Abstract
Wound-induced protein kinase (WIPK) is a tobacco (Nicotiana tabacum) mitogen-activated protein kinase known to play an essential role in defense against wounding and pathogens, although its downstream targets have yet to be clarified. This study identified a gene encoding a protein of 648 amino acids, which directly interacts with WIPK, designated as N. tabacum WIPK-interacting factor (NtWIF). The N-terminal region with approximately 250 amino acids showed a high similarity to the plant-specific DNA binding domain, B3, but no other similarity with known proteins. The C terminus of approximately 200 amino acids appeared to be essential for the interaction with WIPK, and a Luciferase-reporter gene assay using Bright Yellow 2 cells indicated the full-length protein to possess trans-activation activity, located to the middle region of approximately 200 amino acids. In vitro phosphorylation assays indicated that WIPK efficiently phosphorylates the full-length protein and the N terminus but not the C terminus. When full-length NtWIF was coexpressed with WIPK in Bright Yellow 2 cells, the Luciferase transcriptional activity increased up to 5-fold that of NtWIF alone, whereas no effect was observed with a kinase-deficient WIPK mutant. Transcripts of NtWIF began to simultaneously accumulate with those of WIPK 30 min after wounding and 1 h after the onset of hypersensitive response upon tobacco mosaic virus infection. These results suggest that NtWIF is a transcription factor that is directly phosphorylated by WIPK, thereby being activated for transcription of target gene(s) involved in wound and pathogen responses.
Wounded tissues in plants undergo severe cellular changes due to decompartmentalization and release of stored materials and excessive loss of water. A large number of molecules, such as peptides, hormones, and reactive oxygen species, have been implicated in wound responses (Pearce et al., 1991; Ryan, 2000; Leon et al., 2001). The initial response is to activate expression of relevant genes, many of which have been documented as proteinase inhibitors, like the basic pathogenesis-related proteins, defensin and thionin (Pearce et al., 1991; Reymond et al., 2000). However, the processes underlying perception and transduction of the signals that mediate the responses are not completely understood.
The role of protein phosphorylation in plant stress responses was first revealed by the use of inhibitors of kinase and phosphatase (Grosskopf et al., 1990; Felix et al., 1994; Chandra and Low, 1995). The findings were further supported by cloning of genes for kinases, including members of receptor groups and elements mediating signal transfer to the ultimate cellular targets (Ligterink and Hirt, 2001; Romeis, 2001). Among the genes, those for three-tier kinases, mitogen-activated protein kinase (MAPK) extra cellular signal-regulated kinase kinase kinase (MEKK), MAPK extra cellular signal-regulated kinase kinase (MEK), and MAPK, were found to constitute an important signal-mediating pathway, the MAPK cascade. In addition to its contribution to stress responses, this signaling system is also involved in other physiological processes, such as cell growth, cell cycle regulation, and development control (Ligterink, 2000; Ligterink and Hirt, 2001; Tena et al., 2001). It is also implicated in reactions to abiotic stress, such as drought, low or high temperature, and osmotic stress (Jonak et al., 1996; Mizoguchi et al., 1996; Droillard et al., 2000; Iajczyk et al., 2000; Kiegerl et al., 2000). Genetic approaches, including kinase dominant transgenic plants, transient expression in protoplasts, and inducible expression, further supported the positive regulatory role of MAPK cascade in disease defense (Kovtun et al., 2000; Xing et al., 2001; Yang et al., 2001; Zhang and Liu, 2001; Ren et al., 2002).
To date, >50 MAPKs have been documented from a variety of plants, the 23 in the Arabidopsis (Arabidopsis thaliana) genome being typical (Zhang and Klessig, 2001). Tobacco (Nicotiana tabacum) wound-induced protein kinase (WIPK) is one of the best characterized, involved in both wound and pathogen responses (Seo et al., 1995, 1999; Zhang and Klessig, 1997; Romeis et al., 1999; Zhang et al., 2000). Together with salicylic acid-induced protein kinase (SIPK), another tobacco MAPK induced by salicylic acid, WIPK is considered to play a critical role in response to pathogen attack by transducing N-gene signals to downstream genes through the phosphorylation cascade (Zhang and Liu, 2001; Lebrun-Garcia et al., 2002; Liu et al., 2003). Extensive studies with Arabidopsis have indicated the presence of a similar pathway mediated by Arabidopsis thaliana mitogen-activated protein kinase (AtMPK3) and AtMPK6 elicited by fungal attack (Ren et al., 2002). These two MAPKs, which are orthologs of WIPK and SIPK, respectively, are known to be phosphorylated by the MEKs, MKK4 and MKK5, resulting in activation of WRKY-type transcription factors, which are involved in transcription of disease resistance genes (Asai et al., 2002). Furthermore, experiments have revealed that the tobacco MEK2-SIPK/WIPK cascade functions upstream of the WRKY family proteins (Kim and Zhang, 2004). However, whether these MAPKs directly interact with WRKY transcription factors has remained unclear, leaving open the question of how MAPKs function in the signaling relay. This study concerns the identification and properties of a transcriptional activator protein that directly interacts with WIPK.
RESULTS
Isolation of the WIPK-Interacting Protein
To isolate proteins that interact with WIPK, we used the yeast two-hybrid system with full-length WIPK as a bait. Among 105 yeast transformants screened, only one was positive in both His and β-galactosidase complementation assays. The plasmid recovered from this transformant contained a 0.8-kb insert. To confirm the specificity of the interaction, the recovered plasmid was cotransformed with either GBD-WIPK (bait plasmid) or the pBD-GAL4 Cam vector into yeast Y190. Only the transformant containing plasmids encoding both WIPK and its interaction partner were positive in the His and β-galactosidase reporter assays (data not shown), indicating the interaction between them to indeed be specific. Its putative peptide sequence indicated the isolated cDNA clone to be a 5′-truncated product. Consequently, the insert was excised using EcoRI-XhoI digestion and employed as a probe to screen an N. tabacum Uni-ZAP XR phage library, and a positive clone containing a 2.2-kb insert was finally isolated. The identity of the isolated clone was verified by RT-PCR, which produced a PCR product of the expected size predicted from the sequence (data not shown). The clone was designated N. tabacum WIPK-interacting factor (NtWIF), and its nucleotide sequence has been submitted to the GenBank/EMBL/DDBJ data libraries with accession number AB084669.
