Abstract
The ambient-light conditions mediate chloroplast relocation in plant cells. Under the low-light conditions, chloroplasts accumulate in the light (accumulation response), while under the high-light conditions, they avoid the light (avoidance response). In Arabidopsis (Arabidopsis thaliana), the accumulation response is mediated by two blue-light receptors, termed phototropins (phot1 and phot2) that act redundantly, and the avoidance response is mediated by phot2 alone. A mutant, J-domain protein required for chloroplast accumulation response 1 (jac1), lacks the accumulation response under weak blue light but shows a normal avoidance response under strong blue light. In dark-adapted wild-type cells, chloroplasts accumulate on the bottom of cells. Both the jac1 and phot2 mutants are defective in this chloroplast movement in darkness. Positional cloning of JAC1 reveals that this gene encodes a J-domain protein, resembling clathrin-uncoating factor auxilin at its C terminus. The amounts of JAC1 transcripts and JAC1 proteins are not regulated by light and by phototropins. A green fluorescent protein-JAC1 fusion protein showed a similar localization pattern to green fluorescent protein alone in a transient expression assay using Arabidopsis mesophyll cells and onion (Allium cepa) epidermal cells, suggesting that the JAC1 protein may be a soluble cytosolic protein. Together, these results suggest that JAC1 is an essential component of phototropin-mediated chloroplast movement.
Chloroplasts change their position in a cell in response to environmental light conditions (Wada et al., 1993, 2003). Low-fluence rate light induces movement of chloroplasts toward the irradiated area, resulting in chloroplast accumulation at the front face of the cell (accumulation response). Conversely, under high-fluence rate light, chloroplasts move to the anticlinal wall of the cell to avoid photodamage (avoidance response; Kasahara et al., 2002). Chloroplast photorelocation movement is found in several photosynthetic plant species, including yellow and green algae, mosses, ferns, and flowering plants. In most plant species, chloroplast movement is induced by irradiation with blue light, although it is also induced by red light in some cryptogam plants (Wada et al., 1993, 2003). The flowering plant Arabidopsis (Arabidopsis thaliana) has two types of blue-light photoreceptor, cryptochromes (cry1 and cry2) and phototropins (phot1 and phot2). Cryptochrome is a flavoprotein similar to the microbial type-I photolyase and regulates de-etiolation response and entrainment of the circadian clock (Lin and Shalitin, 2003). Phototropin has two light, oxygen, and voltage domains arranged in tandem in the N terminus and a Ser/Thr kinase domain at the C terminus (Briggs et al., 2001). The phot1 protein was identified as a blue-light photoreceptor-mediating phototropism induced by low-fluence rate of blue light (Huala et al., 1997). We previously analyzed chloroplast movement in a cry1cry2 double mutant and a phot1 mutant, but both accumulation and avoidance responses were induced in these photoreceptor mutants comparable to wild-type plants (Kagawa and Wada, 2000).
We screened mutants defective in the avoidance response using white band assay (WBA) in Arabidopsis (Kagawa et al., 2001). To perform the assay, a leaf was detached from the plant at a petiole and irradiated on agar media with strong white light delivered through an open slit of about 1 mm in width. This treatment given to wild-type leaves resulted in a color change from green to pale green as a consequence of a chloroplast avoidance response in the site irradiated through the slit. Using this screening method, we identified defective in chloroplast avoidance movement 1 (cav1) mutants, which showed the color change from green to dark green instead of green to pale green in the irradiated region. It was shown that CAV1 gene is another phototropin gene PHOT2, which is a paralog of PHOT1 (Kagawa et al., 2001). In the phot2 mutant, the accumulation response was observed even under high-fluence rate of blue light (Jarillo et al., 2001; Kagawa et al., 2001). A phot1phot2 double mutant did not show any accumulation response, indicating that phot1 and phot2 redundantly regulate chloroplast accumulation movement (Sakai et al., 2001). In a subsequent analysis of a phot1phot2 double mutant, it was shown the two phototropins also mediate redundantly phototropism, stomatal opening, and leaf expansion (Kinoshita et al., 2001; Sakai et al., 2001; Sakamoto and Briggs, 2002).
Although the photoreceptors for chloroplast photorelocation movement have been identified, the signal transduction pathway is still unknown. Many studies implicate calcium ions in chloroplast movement (Tlalka and Fricker, 1999; Wada et al., 2003), but the assignment of calcium ion as a second messenger in photorelocation movement is controversial. Arabidopsis phototropins mediate blue light-induced calcium influx into the cytoplasm (Baum et al., 1999; Babourina et al., 2002; Harada et al., 2003). In mesophyll cells, phototropins activate calcium-permeable channels on the plasma membrane (Stoelzle et al., 2003). Phototropin-mediated calcium influx is inhibited by application of the calcium channel blockers lanthanum (La3+) and gadolinium (Gd3+; Baum et al., 1999; Harada et al., 2003; Stoelzle et al., 2003). However, both La3+ and Gd3+ are completely ineffective in inhibiting both the light-induced chloroplast accumulation and avoidance responses in protonemal cells of the fern Adiantum capillus-veneris and the moss Physcomitrella patens (Sato et al., 2001, 2003). Therefore, it is unlikely that the influx of extracellular calcium functions as the signal for blue light-mediated chloroplast movement.
