens is mediated by phytochrome as well as by a blue-light receptor. Planta 210: 932–937 [DOI] [PubMed] [Google Scholar]
- Kadota A, Wada M (1992) Photoinduction of formation of circular structures by microfilaments on chloroplasts during intracellular orientation in protonemal cells of the fern Adiantum capillus-veneris. Protoplasma 167: 97–107 [Google Scholar]
- Kagawa T, Kasahara M, Abe T, Yoshida S, Wada M (2004) Function analysis of phototropin2 using fern mutants deficient in blue light-induced chloroplast avoidance movement. Plant Cell Physiol 45: 416–426 [DOI] [PubMed] [Google Scholar]
- Kagawa T, Sakai T, Suetsugu N, Oikawa K, Ishiguro S, Kato T, Tabata S, Okada K, Wada M (2001) Arabidopsis NPL1: a phototropin homolog controlling the chloroplast high light avoidance response. Science 291: 2138–2141 [DOI] [PubMed] [Google Scholar]
- Kagawa T, Wada M (1994) Brief irradiation with red or blue light induces orientational movement of chloroplasts in dark-adapted prothallial cells of the fern Adiantum. J Plant Res 107: 389–398 [Google Scholar]
- Kagawa T, Wada M (1999) Chloroplast-avoidance response induced by high-fluence blue light in prothallial cells of the fern Adiantum capillus-veneris as analyzed by microbeam irradiation. Plant Physiol 119: 917–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa T, Wada M (2000) Blue light induced chloroplast relocation in Arabidopsis thaliana as analyzed by microbeam irradiation. Plant Cell Physiol 41: 84–93 [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, Meagher RB (1999) Actin-organelle interaction: association with chloroplasts in Arabidopsis leaf mesophyll cells. Cell Motil Cytoskeleton 44: 110–118 [DOI] [PubMed] [Google Scholar]
- Kasahara M, Kagawa T, Oikawa K, Suetsugu N, Miyao M, Wada M (2002) Chloroplast avoidance movement reduces photodamage in plants. Nature 420: 829–832 [DOI] [PubMed] [Google Scholar]
- Kim CG, Park D, Rhee SG (1996) The role of carboxyl-terminal basic amino acids in Gqα-dependant activation, particulate association, and nuclear localization of phospholipase C-β1. J Biol Chem 271: 21187–21192 [DOI] [PubMed] [Google Scholar]
- Kitagawa K, Skowyra D, Elledge SJ, Harper JW, Hieter P (1999) SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol Cell 4: 21–33 [DOI] [PubMed] [Google Scholar]
- Kost B, Speilhofer P, Chua N-H (1998) A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J 16: 393–401 [DOI] [PubMed] [Google Scholar]
- Lechowski Z (1974) Chloroplast arrangement as a factor of photosynthesis in multilayered leaves. Acta Soc Bot Pol 63: 531–540 [Google Scholar]
- Locoro B, Papini R, De Michelis MI (1999) N-Ethylmaleimide modifies the conformation of the plasma membrane H+-ATPase, strengthening the inhibitory action of the C-terminal domain. Plant Biol 1: 192–197 [Google Scholar]
- Malec P, Rinaldi RA, Gabrys H (1996) Light-induced movements in Lemna trisulca identification of the motile system. Plant Sci 120: 127–137 [Google Scholar]
- Marchand JB, Moreau P, Paoletti A, Cossart P, Carlier MF, Pantaloni D (1995) Actin-based movement of Listeria monocytogenes-actin assembly results from the local maintenance of uncapped filament barbed ends at the bacterium surface. J Cell Biol 130: 331–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merali Z, Kent P, Anisman H (2002) Role of bombesin-related peptides in the mediation of integration of the stress response. Cell Mol Life Sci 59: 272–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motchoulski KA, Liscum E (1999) Arabidopsis NPH3: a NPH1 photoreceptor-interacting protein essential for phototropism. Science 286: 961–964 [DOI] [PubMed] [Google Scholar]
- Nagalla SR, Barry BJ, Falick AM, Gibson BW, Taylor JE, Dong JZ, Spindel ER (1996) There are three distinct forms of bombesin. J Biol Chem 271: 7731–7737 [DOI] [PubMed] [Google Scholar]
- Niyogi KK (1999) Photoprotection revisited: genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol 50: 333–359 [DOI] [PubMed] [Google Scholar]
- Oikawa K, Kasahara M, Kiyosue T, Kagawa T, Suetsugu N, Takahashi F, Kanegae T, Niwa Y, Kadota A, Wada M (2003) CHLORPLAST UNUSUAL POSITIONING1 is essential for proper chloroplast positioning. Plant Cell 15: 2805–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BM, Quail PH, Hangarter RP (1996) Phytochrome A regulates red-light induction of phototropic enhancement in Arabidopsis. Plant Physiol 110: 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read ND, Shacklock PS, Knight MR, Trewavas AJ (1993) Imaging calcium dynamics in living plant cells and tissues. Cell Biol Int 17: 111–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero S, Le Clainche C, Didry D, Eigle C, Pantaloni D, Carlier MF (2004) Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell 119: 419–429 [DOI] [PubMed] [Google Scholar]