Structural Characterization of NtWIF
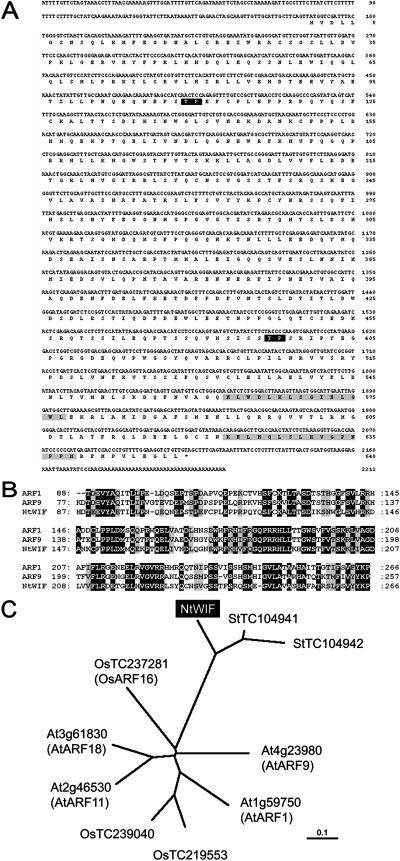
The NtWIF cDNA was found to consist of 2213 bp with a single open reading frame spanning from nucleotide positions 168 to 2114, encoding a polypeptide with 648 amino acids (Fig. 1A). A BLAST homology search revealed NtWIF to contain a conserved B3-type DNA binding domain at the N terminus, with high similarity to those of Arabidopsis auxin response factors (ARFs). Among the known members of Arabidopsis ARFs, NtWIF displays the highest homology to ARF1 and ARF9 (Fig. 1B). Its B3 domain shares 52% and 54% identity with ARF1 and ARF9, respectively, the identity between these two ARFs being in the order of 70% (Fig. 1B). However, two other conserved domains, domains III and IV, found in the C terminus of most ARFs proved to be absent in NtWIF. Homology search indicated that overall sequence of NtWIF best matched with a potato (Solanum tuberosum) expressed sequence tag contig (TC104941) and the C terminus with a tomato (Lycopersicon esculentum) expressed sequence tag derived from nutrient-deficient root tissue (BF097731). The C terminus also contained two putative docking domains, which may be the MAPK-interacting site (Seidel and Graves, 2002; Fig. 1A). Phylogenetic analysis indicated that NtWIF subtypes form a small family distinct from ARFs, which are known from solanaceous plants (Fig. 1C). Since the molecule appeared to consist of three distinct regions, further analyses were performed with domain dissected constructs (Table I).
Figure 1.
Properties of NtWIF. A, Nucleic acid and amino acid sequences. The nucleic acids are presented on the top line and the derived one-letter amino acid sequence is shown below. The stop codon is indicated by an asterisk. The position of the putative phosphorylation site by MAPK is indicated by a black box and that for putative docking domain by a gray box. B, Comparison of the B3 domain sequences among ARF1, ARF9, and NtWIF. Black and gray boxes indicate identical and similar amino acids, respectively. C, Unrooted phylogenetic tree of NtWIF with related proteins. Arabidopsis (At), rice (Os), and potato (St) sequences were obtained from the Munich Information Center for Protein Sequences and The Institute for Genomic Research databases, respectively.
Table I.
NtWIF constructs used in this study
| Designation | Amino Acid Position | Annotation |
|---|---|---|
| NtWIF | 1–648 | Full-length polypeptide |
| ΔNtWIF/N | 1–286 | N-terminal region |
| ΔNtWIF/M | 287–481 | Middle region |
| ΔNtWIF/C | 452–648 | C-terminal region |
| ΔNtWIF/NM | 1–481 | N-terminal and middle regions |
| ΔNtWIF/CM | 287–648 | C-terminal and middle regions |
Interactions between WIPK and NtWIF
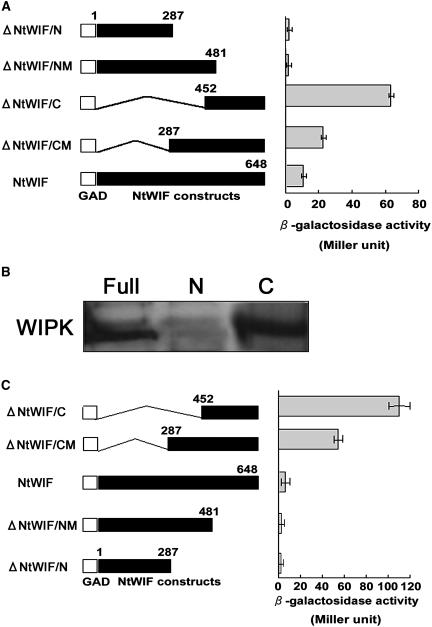
In order to determine the NtWIF domain responsible for interaction with WIPK, a yeast two-hybrid interaction assay was conducted. Different GAD-NtWIF constructs were cotransformed with GBD-WIPK (the bait plasmid) into yeast Y190 and assayed for growth and β-galactosidase activity (Fig. 2A). Among five constructs, each having a different region, two were clearly able to interact with WIPK (Fig. 2A). One, which contained a polypeptide with 196 amino acids at the C terminus (452–648, ΔNtWIF/C), showed a 60-fold higher affinity to WIPK than the control. The other, with 361 amino acids (amino acid positions 287–648, ΔNtWIF/CM), also bound to WIPK, but the binding efficiency was less than half that of the former. The results indicated the region containing 196 amino acids at the C terminus to be sufficient for WIPK interaction, and this was thus designated as the WIPK-interacting domain. It should be noted, however, that the full-length NtWIF (amino acids 1–648) interacted with WIPK rather inefficiently in comparison with those two truncated constructs, showing the binding activity at one-sixth that of ΔNtWIF/C (Fig. 2A).
Figure 2.
Interaction between NtWIF and WIPK. A, Yeast two-hybrid assay. Yeast Y190 cells were cotransformed with GBD-WIPK and various combinations of GAD-NtWIF constructs as indicated on the left. Transformants from the SD/-HTL agar plate were cultured in SD/-TL broth for β-galactosidase enzymatic assays using o-nitrophenyl-β-d-galactopyranoside as substrate (triplicate experiments). B, Pull-down assay of recombinant WIPK and NtWIF. Interaction was tested using GST-fused full-length NtWIF (Full), ΔNtWIF/C (C) and ΔNtWIF/N (N) proteins expressed in Sf9 cells and bacterially expressed His-tagged WIPK protein. NtWIF-GST proteins were bound to a glutathione column, and His-tagged WIPK was added. After elution with reduced glutathione, proteins were separated by SDS-PAGE, and WIPK was detected by anti-His-tag antibodies. C, Intramolecular interaction between the B3 domain and the WIPK-interacting domain. Yeast Y190 was transformed with the indicated GAD-NtWIF constructs and a GBD-ΔNtWIF/N. Yeast transformants were plated on SD/-HTL/+3-amino-1,2,4-triazole agar, and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside filter and β-galactosidase assays were performed as described above. The number beside each column represents the number of amino acids encoded by the respective peptide.