It has been shown that most plants utilize microfilaments for chloroplast movement (Wada et al., 2003). In Arabidopsis, the anti-actin drug Latrunculin B, but not the anti-microtubule drug Oryzalin, induced aberrant aggregation of chloroplasts in mesophyll cells (Kandasamy and Meagher, 1999). Immunolabeling of actin filaments with an anti-actin antibody showed that chloroplasts aligned along the thick actin cables and were enclosed within fine actin filaments (Kandasamy and Meagher, 1999). Recently, we identified a novel mutant, chloroplast unusual positioning 1 (chup1; Oikawa et al., 2003). In chup1 plants, the chloroplasts are unusually positioned, constitutively aggregating on the cell bottom and unable to move in response to light (Kasahara et al., 2002; Oikawa et al., 2003). CHUP1 encodes a novel plant protein capable of interacting with F-actin in vitro (Oikawa et al., 2003). However, the relationship of CHUP1 with microfilaments in vivo remains to be determined. In summary, the signal transduction components for chloroplast photorelocation movement have still not been identified.
Here, we developed a new screening method, the green band assay (GBA; described below), for measuring the chloroplast accumulation response in Arabidopsis. Using this method, we isolated a novel mutant, J-domain protein required for chloroplast accumulation response 1 (jac1), which is defective in the chloroplast accumulation response but not the avoidance response. Moreover, we found that chloroplast accumulation on the cell bottom in darkness is regulated by JAC1 and PHOT2. JAC1 encodes a C-terminal J-domain protein similar to auxilin clathrin-uncoating factor and is the first component identified in the signal transduction pathway for chloroplast photorelocation movement.
RESULTS
Isolation of jac1 Mutants with a New Screening Method, GBA
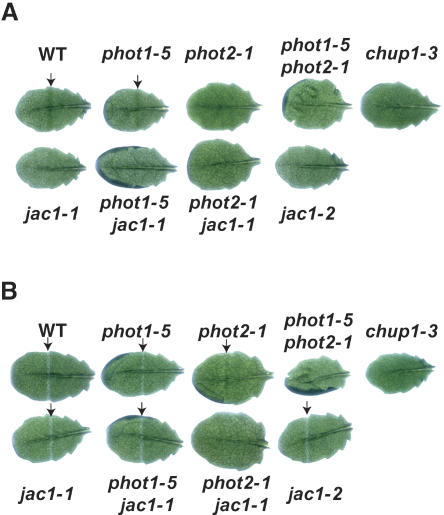
To identify mutations in genes other than PHOT2 and CHUP1 leading to defects in chloroplast avoidance movement, we developed a new screening method to detect the chloroplast accumulation response called the GBA. In this assay, a leaf is detached at the petiole and the whole leaf placed on the surface of an agar plate. The leaf is irradiated with strong white light, which results in a color change in wild-type leaves from green to pale green as a consequence of the avoidance response. Then, the leaf is covered with a black plate with an open slit of 1 mm in width, through which part of the leaf is irradiated with weak blue light. A green band appears in wild-type leaves through the accumulation response, but mutants lacking the accumulation response do not develop the green band. As expected, under whole-leaf irradiation with the strong light, the wild-type leaf showed a green band with GBA, but phot2-1 and chup1-3 mutants did not show a green band due to defect(s) in avoidance response (Fig. 1A). To exclude phot2 and chup1 mutants, WBA was also applied after GBA. As described previously (Kagawa et al., 2001; Sakai et al., 2001), wild-type and phot1-5 leaves showed a white band under WBA, whereas phot2-1 showed a green band. phot1-5 phot2-1 and chup1-3 leaves showed no band (Fig. 1B).
Figure 1.
Detection of chloroplast photorelocation movement using band assays. A, GBA for detection of the chloroplast accumulation response. Green bands are indicated with arrows. B, WBA for detection of the chloroplast avoidance response. White bands are indicated with arrows. Note that phot2-1 showed a green band with the WBA, in contrast to the wild type. WT, Wild type.
Mutants lacking a band under GBA and showing a white band under WBA were selected as candidates for mutants deficient in the chloroplast accumulation response. About 83,000 ethylmethane sulfonate (EMS)-mutagenized seeds, about 13,000 fast-neutron mutagenized seeds, about 26,000 γ-ray mutagenized seeds, and 2,960 T-DNA-tagged lines were screened, and four mutants were obtained. All of them had a single nuclear recessive mutation and fell into two complementation groups (data not shown). In this article, we describe one of the groups, termed jac1.
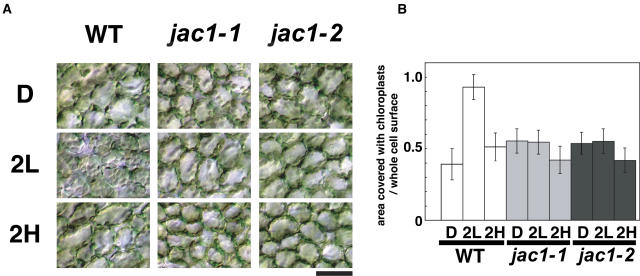
Two independent jac1 alleles, jac1-1 (EMS) and jac1-2 (T-DNA), were isolated. Leaves from these mutants showed a white band under WBA (Fig. 1B) but no green band under GBA (Fig. 1A). To confirm that this band phenotype in jac1 mutants results from the impairment in chloroplast movement, wild-type and jac1 mutant plants were dark adapted for about 12 h and then irradiated with white light of 10 or 100 μmol m−2 s−1 for 2 h (low-fluence [LL] or high-fluence rate white light [HL], respectively). Leaves were then fixed and the distribution of chloroplasts observed (Fig. 2). In wild-type plants, the chloroplasts moved to the cell surface under LL condition in an accumulation response, whereas they moved to the anticlinal wall under HL condition in an avoidance response (Fig. 2A). In jac1 mutants, the distribution of chloroplasts under both LL and HL conditions was similar to that of wild-type plants under HL conditions (Fig. 2A). However, the area of cell surface occupied by chloroplasts under HL was smaller than that seen under LL (Fig. 2B), meaning that jac1 mutants are normal in the avoidance response. To investigate chloroplast photorelocation movement in more detail, part of a jac1 or wild-type mesophyll cell was irradiated with a microbeam of blue light and chloroplast movement recorded using a video camera (Supplemental Movie 1). Under these conditions, wild-type cells responded to low-fluence rate blue light (5.6 μmol m−2 s−1) and the chloroplasts moved toward the light spot. In jac1 mutants, however, the chloroplasts moved away from the beam spot, even at the fluence rate of 5.6 μmol m−2 s−1, and never gathered in the irradiated area, confirming that jac1 mutants are defective in the chloroplast accumulation response but not in the avoidance response.