- Sakai T, Kagawa T, Kasahara M, Swartz TE, Christie JM, Briggs WR, Wada M, Okada K (2001) Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc Natl Acad Sci USA 98: 6969–6974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Wada M, Ishiguro S, Okada K (2000) RPT2: a signal transducer of the phototropic response in Arabidopsis. Plant Cell 12: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Briggs WR (2002) Cellular and subcellular localization of phototropin 1. Plant Cell 14: 1723–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Wada M, Kadota A (2000) Choice of tracks, microtubules and/or actin filaments for chloroplast photo-movement is differentially controlled by phytochrome and a blue light phototreceptor. J Cell Sci 114: 269–279 [DOI] [PubMed] [Google Scholar]
- Sato Y, Wada M, Kadota A (2001) External Ca2+ is essential for chloroplast movement induced by mechanical stimulation but not by light stimulation. Plant Physiol 127: 497–504 [PMC free article] [PubMed] [Google Scholar]
- Seitz K (1972) Primary process controlling light induced movement of chloroplasts. Acta Protozool 11: 225–235 [Google Scholar]
- Shimazaki K, Gho CH, Kinoshita T (1999) Involvement of intracellular Ca2+ in blue light-dependent proton pumping in guard cell protoplasts from Vicia fab. Physiol Plant 105: 554–561 [Google Scholar]
- Shimmen T, Yakota E (1994) Physiological and biochemical aspects of cytoplasmic streaming. Int Rev Cytol 155: 97–139 [Google Scholar]
- Southwick FS, Purich DL (1994) Arrest of Listeria movement in host cells by a bacterial ActA analogue: implications for actin-based motility. Proc Natl Acad Sci USA 91: 5168–5172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger CJ (2000) Signaling to the actin cytoskeleton in plants. Annu Rev Plant Physiol Plant Mol Biol 51: 257–288 [DOI] [PubMed] [Google Scholar]
- Stoelzle S, Kagawa T, Wada M, Hedrich R, Dietrich P (2003) Blue light activates calcium-permeable channels in Arabidopsis mesophyll cells via the phototropin signaling pathway. Proc Natl Acad Sci USA 100: 1456–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Yamaguchi M (1999) Role of regucalcin as an activator of Ca2+-ATPase activity in rat liver microsomes. J Cell Biochem 74: 663–669 [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) ClustalW—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tlalka M, Fricker M (1999) The role of calcium in blue-light-dependant chloroplast movement in Lemna trisulca L. Plant J 20: 461–473 [DOI] [PubMed] [Google Scholar]
- Tlalka M, Gabrys H (1993) Influence of calcium on blue-light-induced chloroplast movement in Lemna trisulca L. Planta 189: 491–498 [Google Scholar]
- Trojan A, Gabrys H (1996) Chloroplast distribution in Arabidopsis thaliana (L.) depends on light conditions during growth. Plant Physiol 111: 419–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada M, Suetsugu N (2004) Plant organelle positioning. Curr Opin Plant Biol 7: 626–631 [DOI] [PubMed] [Google Scholar]
- Wang T, Pentyala S, Elliott JT, Dowal L, Gupta E, Rebecchi MJ, Scarlata S (1999) Selective interaction of the C2 domains of phospholipase C-β1 and -β2 with activated Gαq subunits: an alternative function for C2-signaling modules. Proc Natl Acad Sci USA 96: 7843–7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Pesacreta TC (2004) A subclass of myosin XI is associated with mitochondria, plastids, and the molecular chaperone subunit TCP-1 alpha in maize. Cell Motil Cytoskeleton 57: 218–232 [DOI] [PubMed] [Google Scholar]
- Wolenski J (1995) Regulation of calmodulin-binding myosins. Trends Cell Biol 5: 311–315 [DOI] [PubMed] [Google Scholar]
- Xu P, Lloyd CW, Staiger CJ, Drøback BK (1992) Association of phophatidylinositol 4-kinase with the plant cytoskeleton. Plant Cell 4: 941–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurzycki J (1955) Chloroplasts arrangement as a factor in photosynthesis. Acta Soc Bot Pol 24: 27–63 [Google Scholar]
- Zurzycki J (1961) The influence of chloroplast displacements on the optical properties of leaves. Acta Soc Bot Pol 30: 503–527 [Google Scholar]
- Zurzycki J (1972) Primary reactions in chloroplast rearrangements. Acta Protozool 11: 189–199 [Google Scholar]