The observed interaction of WIPK and NtWIF in the yeast two-hybrid system was confirmed in an in vitro pull-down assay (Fig. 2B). His-tagged WIPK was applied to a glutathione-Sepharose column containing full or truncated NtWIF-glutathione S-transferase (GST) proteins, eluted with a buffer containing reduced glutathione, separated by SDS-PAGE, and subjected to western-blot hybridization with anti-His-tag antibodies. Results clearly showed that WIPK bound to ΔNtWIF/C but not to ΔNtWIF/N (Fig. 2B), being consistent with results from yeast two-hybrid assays (Fig. 2A). However, in contrast to the yeast assay, WIPK was found to bind to the full-length NtWIF at equal efficiency with that of ΔNtWIF/C (Fig. 2B). This discrepancy might due to different assay methods: an in vivo system using yeast cells and an in vitro system using large amounts of purified proteins.
As the full-length NtWIF fusion construct scarcely bound to WIPK in the yeast two-hybrid system, it was speculated that the B3 domain (N terminus) and the WIPK-interacting domain (C terminus) might interact with each other in vivo, resulting in a change of molecular conformation. A yeast two-hybrid interaction assay was performed to test this hypothesis. The GBD-ΔNtWIF/N construct was cotransformed with different GAD-NtWIF constructs into yeast Y190. The results showed that, among five constructs, two containing the WIPK-interacting domain (ΔNtWIF/C and ΔNtWIF/CM) were able to interact with GBD-ΔNtWIF/N (Fig. 2C). A notable feature, however, was that the construct containing the full-length NtWIF did not show any interaction with the B3 domain. These results support the idea that the B3 domain interacts with the WIPK-interacting domain in the NtWIF molecule and suggest that such intramolecular and/or intermolecular interactions may affect its interaction with WIPK in vivo.
Trans-Activation
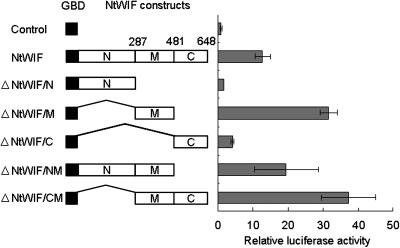
Since the presence of a B3 DNA binding domain at the N terminus suggested NtWIF to be a transcription factor, in vivo trans-activation assays were performed (Fig. 3). Various combinations of GBD-NtWIF effector constructs were cobombarded with a reporter plasmid and a reference plasmid into Bright Yellow 2 (BY2) cells. The full-length effector construct activated luciferase 10-fold over the control. Other effector constructs (ΔNtWIF/M and ΔNtWIF/CM), both containing a region of 194 amino acids (middle region), were also able to confer trans-activation activity >2-fold that for the full-length construct. However, in comparison with these constructs, the trans-activation activity was less than half in the construct containing the middle and the B3 regions (ΔNtWIF/NM). The effector construct containing only C or N terminus (ΔNtWIF/C or ΔNtWIF/N) was inactive. These results suggested that the middle region is responsible for transactivation and that the B3 domain (N terminus) might have a negative regulatory function on the transcriptional activity of NtWIF.
Figure 3.
Trans-activation assay. Effector constructs encoding different GBD-NtWIF fusion proteins as indicated were cobombarded with the reporter and reference plasmid into 7-d-old BY2 cells. Luciferase activity of each type of transformant was normalized to the respective R-luciferase activity. Relative luciferase activity was calculated by dividing the luciferase activity of each clone by that of the clone containing the GAD4 DBD effector construct (triplicate experiments). The number beside each column represents the number of amino acids encoded by the respective peptide.
Phosphorylation
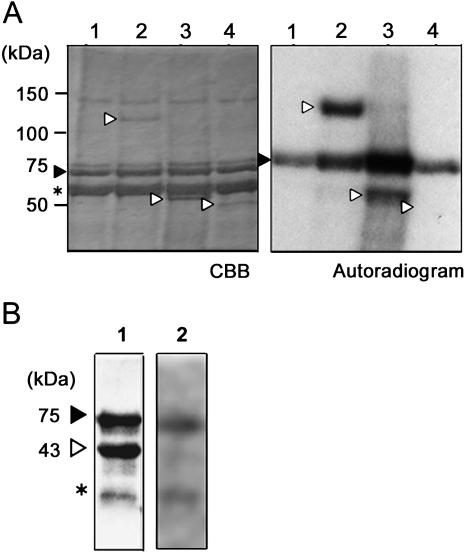
The observed preferential interaction suggested that NtWIF can possibly be phosphorylated by WIPK. In order to examine this possibility, in vitro phosphorylation assays were performed using recombinant proteins. As substrates, three constructs, full-length NtWIF, ΔNtWIF/C, and ΔNtWIF/N, were prepared in insect cells as the expression system. Using these protein preparations, kinase reactions with bacterially expressed WIPK were performed. The results clearly showed the full-length protein to be phosphorylated (Fig. 4A, lane 2). The N terminus (ΔNtWIF/N) was also phosphorylated, but the C terminus (ΔNtWIF/C) was not at all (Fig. 4A, lanes 3 and 4, respectively). The inability of the C terminus to be phosphorylated was confirmed with bacterially expressed protein (Fig. 4B). Under these experimental conditions, autophosphorylation of WIPK was strongly apparent (Fig. 4A, lane 1), and control myelin basic protein was efficiently phosphorylated (see Fig. 6A). The results thus indicated that differential phosphorylation activity toward NtWIF is an inherent property of WIPK.
Figure 4.
Phosphorylation of NtWIF by WIPK. A, In vitro kinase assays. GST-tagged full-length (NtWIF) or truncated (ΔNtWIF/C or ΔNtWIF/N) proteins expressed in Sf9 cells were subjected to phosphorylation reaction with bacterially expressed GST-WIPK (75 kD) protein. After electrophoresis on 7.5% polyacrylamide, the gel was stained by Coomassie Brilliant Blue (CBB; left), dried, and used for exposure to x-ray film (right). Samples are GST-WIPK (lane 1, closed arrowhead), full-length NtWIF (110 kD; lane 2, open arrowhead), ΔNtWIF/N (55 kD; lane 3, open arrowhead), and ΔNtWIF/C (43 kD; lane 4, open arrowhead). The sizes of proteins are indicated on the left, and the impurity is indicated by an asterisk. B, In vitro kinase assay using bacterially expressed ΔNtWIF/C (open arrowhead) and WIPK (closed arrowhead). After Coomassie Brilliant Blue staining (lane 1), gel was autoradiographed (lane 2).
Figure 6.