Figure 2.
Characterization of chloroplast movement in jac1 mutants. A, Chloroplast positioning before and after irradiation with white light for 2 h. After dark adaptation for about 12 h (D), plants were irradiated with white light at 10 (2L) or 100 μmol m−2 s−1 (2H) for 2 h. Bar = 50 μm. B, Comparison of the ratio of the area occupied with chloroplasts to the area of whole cell surface of wild-type and jac1 mutant plants. Error bars are sds, where n = 144 mesophyll cells. In one experiment, eight leaves from approximately three to four plants were sampled and six cells per a leaf were analyzed. Data from three independent experiments are combined. WT, Wild type.
The Chloroplast Accumulation on the Cell Bottom Is Dependent on phot2 and on JAC1 in Dark-Adapted Cells
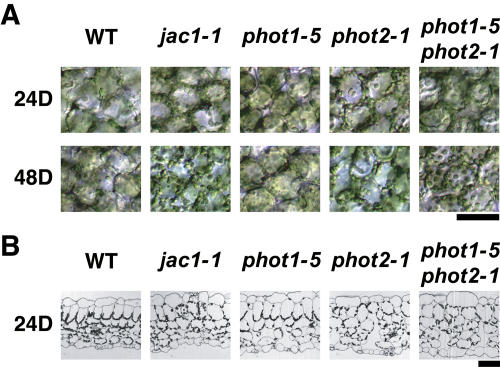
In cells of wild-type plants dark adapted for about 12 h prior to irradiation, we found that most of the chloroplasts were at the bottoms of cells and few chloroplasts were visible on the upper cell surfaces (Figs. 2A and 3). Interestingly, the chloroplast positioning in dark-adapted jac1 mutant cells resembled that of wild-type cells irradiated with HL (Figs. 2A and 3). Dark adaptation for longer time periods (for 24 h or 48h) did not induce chloroplast accumulation on the cell bottom in jac1 mutants (Fig. 3A). In cross sections of wild-type and mutant leaves, chloroplasts in the wild-type cells showed accumulation on the cell bottom, whereas chloroplasts in jac1 cells did not sediment and were distributed randomly (Fig. 3B). Surprisingly, phot2-1 mutant cells but not phot1-5 mutant cells were also defective in chloroplast accumulation on the cell bottom. The chloroplast distribution was similar to that of jac1 mutants (Fig. 3). The phot1-5 phot2-1 double mutant also lacked the dark-accumulation response, and some chloroplasts were found on the upper cell surface. Other alleles of phot2 were also defective in the dark accumulation response (data not shown). Given that the dark-accumulation response is normal in phot1-5, a null allele (Huala et al., 1997), and is impaired in several phot2 alleles other than phot2-1, it is likely that only phot2 mediates this response, possibly via JAC1.
Figure 3.
Dark-induced chloroplast accumulation response on the cell bottom. A, Upper view of dark-adapted cells. Three-week-old plants were irradiated with white light at 10 μmol m−2 s−1 for 24 h. Plants were then dark adapted for the indicated times. B, Cross sections of dark-adapted leaves. Leaves were fixed after dark adaptation for 24 h. Sections were stained with toluidine blue. WT, Wild type. Bar = 50 μm.
Analyses of Double or Triple Mutants between jac1-1 and Phototropin Mutants
To further investigate the role of the JAC1 gene in phototropin-mediated chloroplast photorelocation movement, the phenotypes of phot1-5 jac1-1, phot2-1 jac1-1, and phot1-5 phot2-1 jac1-1 mutants were analyzed and compared with that of phot1-5, phot2-1, and phot1-5 phot2-1, respectively (Supplemental Fig. 1). The phot1-5 jac1-1 double mutant has a similar phenotype to the jac1-1 single mutant (Supplemental Fig. 1). In this experiment, the accumulation response in phot1-5 was very weak, and chloroplasts accumulated less on cell surface (Supplemental Fig. 1) compared to the wild type. The phot2-1 jac1-1 double mutant showed no band under either WBA or GBA (Fig. 1), and was defective in the dark-sedimentation and avoidance responses, although some chloroplasts were found on the cell surface under both LL and HL conditions (Supplemental Fig. 1). However, the chloroplast density on the surface was constant for at least 2 h under LL or HL conditions, unlike in wild type or the phot2-1 single mutant. Experiments with microbeam irradiation revealed that the phot2-1 jac1-1 mutant cells failed to undergo both accumulation and avoidance movements (Supplemental Movie 2), indicating that the chloroplast distribution on the cell surface under LL and HL conditions is not the result of light-induced directional movements. The difference in chloroplast distribution between light and dark condition was not found in phot1-5 phot2-1 or phot1-5 phot2-1 jac1-1 mutants (Supplemental Fig. 1). The chloroplast densities in phot1-5 phot2-1 or phot1-5 phot2-1 jac1-1 mutant were constant regardless of the light conditions (Supplemental Fig. 1). The phot1-5 phot2-1 and phot1-5 phot2-1 jac1-1 mutant plants lacked all three types of chloroplast movement (dark positioning, accumulation movement, and avoidance movement). Altogether, phot2 mediates three different types of chloroplast movement under the three light conditions tested (darkness, LL, and HL), but phot1 regulates only the accumulation response under LL and HL. Although chloroplasts were found at the cell surfaces of the periclinal wall in phot1-5 phot2-1 plants, the chloroplasts of phot1-5 phot2-1 jac1-1 plants tended to be at the anticlinal wall, similar to the chloroplast distribution in jac1 mutants (Supplemental Fig. 1). Taken together, these results suggest that JAC1 must be an indispensable component for chloroplast accumulation movement with the exception of the avoidance response.