Transcriptional activation by WIPK in planta. A, In vitro kinase assays using GST-WIPK (WIPK) or GST-mutated WIPK (K73R) fusion proteins. Five micrograms of myelin basic protein was used as the substrate, and after incubation at 30°C for 30 min, reaction products were separated on a 12% SDS-polyacrylamide gel, stained with Coomasie Brilliant Blue (left), and applied to an x-ray film (right). The position of GST-WIPK (75 kD) is indicated by a closed arrowhead and that of myelin basic protein (12 kD) by an open arrowhead. B, Schematic illustration of plasmids used in the WIPK-mediated trans-activation assay. PNOS, nopaline synthase promoter; TNOS, nopaline synthase terminator; P35S, 35S promoter of cauliflower mosaic virus (CaMV); GAL4UAS, GAL4 binding sequence; P35Smini, CaMV35S minimal promoter. C, WIPK-mediated trans-activation assay. Trans-activation assay with BY2 cells was performed with plasmids containing indicated constructs. Indicated plasmids were mixed and used for coating gold particles, which were introduced into BY2 cells using particle bombardment. Dual-luciferase activity was assayed in nine independent experiments by a luminometer (Lumat LB9507; EG & G Berthold) as described in the legend for Figure 3.
Subcellular Localization
The results described above suggested the possibility that NtWIF is transcriptionally activated upon phosphorylation by WIPK in vivo. In order to confirm this idea, the localization of NtWIF and WIPK was examined using fusion proteins with green fluorescent protein (GFP) and/or cyan fluorescent protein (CFP). Initial experiments with the full-length NtWIF fused to GFP failed to show fluorescence signals upon introduction into onion epidermal cells (data not shown). Subsequently, an N-terminal truncated protein was constructed by deleting the first 281 amino acids (ΔNtWIF/CM:GFP) and analyzed. Onion cells transformed with the plasmid expressing GFP alone showed fluorescence throughout the cell. In contrast, fluorescence was detected predominantly in the nucleus in cells transformed with the plasmid expressing ΔNtWIF/CM:GFP fusion protein (Fig. 5A). The WIPK protein was fused to the C terminus of CFP, yielding a CFP:WIPK fusion protein with a molecular mass of approximately 70 kD. As a control, the NtAAA1:GFP protein was used because of its molecular size of 80 kD, similar to that of CFP:WIPK (Sugimoto et al., 2004). When assayed in onion epidermal cells, the control was clearly localized to cytoplasm, while the CFP:WIPK protein was found to localize to both nucleus and cytoplasm (Fig. 5B). Since a protein with >60 kD cannot diffuse through nuclear pores (Haasen et al., 1999), these observations indicate that the CFP:WIPK protein might be actively incorporated into the nucleus with the aid of other factor(s). Results suggested that both NtWIF and WIPK were localized to nucleus and, therefore, that both might be able to interact with each other.
Figure 5.
Intracellular localization of NtWIF and WIPK. A, Intracellular localization of NtWIF. Plasmids containing genes encoding GFP or ΔNtWIF/CM:GFP fusion protein were introduced into onion epidermal cells by bombardment, and individual cells were observed by epifluorescence. B, Intracellular localization of WIPK. Plasmids containing genes encoding indicated fusion proteins were introduced into onion epidermal cells by bombardment, which were then subjected to fluorescence microscopy. The control was CFP alone. The molecular sizes of CFP:WIPK and NtAAA1:GFP are 70 and 80 kD, respectively.
WIPK-Mediated Activation of NtWIF
Interaction between NtWIF and WIPK was further examined by Luc assay in BY2 cells. First, we generated a kinase-deficient WIPK by PCR mutagenesis as the reference. In this mutant protein, the essential Lys at position 73, which is critical for ATP binding, was substituted with a nonactive Arg residue (WIPK/K73R). In vitro assays confirmed the kinase activity of this mutant to be abolished, as shown by the inability to phosphorylate itself and also a synthetic MAPK substrate myelin basic protein (Fig. 6A). Second, we generated several constructs for transformation, which could be temporarily expressed in BY2 cells (Fig. 6B). Using these constructs, we determined whether transcriptional activity of NtWIF is activated by WIPK in planta. Upon introduction into cells, NtWIF alone could induce transcription of the reporter gene to a level 10-fold that of the control (Fig. 6C). However, with WIPK, the transcriptional activity increased up to 50-fold. This increase was clearly due to phosphorylation, since kinase-deficient WIPK failed to exhibit such activation, leaving the activity at the same level as NtWIF alone. It was concluded that, in BY2 cells, both WIPK and NtWIF become imported into nucleus and that the former actively phosphorylates the latter, thereby enhancing the transcriptional activity.
Induction Profile of NtWIF Transcripts
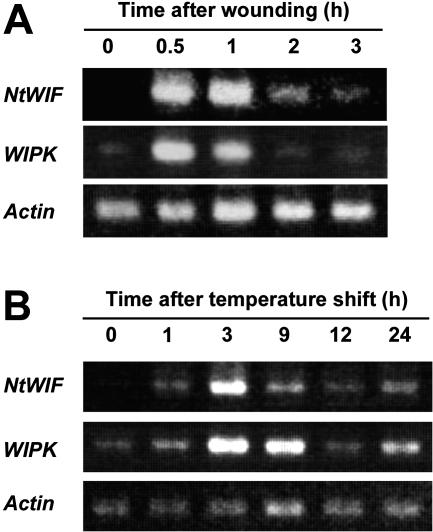
RT-PCR experiments showed that NtWIF transcripts were not accumulated in healthy tobacco leaves but were induced transiently from 30 min after wounding and then declined to the basal level about 2 h later, closely resembling the observed transient increase of WIPK transcripts (Fig. 7A). RNA-blot hybridization assay showed the same profile (data not shown). Accumulation of NtWIF transcripts in response to pathogen attack was also examined. When tobacco plants carrying the N gene are inoculated with tobacco mosaic virus (TMV) and incubated at 30°C, at which temperature the N gene does not function, viral particles multiply. Upon transfer to 20°C, the N gene is activated, resulting in lethal hypersensitive response (Gianinazzi, 1970). Results using this experimental system showed that NtWIF transcripts were induced 1 h after onset of hypersensitive response by temperature shift, reaching a maximum 3 h later (Fig. 7B). The induction profile of NtWIF during the hypersensitive response was identical to that of WIPK, strongly suggesting some common role in both wounding and pathogen signal transduction pathways.
Figure 7.
Induction of NtWIF transcripts by wounding and upon activation of hypersensitive response. RNA was isolated from leaves punched with a paper puncher or from those showing hypersensitive response upon temperature shift after TMV infection. Five micrograms of total RNA from each wounded (A) or TMV-infected (B) leaf sample was first reverse-transcribed with oligo(dT)15, then amplified with specific primers and separated by agarose gel electrophoresis.