Cloning of the JAC1 Gene
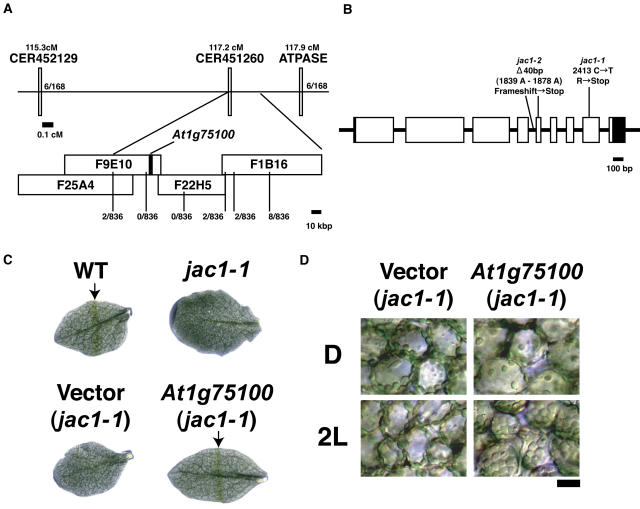
Initially, the jac1 mutation was mapped close to the simple sequence length polymorphism (SSLP) marker ATPase on the lower arm of chromosome 1 (Fig. 4A). To narrow down the map position of the jac1 mutation further, fine structure mapping was performed using the Cereon Arabidopsis polymorphism collection data (Jander et al., 2002), which contains new SSLP and cleaved amplified polymorphic sequence (CAPS) markers. Analysis of 836 chromosomes derived from the segregating F2 mutant plants from crosses between the jac1-1 Columbia (Col-0) background and Landsberg erecta narrowed the position of JAC1 to two bacterial artificial chromosomes (BACs), F9E10 and F22H5 (Fig. 4A). Since the genomic sequences of these BACs are available, the annotated genes within this interval were amplified by PCR in both jac1-1 and jac1-2, and then sequenced. Both mutants contained a lesion in the putative open reading frame of one gene, At1g75100 (Fig. 4B). Sequences of several expressed sequence tags (ESTs) have been deposited in GenBank. Although some ESTs contain a 3′ polyadenylation site, both 5′ and 3′ end sequences were determined by RACE-PCR. Recently, two full-length cDNA clones corresponding to this gene (AY057504 and AY103303) were deposited in GenBank. Comparison between the two cDNA sequences and our cDNA sequence revealed that the complete coding region is present in all cDNA clones. The gene consists of nine exons interrupted by eight introns (Fig. 4B). The jac1-1 mutation is a C-to-T transition at the 2,413 nucleotide from the start codon. The jac1-2 mutation is 40-bp deletion between nucleotides 1,839 and 1,878, which is predicted to truncate a splice site between intron 4 and exon 5. Transgenic jac1-1 mutants carrying a T-DNA insertion containing a 4.5-kb At1g75100 genomic fragment recovered not only the chloroplast accumulation response but also the dark positioning (Fig. 4, C and D). However, the jac1-1 plants transformed with vector alone did not recover these responses, confirming that At1g75100 corresponds to the JAC1 gene.
Figure 4.
Cloning of the JAC1 gene and complementation of the jac1-1 mutant. A, Mapping of the JAC1 locus on the distal arm chromosome 1. The genetic linkage map is shown at the top. ATPASE indicates an SSLP marker. CER452129 and CER451260 are SSLP markers published by Cereon. The physical linkage map is shown below. BACs are shown as white rectangles. The At1g75100 gene is shown as a black rectangle. Recombination rates are shown underneath. B, The JAC1 gene (At1g75100) structure. The JAC1 gene has nine exons (white rectangle; black area indicates the untranslated regions) and eight introns (lines between white rectangles). Positions and identities of two jac1 alleles are indicated. C, GBA in wild type, jac1-1, and jac1-1 transformed with the vector only (vector) or a JAC1 genomic clone (At1g75100). Green bands are indicated by arrows. D, Complementation of a jac1-1 mutant. Wild type, jac1-1, and jac1-1 transformed with the vector only (vector) or a JAC1 genomic clone (At1g75100) was dark adapted (D) and then irradiated with 10 μmol m−2 s−1 light (2L) for 2 h. Plants transformed with the JAC1 gene were rescued, but plants transformed with the vector alone were not (compare to Fig. 2). Bar = 50 μm.
The JAC1 Gene Encodes a C-Terminal J-Domain Protein Similar to Auxilin
The JAC1 gene is predicted to encode a 651-amino acid polypeptide (Fig. 5A). Domain homology searches show that the JAC1 protein contains a J-domain at the C terminus (Fig. 5A). The J-domain, which contains the highly conserved His/Pro/Asp tripeptide, plays an important role in organizing interactions with its Hsp70 chaperone partner(s) (Kelley, 1998). The JAC1 J-domain is similar to the J-domain of auxilin, a clathrin-uncoating factor in cow, yeast, and worm (Lemmon, 2001). Auxilin has been shown to uncoat clathrin-coated vesicles (Ungewickell et al., 1995). The auxilin J-domain interacts with HSC70 protein, recruiting HSC70 to the clathrin heavy chain. Across the C-terminal domain, JAC1 shares between 25% and 31% amino acid similarity with animal and yeast auxilins (Fig. 5B). A BLAST search (The Arabidopsis Information Resource; http://www.arabidopsis.org/Blast/) revealed the existence of six other Arabidopsis proteins similar to auxilin. The C-terminal domain of JAC1 shares between 47% and 57% amino acid similarity with that of the six Arabidopsis proteins (Fig. 5B). A His/Pro/Asp motif is completely conserved among all these proteins. However, their N-terminal domains have no homology to other nonplant auxilins and no other recognizable domain, but a short stretch of conserved amino acids is found within a 100-amino acid region of the N termini (Fig. 5C).