DISCUSSION
NtWIF was found to consist of three distinct regions: a B3 DNA binding domain at the N terminus, a middle transactivation domain, and a WIPK-interacting domain at the C terminus. These findings are consistent with the results of amino acid sequence analysis, indicating the presence of a docking domain in the C terminus and a Thr-Pro sequence in the N-terminal B3 domain, each being possible sites for binding of, and phosphorylation by, MAPK (Sharrocks et al., 2000; Yang et al., 2003). However, domain dissection revealed that the full-length NtWIF is weak in binding to WIPK in a yeast two-hybrid system, although the in vitro pull-down assay confirmed a specific interaction between the two. Such a weak interaction in vivo may be explained by intramolecular interference, since the B3 domain tightly binds to the C-terminal WIPK-interacting domain, thereby possibly forcing the full-length protein to form a specific conformation that may attenuate the interaction with WIPK. Since an in vitro kinase assay showed efficient phosphorylation of full-length NtWIF by WIPK, tight binding between these two may not be a prerequisite for the phosphorylation reaction. Perhaps WIPK is able to recognize the C terminus of NtWIF despite its higher conformation and catalyze phosphorylation of the N terminus. This is consistent with suggestions that MAPKs phosphorylate many proteins without forming tight complexes (Sharrocks et al., 2000).
Sequence analysis of NtWIF has revealed two Thr-Pro sites, which possibly serve as the targets for MAPK (Yang et al., 2003): one at position 109 and the other at position 382, located in the N- and C-terminal regions, respectively. Since the C-terminal region was not phosphorylated in our in vitro assay, the N terminus is likely the correct phosphorylation site of WIPK. A preliminary experiment indeed supported this idea, showing that, when the Thr-Pro site in the N terminus was mutated into other amino acids, trans-activation activity was completely lost, even in the presence of the active form of WIPK (K.-M. Chung and H. Sano, unpublished data). Specificity of MAPKs has been suggested to be determined by the docking domain and phosphoacceptor sites. However, the former does not appear to be directly involved in phosphorylation but rather to increase phosphorylation efficiency by recruiting target proteins (Kallunki et al., 1996; Holland and Cooper, 1999; Sharrocks et al., 2000). Our results are consistent with this idea, showing that the N terminus fragment was phosphorylated to a certain extent, despite the lack of docking domains. We thus speculated that a conformational change of the full-length NtWIF is induced by the N terminus phosphorylation, which is catalyzed by WIPK, preferentially interacting with the C-terminal docking domain.
If this is indeed the case, the active form of WIPK appears to be critical. It has been established that phosphorylation of the Thr-Glu-Tyr (TEY) site in MAPK is required for enzymatic activation (Pelech and Sanghera, 1992). Usually this phosphorylation is catalyzed by MAPKK (MEK) but also can be accomplished by MAPK itself (Craig et al., 1991). Bacterially expressed MAPK (ERK) exhibits distinct autophosphorylation at the TEY site and concomitant enzymatic activity (Boulton et al., 1991; Huang et al., 2000). Analysis of AtMPK4, an Arabidopsis WIPK homolog, has also indicated that phosphorylation of the TEY site by MEK might be required for full activation (Huang et al., 2000). Since our bacterially expressed WIPK showed clear autophosphorylation activity, it is evident that NtWIF/WIPK interactions are dependent on the active form of WIPK itself.
In vivo trans-activation assays clearly indicated NtWIF to be a novel type of transcription factor. This was confirmed by the in planta transcription activity of NtWIF. When expressed in BY2 cells, NtWIF activated the reporter gene 10-fold over the control, this probably being mediated through endogenous WIPK activated by the bombardment. However, this activity increased up to 50-fold with additional expression of WIPK. Since inactive mutant WIPK failed to activate NtWIF, phosphorylation evidently was responsible for the enhanced activity. To date, intracellular localization of WIPK has not necessarily been clear, although MPK3, a WIPK ortholog from Arabidopsis, was shown to normally localize to cytoplasm and, upon activation, become transferred to the nucleus (Ahlfors et al., 2004). In contrast, we found WIPK to simultaneously locate in nucleus and cytoplasm in onion epidermal cells. This may due to our experimental system using particle bombardment, by which cell injury is unavoidable. When introduced WIPK was expressed and translated in cytoplasm, the product was possibly activated by wounding and translocated to the nucleus. Since neither WIPK nor NtWIF possess nuclear localization signals, a third factor(s) might have functioned in this process.
Taken all the available evidence together, we propose the following model for WIPK/NtWIF interaction. NtWIF is a transcription factor, but under ordinary conditions is inactive because of a specific conformation due to interaction between the C- and N-terminal regions. Upon external stresses, WIPK is activated through phosphorylation by MEK and also by itself and preferentially interacts with the C-terminal region of NtWIF. WIPK then phosphorylates the Thr-Pro target site located in the N-terminal region, resulting in activation of NtWIF to function as a transcriptional activator.
If NtWIF modulates expression of wound and pathogen responsive genes in combination with WIPK, a question arises as to the identity of its downstream targets. There is currently no clear answer, but some clues are available. First, the presence of the B3 domain is indicative. It was initially reported in maize (Zea mays) viviparous 1 (McCarty et al., 1991), in Arabidopsis ABI3 (Giraudat et al., 1992), and in members of the ARF family of transcription factors (Ulmasov et al., 1999). These transcription factors have been implicated in either abscisic acid or auxin signal transduction, but so far have not been reported to be related to wounding- or pathogen-associated signaling. However, it was proposed that MPK3/6 and WIPK/SIPK are involved in the suppression of auxin responsive genes (Ren et al., 2002). Our findings may provide a molecular basis for this assumption and suggest that NtWIF could plays multiple roles in auxin signaling as well as in defense responses. Alternatively, crosstalk between auxin and defense signaling might exist, mediated by MAPKs, since NPK1, a tobacco MEKK, and its Arabidopsis ortholog, ANP1, contribute to both auxin- and stress-mediated responses (Kovtun et al., 1998, 2000). Second, recent research on the plant MAPK cascade has identified a series of genes that are located upstream and downstream (Asai et al., 2002; Kim and Zhang, 2004). For example, NtMEK2 specifically phosphorylates WIPK and SIPK, and its overexpression induces transcripts for HMGR and PAL, both critical for the defense response (Yang et al., 2001). In conditional gain-of-function NtMEK2 transgenic tobacco, ethylene production has been found to be increased (Kim et al., 2003) and WRKY genes transcriptionally activated (Kim and Zhang, 2004). It was suggested that the SIPK/WIPK cascade controls defense genes through WRKY-type transcription factors (Kim and Zhang, 2004). In Arabidopsis, MPK6, an ortholog of tobacco SIPK, was recently shown to directly phosphorylate aminocyclopropane carboxylic acid synthase, thereby inducing constitutive ethylene production (Liu and Zhang, 2004).
Although >800 MAPKs have so far been reported from eukaryotes, clarification of the identity of targets that are phosphorylated by them has been limited. In mammalian cells, transcription factors, including the MADS box, zinc finger, bZIP, and bHLH proteins, have been shown to be targeted by MAPKs (Yang et al., 2003). In plants, few examples have so far been reported. A rice (Oryza sativa) MAPK was shown to specifically phosphorylate and activate a transcription factor, which recognizes a GCC box element present in several pathogenesis-related gene promoters (Cheong et al., 2003). Our finding is compatible with this observation and further supports the idea that MAPKs regulate transcription of a variety of stress responsive genes through modulation of corresponding plant transcription factors and that this is common among organisms, although the target transcription factors are unique to each phylum of living things.