Figure 5.
The JAC1 protein has a C-terminal J-domain similar to auxilin. A, Alignment of the amino acid sequences of the Arabidopsis JAC1 protein and the rice homolog, OsJAC1. The auxilin-like C-terminal domain including a J-domain is underlined. Short blocks conserved between Arabidopsis and rice are double underlined. B, Alignment of the deduced amino acid sequences of C-terminal domains of Arabidopsis auxilin-like protein and auxilin proteins. Black boxes represent residues identical to Arabidopsis JAC1. Bt, Bos taurus (Ungewickell et al., 1995); Ce, C. elegans (Greener et al., 2001); Sc, Saccharomyces cerevisiae (Gall et al., 2000; Pishvaee et al., 2000). C, Short conserved region at the N terminus of JAC1 and Arabidopsis auxilin-like proteins. Black boxes represent identical residues in more than three proteins. Black circles indicate an FxDxF motif.
Recently, sequences of 28,469 full-length cDNA clones from rice (Oryza sativa L. ssp. japonica cv Nipponbare) were published (Kikuchi et al., 2003). We searched the full-length cDNA database of the Rice Genome Resource Center (http://cdna01.dna.affrc.go.jp/cDNA/) and identified two cDNA clones (AK071995 and AK099437) as putative rice JAC1 homologs (tentatively OsJAC1). Comparison of genomic and cDNA sequences revealed that the OsJAC1 gene has nine exons and eight introns, the same as the Arabidopsis JAC1 gene. The OsJAC1 gene is predicted to encode a 607-amino acid polypeptide, and the amino acid similarity of the C-terminal J-domain with that of the Arabidopsis JAC1 protein is 69.2% (Fig. 5,A and B). OsJAC1 protein has a higher similarity to JAC1 than to the other Arabidopsis auxilin-like proteins, suggesting that OsJAC1 is the rice ortholog of JAC1 (Fig. 5B). Besides the J-domain, the JAC1 and OsJAC1 proteins, but not other auxilin-like proteins, share two blocks of conserved amino acids, ExPVFGxxTSSxRRR and VKGKVxDFxKIFS (Fig. 5A, double underlined). Additional cDNA sequences highly similar to OsJAC1 were found in the maize (Zea mays), wheat (Triticum aestivum), and barley (Hordeum vulgare) EST collections in database searches (data not shown). However, it remained to be determined whether these monocot JAC1-like genes are also involved in chloroplast movement.
Analyses of JAC1 Gene Expression and JAC1 Protein Abundance
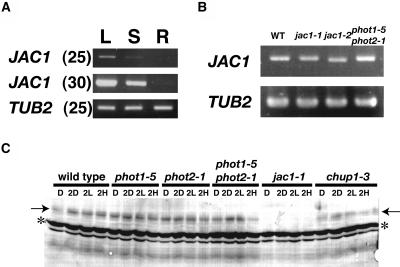
Since we could not detect JAC1 transcripts by RNA gel-blot analysis (data not shown), expression of the JAC1 gene was determined by reverse transcription (RT)-PCR (Fig. 6). JAC1 gene was found to be expressed in leaves and stems but not in root tissues of 7-week-old plants (Fig. 6A), and expression was higher in leaves than in stems. In jac1-1 and jac1-2 mutants, the JAC1 transcripts accumulated at levels similar to wild type (Fig. 6B). However, jac1-1 transcripts contain a C-to-U transition 1,807 nucleotides from the start codon, corresponding to the jac1-1 mutation in JAC1 gene. This base change introduces a stop codon. In jac1-2, amplified JAC1 fragments were thus slightly smaller than the others (Fig. 6B). The jac1-2 transcripts had no exon 5 sequence and contained premature stop codon. Thus, the two jac1 alleles were deduced to encode prematurely truncated polypeptides. In phot1-5 phot2-1 double mutants, JAC1 transcripts accumulated at similar levels to wild type (Fig. 6), indicating that the defect in the chloroplast accumulation response is not a defect in the accumulation of JAC1 transcripts.
Figure 6.
RT-PCR analysis of expression of JAC1. The TUB2 gene was amplified as a quantitation control. The amplified products of JAC1 are 1,946 bp (2–1,947) and of TUB2 are 664 bp (741–1,304). WT, Wild type. A, JAC1 gene expression in leaves (L), stems (S), and roots (R). The number in parenthesis indicates PCR amplification cycle number. B, JAC1 gene expression in wild type, jac1 mutants, and phot1-5 phot2-1 double mutants. PCR products amplified for 25 cycles were used. C, Immunoblot analysis of the JAC1 protein. After dark adaptation for about 12 h (D), plants were kept in darkness (2D) or were irradiated with white light at 10 (2L) or 100 μmol m−2 s−1 (2H) for 2 h. Arrows indicate bands corresponding to JAC1 protein. This band is not found in jac1-1 samples. Asterisks indicate nonspecific bands. This is a representative result from three separate experiments.
To investigate accumulation of JAC1 protein in wild-type, jac1, and phototropin mutant plants, endogenous JAC1 protein was analyzed by western blotting with polyclonal antisera against the C terminus of JAC1 (Fig. 6C). The JAC1 antisera recognized a protein band of around 85 kD, although it was larger than the predicted molecular mass (approximately 75 kD). This protein band was not observed in jac1-1 (Fig. 6C) and jac1-2 (data not shown) mutants, indicating that these two mutants are null alleles. JAC1 protein abundance in wild-type plants was not affected by dark adaptation for about 12 h (D) followed by irradiation with white light of 10 or 100 μmol m−2 s−1 for 2 h (2L or 2H) or darkness (2D; Fig. 6C). Moreover, the amount of JAC1 protein did not change in phot1-5, phot2-1 and phot1-5 phot2-1 double mutant plants (Fig. 6C). Therefore, the defect in chloroplast movement in phototropin mutants is not caused by the lack of JAC1 protein accumulation. Since chup1-3 plants also contain amounts of JAC1 protein comparable to wild-type plants (Fig. 6C), the unusual positioning of chloroplasts in chup1-3 plants (Oikawa et al., 2003) must not result from deficiency of JAC1 protein.
GFP-JAC1 Fusion Protein Has a Similar Localization Pattern to Green Fluorescent Protein
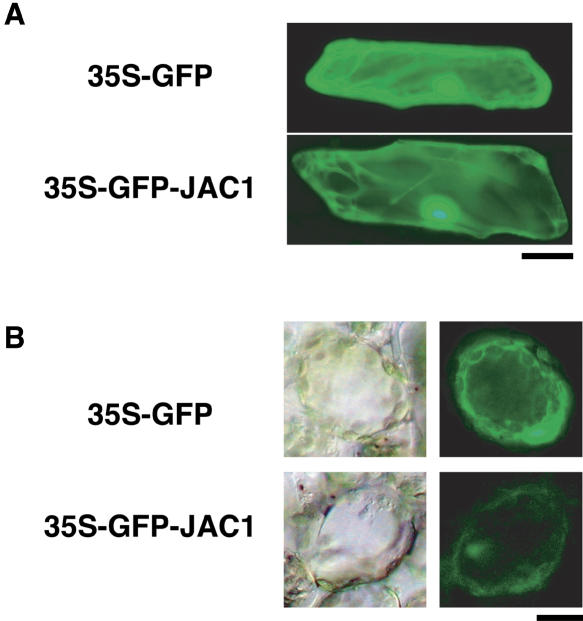
To investigate JAC1 protein localization, a cauliflower mosaic virus (CaMV) 35S promoter-driven fusion between the green fluorescent protein (GFP) gene and JAC1 cDNA (35S-GFP-JAC1) was expressed transiently in onion (Allium cepa) epidermal cells (Fig. 7A) and in Arabidopsis mesophyll cells (Fig. 7B). A 35S-GFP vector alone was used as the control. In both type of cells transformed with the fusion protein, GFP fluorescence was observed all over the cells, including the nuclei (Fig. 7). Organelle-specific fluorescence was not detected, but in the onion epidermal cells (Fig. 7A) fluorescence of cytoplasmic strands was clearly visible, suggesting that both GFP-JAC1 and GFP itself must be distributed in the cytosol.
Figure 7.
Subcellular localization of GFP-JAC1. A, Transient expression of GFP and GFP-JAC1 fusion protein in onion epidermal cells. GFP or GFP-JAC1 fusion genes driven by CaMV 35S promoter were delivered by particle bombardment to onion epidermal cells. Bar = 50 μm. B, Transient expression of GFP and GFP-JAC1 fusion protein in the mesophyll cells of Arabidopsis. The transmission images of GFP-expressing cells are also shown (left photos). Bar = 20 μm.
DISCUSSION
jac1 Mutants Define Three Signaling Pathways for Phototropin-Mediated Chloroplast Movement in Arabidopsis
We found that jac1 mutants are defective in the accumulation response but not in the avoidance response (Fig. 2), suggesting that phot1- and phot2-mediated accumulation response depends on JAC1, but phot2-mediated avoidance response under high-fluence rate light does not. Kagawa and Wada (1996, 1999) postulated that the signal transduction pathways for the accumulation and avoidance responses must differ. By examining chloroplast movement in Adiantum prothalial cells treated with brief blue-light microbeam irradiation, they found that (1) the signal for the accumulation response can be transferred long distances away from the irradiated area to chloroplasts but the signal of avoidance response cannot, and that (2) the lifetimes of the both signals are different from each other (30 to approximately 40 min for the accumulation response and less than 10 min for the avoidance response; Kagawa and Wada, 1996, 1999). Their hypothesis is consistent with the results described here: The accumulation response is dependent on JAC1, but the avoidance response is not. In jac1 mutants, the avoidance response was induced by irradiation with lower intensity blue light than in wild type, indicating that jac1 mutants are more sensitive to blue light in the avoidance response. Moreover, in phot2 mutants, accumulation response is induced even under strong-light conditions (Jarillo et al., 2001; Kagawa et al., 2001). Therefore, both signals could be generated under low- and high-intensity of light in wild-type cells. Under the weak-light condition, JAC1 mediates the chloroplast accumulation response via two phototropins. Under the strong-light condition, the signal for the phot2-mediated avoidance response overrides the pathways for the JAC1-mediated accumulation response, although phot1 activates the JAC1 pathway for the accumulation response at the same time.