MATERIALS AND METHODS
Plant Materials and Treatments
Tobacco plants (Nicotiana tabacum cv Xanthi nc) were grown in a growth cabinet at 23°C under a 14/10-h light/dark cycle. For wounding, intact leaves from 3-month-old plants were punched with a paper puncher, and samples were collected at appropriate time points. TMV inoculation was performed adopting the temperature shift method as described by Yap et al. (2002). Briefly, healthy leaves were inoculated with TMV using Carborundum and kept at 30°C for 48 h, and then at 20°C, allowing plants to initiate the hypersensitive response. All harvested leaf samples were immediately frozen in liquid nitrogen before RNA extraction.
Gene Isolation
A cDNA library was constructed with the HybriZAP-2.1 Two-Hybrid predigested vector system (Stratagene) using RNA mixtures isolated from unwounded and wounded leaves. The bait plasmid was constructed by in-frame fusion of GBD-WIPK containing a 1.2-kb WIPK full-length cDNA (Seo et al., 1995) to the GAL4 DNA binding domain in a pBD-GAL4 Cam vector (Stratagene). The bait plasmid and cDNA library clones were then transformed sequentially into the yeast strain, Y190 (CLONTECH). A total of 5 × 105 transformants were screened for complementation of growth on SD agar supplemented with 60 mm 3-amino-1,2,4-triazole and a mixture of appropriate amino acids, which was depleted of Trp, Leu, and His. Secondary screening was conducted by 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside filter lift assay as described by the manufacturer (CLONTECH). Quantitation of binding was estimated by β-galactosidase activity according to the manufacturer's instructions (CLONTECH).
cDNA Isolation and RT-PCR
A 0.8-kb EcoRI-XhoI cDNA fragment from the yeast two-hybrid positive clone was used to screen a Uni-ZAP XR phage library prepared from N. tabacum cDNAs. RNA extraction and northern hybridization were performed as described (Yap et al., 2002). The NtWIF probe was a 1-kb cDNA fragment spanning nucleotides 1030 to 2114. RT-PCR was performed using SuperScript II, 5 μg of total RNA from each sample, and oligo(dT)12 to 18 primers under the conditions described by the manufacturer (Invitrogen). First-strand cDNA synthesized was then amplified with different sets of specific primers to estimate the transcript levels of NtWIF, WIPK, and actin genes.
Fusion Proteins
GST-WIPK and His-tagged WIPK were constructed according to the described method (Seo et al., 1995). GST-WIPK/K73R was obtained by subcloning a 1.2-kb mutated WIPK cDNA fragment encoding the full-length protein with a Lys (amino acid 73) substituted with Arg in the pGEX2T vector. The resulting plasmids were transformed into Escherichia coli JM109, and expression of fusion proteins was induced at 25°C for 3 h with 0.5 mm isopropyl-β-d-thiogalactoside. Full-length and deletion constructs of NtWIF fused to GST (Table I) were prepared by cloning corresponding NtWIF cDNA fragments into the GATEWAY cloning system (Invitrogen) according to the manufacturer's instructions. The final fragment was cloned into the pDEST20 vector plasmid and transformed into E. coli together with bacumid (genome of a baculovirus) to allow in vivo recombination, and resulting single plaques were picked up and further propagated. Sf9 cells (6 × 109), maintained in Grace's insect medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) and 500 μg mL−1 gentamycin, were infected with the recombinant baculovirus (500 μL) and incubated at 27°C for 4 d (Wada et al., 2003). Cells from one dish were suspended in 1 mL of lysis buffer (20 mm Tris-HCl, pH 7.5, 5 mm EDTA, 1% Nonidet P-40, 25% [v/v] glycerol, 1 mm DTT, 1 mm phenylmethylsulfonyl fluoride, and 100 μg mL−1 aprotinin) and sonicated for 10 s twice. Fusion proteins were purified through a glutathione-sepharose column according to the manufacturer's instructions (Amersham Biosciences). Protein concentrations were estimated by the Bradford method.
Pull-Down Binding Assay
Full length (NtWIF) and truncated (ΔNtWIF/N or ΔNtWIF/C) proteins were expressed in Sf9 cells as the GST fusion as described above, and His-tagged WIPK was expressed in E. coli. Approximately 5 μg of GST fusion protein was bound to a glutathione Sepharose4B column (MicroSpin GST Purification Module; Amersham Pharmacia Biotech). The binding reaction was performed with 0.5 μg of His-tagged WIPK at 4°C for 1 h (Swaffield and Johnston, 1996). After three washes with PBS buffer (140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, and 1.8 mm KH2PO4, pH 7.3), GST fusion proteins bound to the columns were eluted with a buffer containing 10 mm reduced glutathione in 50 mm Tris-HCl at pH 8. Eluted proteins were fractionated on 7.5% SDS-PAGE and subjected to western-blot hybridization using rabbit anti-His-tag antibodies (Santa Cruz Biotechnology) and anti-rabbit HRP conjugate antibodies (Bio-Rad).
Trans-Activation Assay
The GBD-NtWIF effector plasmids were constructed by fusing cDNA fragments encoding different domains of NtWIF (Table I) with the GAL4 DNA binding domain in a yy64 vector, a derivative of pMA560 (Yamamoto and Deng, 1998). As the negative control, yy44 encoding the GAL4 DNA binding domain was used (Yamamoto and Deng, 1998). The reporter plasmid contained a luciferase gene placed under the control of the GAL4 binding site (Yamamoto and Deng, 1998). An internal control (reference) plasmid, containing an R-luciferase gene placed under the control of a CaMV 35S-promoter was used to normalize for differences in bombardment efficiency. The WIPK was cloned into pBI221 driven by a 35S-CaMV promoter. Seven-day-old BY2 suspension cells were plated on half-strength Murashige and Skoog agar and bombarded with plasmids (effector:reporter:reference = 2:2:1) coated on a 1.0-μm microcarrier in a vacuum of 28 inches of mercury using a helium pressure of 1,100 psi (PDS 1000; Bio-Rad). Cells were placed 9 cm from the stopping screen. After bombardment, they were incubated in the dark at 25°C for 24 h. Luciferase and R-luciferase activities were assayed using a dual-luciferase assay kit (Promega) according to the manufacturer's instructions. Chemical luminescence was measured using a luminometer (Lumat LB9507; EG & G Berthold). Protein concentrations were determined with Bradford reagent (Bio-Rad) using bovine serum albumin as standard.
Phosphorylation Assay
In vitro kinase assays were performed with 5 μg of purified GST-WIPK or GST-WIPK/K73R fusion protein in a buffer containing 20 mm HEPES-KOH, pH 7, 0.5 mm EGTA, 1 mm DTT, 20 mm MgCl2, and 5 μm [γ-32P]ATP (New England Nuclear). Five micrograms of myelin basic protein or GST fusion proteins (full-length NtWIF, ΔNtWIF/C, or ΔNtWIF/N) were added as substrates in separate reactions. Mixtures were incubated at 30°C for 30 min, and an appropriate volume of SDS-PAGE dye was added to terminate the reactions. Proteins were separated on SDS-polyacrylamide gels (7.5%), stained with 0.25% Coomassie Brilliant Blue, and subjected to autoradiography for kinase activity, which was detected by applying BioMax film (Kodak) to the gels at −80°C for 72 h.
Epifluorescence Analysis
Appropriate full-length or partial cDNA regions of NtWIF were subcloned into the SalI/NcoI site of the CaMV35S-sGFP(S65T)-nos vector, harboring a synthetic gene for improved GFP, sGFP(S65T), driven by the CaMV35S promoter and NOS terminator (Chiu et al., 1996). The CFP:WIPK fusion was similarly generated by subcloning a 1.2-kb WIPK cDNA fragment into the pAVA574 vector. The GFP:NtAAA was prepared as described (Sugimoto et al., 2004). Onion epidermal cell layers were bombarded with gold particles (Bio-Rad) coated with CaMV35S-sGFP(S65T)-nos or different full-length or truncated pNtWIF::GFP constructs (Hara et al., 2000). After 6 to 18 h incubation at 25°C in darkness, the epidermal cell layers were viewed under a microscope (Olympus Provis AX70) equipped with a fluorescence module.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AB084669.
Acknowledgments
The authors thank Drs. Yasuo Niwa (Shizuoka University, Shizuoka, Japan) and Albrecht von Arnim (University of Tennessee Knoxville, TN) for generous gifts of psGFP(S65T) and plasmid 574 (ECFP), respectively. We also thank Ms. Yumi Yoshida (Nara Institute of Science and Technology) and Dr. Malcolm Moore (Intermal, Nagoya, Japan) for preparation and critical reading of the manuscript, respectively.
This work was supported by a grant for the Research for the Future from the Japan Society for the Promotion of Science Program (grant no. JSPS–RFTF 00L01604).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.065656.
References
- Ahlfors R, Macioszek V, Rudd J, Brosché M, Schlichting R, Scheel D, Kangasjärvi J (2004) Stress hormone-independent activation and nuclear translocation of mitogen-activated protein kinase in Arabidopsis thaliana during ozone exposure. Plant J 40: 512–522 [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signaling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Boulton TG, Nye SH, Robbins DJ, Ip NY, Radziejewska E, Morgenbesser SD, DePinho RA, Panayotatos N, Cobb MH, Yankopoulos GD (1991) ERKs: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell 65: 663–675 [DOI] [PubMed] [Google Scholar]
- Chandra S, Low PS (1995) Role of phosphorylation in elicitation of the oxidative burst in cultured soybean cells. Proc Natl Acad Sci USA 92: 4120–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Moon BC, Kim JK, Kim CY, Kim MC, Kim IH, Park CY, Kim JC, Park BO, Koo SC, et al (2003) BWMK1, a rice mitogen-activated protein kinase, locates in the nucleus and mediates pathogenesis-related gene expression by activation of a transcription factor. Plant Physiol 132: 1961–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WL, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Craig CM, Alessandrini AA, Erikson RL (1991) Mouse Erk-1 gene product is a serine/threonine protein kinase that has the potential to phosphorylate tyrosine. Proc Natl Acad Sci USA 88: 8845–8849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droillard MJ, Thibivilliers S, Cazale AC, Barbier-Brygoo H, Lauriere C (2000) Protein kinases induced by osmotic stresses and elicitor molecules in tobacco cell suspensions: two crossroad MAP kinases and one osmoregulataion-specific protein kinase. FEBS Lett 474: 217–222 [DOI] [PubMed] [Google Scholar]
- Felix G, Regenass M, Spanu P, Boller T (1994) The protein phosphatase inhibitor calyculin A mimics elicitor action in plant cells and induces rapid hyperphosphorylation of specific proteins as revealed by pulse labeling with [33P]phosphate. Proc Natl Acad Sci USA 91: 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianinazzi S (1970) Hypersensibilite aux virus, temperatures et proteines solubles chez le Nicotoana tabacum cv. xanthi-nc. Comptes rendus de l' Academie de Paris D 270: 2382–2386 [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskopf DG, Felix G, Boller T (1990) K-252a inhibits the response of tomato cells to fungal elicitors in vivo and their microsomal protein kinase in vitro. FEBS Lett 275: 177–180 [DOI] [PubMed] [Google Scholar]
- Haasen D, Köhler C, Neuhaus G, Merkle T (1999) Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana. Plant J 20: 695–705 [DOI] [PubMed] [Google Scholar]
- Hara K, Yagi M, Koizumi N, Kusano T, Sano H (2000) Rapid systemic accumulation of transcripts for the tobacco WRKY transcription factor on wounding. Mol Gen Genet 263: 30–37 [DOI] [PubMed] [Google Scholar]
- Holland PM, Cooper JA (1999) Protein modification: docking sites for kinases. Curr Biol 9: R329–R331 [DOI] [PubMed] [Google Scholar]
- Huang Y, Li H, Gupta R, Morris P, Luan S, Kieber JJ (2000) ATMPK4, an Arabidopsis homolog of mitogen activated protein kinase is activated in vitro by AtMEK1 through threonine phosphorylation. Plant Physiol 122: 1301–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iajczyk MM, Awotunde OS, Muszynska G, Klessig DF, Dobrowolska G (2000) Osmotic stress induces rapid activation of a salicyclic acid-induced protein kinase and a homolog of protein kinase ASK1 in tobacco cells. Plant Cell 12: 165–178 [PMC free article] [PubMed] [Google Scholar]
- Jonak C, Kiegerl S, Ligterink W, Barker PJ, Huskisson NS, Hirt H (1996) Stress signaling in plants: A mitogen-activated protein kinase pathway is activated by cold and drought. Proc Natl Acad Sci USA 93: 11274–11279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallunki T, Deng T, Hibi M, Karin M (1996) c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell 87: 929–939 [DOI] [PubMed] [Google Scholar]
- Kiegerl S, Cardinale F, Siligan C, Gross A, Baudouin E, Liwosz A, Eklof S, Till S, Bogre L, Hirt H, Meskiene I (2000) SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress-induced MAPK, SIMK. Plant Cell 12: 2247–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CY, Yidong L, Thorne ET, Yang H, Fukushige H, Gassmann W, Hildebrand D, Sharp RE, Zhang S (2003) Activation of a stress-zresponsive mitogen-activated protein kinase cascade induces the biosynthesis of ethylene in plants. Plant Cell 15: 2707–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CY, Zhang S (2004) Activation of a mitogen-activated protein kinase cascade induces WRKY family of transcription factors and defense genes in tobacco. Plant J 38: 142–151 [DOI] [PubMed] [Google Scholar]