In dark-adapted Adiantum and Physcomitrella protonemal cells, chloroplasts are found to be distributed evenly along the cell lengths (Sato et al., 2001; Wada and Kagawa, 2001). In two-dimensional gametophytes of Adiantum, the chloroplasts accumulate along the anticlinal walls in darkness, except at the marginal cell wall of the prothallus (Wada and Kagawa, 2001). In dark-adapted Arabidopsis, the chloroplasts sediment on the bottoms of mesophyll cells (Fig. 3). In general, leaves consist of several layers of cells, whereas fern prothallia have one layer of cells, and protonemal cells are filamentous. This anatomical difference may bring about the differences in the type of dark positioning of chloroplasts among plant species and/or tissues. Dark-induced accumulation of chloroplasts was not found in jac1 mutants (Fig. 3). Hence, JAC1 may receive the signal for chloroplast accumulation on the cell bottom in the dark. Interestingly, phot2 mutants but not phot1 mutants also lacked this response. Therefore, phot2 may generate the signal for chloroplast movement in darkness. Given that phot1 mediates only accumulation movement, it is likely that the signal for dark positioning and that for accumulation response are different.
Given the results described above, there must be three signaling pathways for chloroplast movement in Arabidopsis: (1) the pathway for the accumulation response dependent on JAC1, which is activated by phot1 and phot2 in the presence of light; (2) the pathway for the avoidance response independent of JAC1, which is activated by phot2 only under high-light condition; and (3) the pathway for the dark positioning dependent on PHOT2 and JAC1 in darkness. Although phot1 and phot2 have very different functions in chloroplast movement, PHOT1 and PHOT2 proteins have overall 58% amino acid identity and 67% amino acid similarity, and the two light, oxygen, and voltage domains and C-terminal Ser/Thr kinase domain are highly conserved. At present, we cannot say what difference between PHOT1 and PHOT2 brings about the functional divergence in chloroplast movement. The pair of PHOT1 and PHOT2 genes was found not only in the dicot Arabidopsis, but also in the monocot rice and the fern A. capillus-veneris. Therefore, the functional divergence between phot1 and phot2 may be evolutionally conserved.
JAC1 Encodes an Auxilin-Like Protein Bearing J-Domain at the C Terminus
The JAC1 protein has a C-terminal J-domain and resembles the clathrin-uncoating factor auxilin (Fig. 5B). Clathrin constitutes clathrin-coated vesicles together with the adaptor protein complexes during endocytosis (Schmid, 1997). The J-domain of auxilin binds to HSC70 and targets it to clathrin, which then interacts with the amino terminus of auxilin (Ungewickell et al., 1995). Although the J-domain of JAC1 is highly similar to auxilin, the complete amino terminus is not (Fig. 5). Auxilins have nonconserved clathrin-binding regions at the N terminus and all except the worm auxilin contain an Asp-Pro-Phe/Trp (DPF/W) adaptor protein-binding motif in this region (Owen et al., 1999; Traub et al., 1999; Lemmon, 2001). This domain is known to bind to the appendage domain of α- or β-subunits of an adaptor protein complex AP-2, which constitutes a heterotetramer essential for clathrin-coated vesicle formation. One DPF motif was found in four Arabidopsis auxilin-like proteins (At1g21660, At1g30280, At4g12770, and At4g12780), but JAC1 does not have this motif. Recently, At4g12770 was shown to be capable of uncoating clathrin in vitro, suggesting that At4g12770 is one of the Arabidopsis auxilins (Lam et al., 2000). At1g12770 was identified as an interacting partner of Arabidopsis SH3-containing protein 1 (AtSH3P1), involved in trafficking of clathrin-coated vesicles (Lam et al., 2000). Interaction between At4g12770 and AtSH3P1 is mediated through a Pro-rich domain. This Pro-rich domain is not present in the JAC1 protein sequence, suggesting that JAC1 may not function in AtSH3P1-dependent clathrin uncoating. Besides the DPF/W motif, the Phe-x-Asp-x-Phe (FxDxF; x is any amino acid) motif is also defined as an AP-2 α appendage-binding motif and is found in several accessory proteins indispensable for assembly of clathrin-coated vesicles (Brett et al., 2002). Interestingly, Arabidopsis auxilin-like proteins (including JAC1 and At4g12770) and OsJAC1 include this motif within the short conserved segment at the N terminus (Fig. 5C), although cow, worm, and yeast auxilins do not have this motif. At present, it is not clear whether the FxDxF motif and also the DPF/W motif function as AP-2 α appendage-binding domain in plants.
Mutants lacking auxilin in yeast (Gall et al., 2000; Pishvaee et al., 2000) and Caenorhabditis elegans (Greener et al., 2001) show a growth defect as a result of impairment of clathrin uncoating. However, growth and development of plant jac1 mutants are not different from those of wild type under our growth condition (data not shown). Further, a relationship between clathrin-mediated endocytosis and chloroplast movement has never been reported, to our knowledge, in any plant species. Therefore, JAC1 may not have a role in clathrin uncoating in Arabidopsis and may regulate chloroplast accumulation movement by a mechanism other than clathrin uncoating. However, we cannot exclude the possibility that JAC1 also functions as clathrin-uncoating factor. The J-domain presents a specific substrate(s) for an HSP70 partner (Kelley, 1998). Identification of JAC1-interacting proteins may clarify the role of JAC1 in chloroplast movement.
Regulation of JAC1 by Phototropins
How do two phototropins regulate JAC1 protein function in the mediation of chloroplast movement? When fern A. capillus-veneris protonemal cells were truncated with a thin string to cut off the nuclear-localizing portions, chloroplast accumulation and avoidance responses could still be induced in the remaining enucleated cells, indicating that nuclear gene expression at the level of transcription does not contribute to chloroplast photorelocation movement (Wada, 1988). Given that both PHOT1 and PHOT2 are plasmamembrane-localized proteins (Sakamoto and Briggs, 2002; Harada et al., 2003), it is unlikely that phototropins mediate chloroplast movement by regulating nuclear gene expression. Since chloroplast movement in A. capillus-veneris is also regulated by phototropins (Kagawa et al., 2004), phototropins may regulate chloroplast photorelocation movement without nuclear gene expression in Arabidopsis. Actually, JAC1 transcript levels are unaffected in a phot1phot2 double mutant (Fig. 6B). Since JAC1 protein levels in phototropin mutant plants were also comparable to those of wild-type plants under all light conditions (Fig. 6C), the defect of phototropin mutants in chloroplast movement must not result from an impairment in JAC1 protein accumulation.