- Kovtun Y, Chiu WL, Tena G, Sheen J (2000) Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA 97: 2940–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun Y, Chiu WL, Zeng W, Sheen J (1998) Suppression of auxin signal transduction by a MAPK cascade in higher plants. Nature 395: 716–720 [DOI] [PubMed] [Google Scholar]
- Lebrun-Garcia A, Chiltz A, Gout E, Bligny R, Pugin A (2002) Questioning the role of salicylic acid and cytosolic acidification in mitogen-activated protein kinase activation induced by cryptogein in tobacco cells. Planta 214: 792–797 [DOI] [PubMed] [Google Scholar]
- Leon J, Rojo E, Sanchez-Serrano JJ (2001) Wound signaling in plants. J Exp Bot 52: 1–9 [DOI] [PubMed] [Google Scholar]
- Ligterink W (2000) MAP kinases in plant signal transduction: how many, and what for? Results Probl Cell Differ 27: 11–27 [DOI] [PubMed] [Google Scholar]
- Ligterink W, Hirt H (2001) Mitogen-activated protein (MAP) kinase pathways in plants: versatile signaling tools. Int Rev Cytol 201: 209–275 [DOI] [PubMed] [Google Scholar]
- Liu Y, Jin H, Yang K-Y, Kim CY, Baker B, Zhang S (2003) Interaction between two mitogen-activated protein kinases during tobacco defense signaling. Plant J 34: 149–160 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang S (2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR, Hattori T, Carson CB, Vasil V, Lazar M, Vasil IK (1991) The viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66: 895–905 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Irie K, Hirayama T, Hayashida N, Yamaguchi-Shinozaki K, Matsumoto K, Shinozaki K (1996) A gene encoding a mitogen-activated protein kinase kinase kinase is induced simultaneously with genes for a mitogen-activated protein kinase and an S6 ribosomal protein kinase by touch, cold and water stress in Arabidopsis thaliana. Proc Natl Acad Sci USA 93: 765–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G, Strydom D, Johnson S, Ryan CA (1991) A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253: 895–898 [DOI] [PubMed] [Google Scholar]
- Pelech SL, Sanghera JS (1992) MAP kinase: charting the regulatory pathways. Science 257: 1355–1356 [DOI] [PubMed] [Google Scholar]
- Ren D, Yang H, Zhang S (2002) Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J Biol Chem 277: 559–565 [DOI] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 709–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T (2001) Protein kinases in the plant defence response. Curr Opin Plant Biol 4: 407–414 [DOI] [PubMed] [Google Scholar]
- Romeis T, Piedras P, Zhang S, Klessig DF, Hirt H, Jones JDG (1999) Rapid Avr9- and Cf-9-dependent activation of MAP kinases in tobacco cell cultures and leaves: convergence of resistance gene, elicitor, wound and salicylate responses. Plant Cell 11: 273–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CA (2000) The systemin signaling pathway: differential activation of plant defensive genes. Biochim Biophys Acta 1477: 112–121 [DOI] [PubMed] [Google Scholar]
- Seidel JJ, Graves BJ (2002) An ERK2 docking site in the pointed domain distinguishes a subset of ETS transcription factors. Genes Dev 16: 127–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Okamoto M, Seto H, Ishizuka K, Sano H, Ohashi Y (1995) Tobacco MAP kinase: a possible mediator in wound signal transduction pathways. Science 270: 1988–1992 [DOI] [PubMed] [Google Scholar]
- Seo S, Sano H, Ohashi Y (1999) Jasmonate-based wound signal transduction requires activation of WIPK, a tobacco mitogen-activated protein kinase. Plant Cell 11: 289–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrocks AD, Yang SH, Galanis A (2000) Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem Sci 25: 448–453 [DOI] [PubMed] [Google Scholar]
- Sugimoto M, Yamaguchi Y, Nakamura K, Tatsumi Y, Sano H (2004) Hypersensitive response-induced ATPase associated with various cellular activities (AAA) protein from tobacco plants. Plant Mol Biol 56: 973–985 [DOI] [PubMed] [Google Scholar]
- Swaffield JC, Johnston SA (1996) Affinity purification of proteins binding to GST fusion proteins. In FM Ausubel, R Brent, RE Kingston, DD Moore, JG Seidman, JA Smith, K Struhl, eds, Current Protocols in Molecular Biology (Suppl 33). John Wiley & Sons, New York, pp 20.2.1–20.2.10 [DOI] [PubMed]
- Tena G, Asai T, Chiu WL, Sheen J (2001) Plant mitogen-activated protein kinase signaling cascades. Curr Opin Plant Biol 4: 392–400 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ (1999) Dimerization and DNA binding of auxin response factors. Plant J 19: 309–319 [DOI] [PubMed] [Google Scholar]
- Wada Y, Ohya H, Yamaguchi Y, Koizumi N, Sano H (2003) Preferential de novo methylation of cytosine residues in non-CpG sequences by a domains rearranged DNA methyltransferase from tobacco plants. J Biol Chem 278: 42386–42393 [DOI] [PubMed] [Google Scholar]
- Xing T, Malik K, Martin T, Miki BL (2001) Activation of tomato PR and wound-related genes by mutagenized tomato MAP kinase kinase through divergent pathways. Plant Mol Biol 46: 109–120 [DOI] [PubMed] [Google Scholar]
- Yamamoto YY, Deng XY (1998) A new vector set for GAL-4-dependent transactivation assay in plants. Plant Biotechnol 15: 217–220 [Google Scholar]
- Yang KY, Liu Y, Zhang S (2001) Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc Natl Acad Sci USA 98: 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Sharrocks AD, Whitmarsh AJ (2003) Transcriptional regulation by MAP kinase signaling cascades. Gene 320: 3–21 [DOI] [PubMed] [Google Scholar]
- Yap Y-K, Kakamu K, Yamaguchi Y, Koizumi N, Sano H (2002) Promoter analysis of WIPK, a gene encoding a tobacco MAP kinase, with reference to wounding and tobacco mosaic virus infection. J Plant Physiol 159: 77–83 [Google Scholar]
- Zhang S, Klessig DF (1997) Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell 9: 809–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Klessig DF (2001) MAPK cascades in plant defense signaling. Trends Plant Sci 11: 520–527 [DOI] [PubMed] [Google Scholar]
- Zhang S, Liu Y (2001) Activation of salicyclic acid-induced protein kinase, a mitogen-activated protein kinase, induces multiple defense responses in tobacco. Plant Cell 13: 1877–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Liu Y, Klessig DF (2000) Multiple levels of tobacco WIPK activation during the induction of cell death by fungal elicitins. Plant J 23: 339–347 [DOI] [PubMed] [Google Scholar]