It is well known that phototropins are Ser/Thr kinases and that autophosphorylation is induced by irradiation with blue light (Christie et al., 1998; Sakai et al., 2001). However, the substrates of phototropins have not yet been identified. Phototropins mediate a variety of responses including chloroplast photorelocation, phototropism, and stomatal opening (Jarillo et al., 2001; Kagawa et al., 2001; Kinoshita et al., 2001; Sakai et al., 2001). A screen for nonphototropic mutants yielded nph3 (Liscum and Briggs, 1995). The NPH3 gene encodes a protein that interacts with the PHOT1 N-terminal domain in vitro and the interaction was also shown by yeast two-hybrid assay, suggesting that NPH3 functions downstream of phot1 in phototropism (Motchoulski and Liscum, 1999). In Arabidopsis guard cells, phototropins activate a plasma membrane H+-ATPase (Kinoshita et al., 2001). Blue light causes phosphorylation and subsequent activation of H+-ATPase in Vicia fava guard cells (Kinoshita and Shimazaki, 1999), but whether phototropin directly phosphorylates H+-ATPase remained to be determined. Although JAC1 is a Ser-rich protein, in western-blot analyses with or without light treatment, an electrophoretic mobility shift of JAC1 protein, indicative of phosphorylation, has not been detectable to date. Moreover, the interaction of JAC1 with both PHOT1 and PHOT2 has not been detected in yeast two hybrid assays (data not shown).
We found that GFP-JAC1 protein localized in the cytosol in a transient expression assay in onion epidermal cells and the Arabidopsis mesophyll cells, although nuclear localization was also visible (Fig. 7). Therefore, the unidentified signal from phototropins localized on plasmamembrane must be received by cytosolic JAC1 protein, then passed to chloroplasts to regulate the direction of their movement.
In conclusion, we have identified the JAC1 gene as an essential component for phot1- and phot2-mediated chloroplast accumulation movement, and the phot2-mediated dark sedimentation response. Further, we have shown that JAC1 is dispensable for phot2-mediated chloroplast avoidance response induced by high-light irradiation. The JAC1 protein is similar to auxilin, functioning as a clathrin-uncoating factor, and has a J-domain at the C terminus. However, the regulation of JAC1 protein function by phototropins remained to be shown. Since rice and other monocot species have JAC1 genes, dicots and monocots may share a similar mechanism for regulating chloroplast movement. Further analyses are necessary to clarify the function of JAC1 in phototropin-mediated chloroplast movement.
MATERIALS AND METHODS
Plant Growth and Mutant Screening
The culture medium for plant cultivation was 0.8% agar plate containing one-third-strength Murashige and Skoog inorganic salt described previously (Kagawa and Wada, 2000), but supplemented with 1% Suc. Seeds were sown on the culture medium, treated at 4°C for 2 d, and grown under white light at approximately 100 μmol m−2 s−1 (16 h)/dark (8 h) cycle at 25°C in an incubator (Biotron LH300-RPSMP; Nippon Medical & Chemical). Plants were cultured for at least 2 weeks for mutant screening and mapping, and for about 3 weeks for other experiments. EMS-, γ-ray-, and fast neutron-mutagenized seeds (Lehle Seeds) and T-DNA-tagged lines (Arabidopsis Biological Resource Center) were screened by band assay. For the GBA, excised leaves were placed on a 0.8% agar plate with their adaxial sides up, covered with a transparent film, and irradiated with strong cool-white light (approximately 800 μmol m−2 s−1) for 1 h. Thereafter, the plate was covered with a black box with open slits and the leaves were irradiated with weak white light (about 47 μmol m−2 s−1) through the slits for 30 min. Appearance of a green band was assessed for each leaf. For WBA, excised leaves were set on agar plates as for GBA and then irradiated with strong cool-white light through the slits for 1 h (Kagawa et al., 2001). Mutants were backcrossed at least three times with wild-type Col-0 gl1 background (Lehle Seeds). To obtain double or triple mutants, jac1-1 plants were backcrossed with pollen of phot1-5 (Huala et al., 1997) or phot2-1 (Kagawa et al., 2001). The phot1-5 jac1-1 and phot2-1 jac1-1 double mutants were confirmed in no green band seedlings in the F2 progeny from each cross using the polymorphism in each gene. phot1-5 phot2-1 double mutant was described previously (Kinoshita et al., 2001). phot1-5 phot2-1 jac1-1 triple mutant was constructed by crossing between phot1-5 jac1-1 and phot2-1 jac1-1. The chup1-3 mutant was described previously (Oikawa et al., 2003).
Analysis of Chloroplast Movement
Three-week-old seedlings, grown on 0.8% agar plates containing one-third-strength Murashige and Skoog salt and 1% Suc, were dark adapted for about 12 h and then irradiated with white light at 10 or 100 μmol m−2 s−1 for 2 h. White light was obtained from 40-W white fluorescent tubes (FLR40SW; Mitsubishi). Leaves of dark-adapted and white light-treated plants were cut at the petioles and fixed with 2.5% glutaraldehyde in fixation buffer (20 mm PIPES, 5 mm MgCl2, 5 mm EGTA, 0.5 mm phenylmethylsulfonyl fluoride, 1% dimethyl sulfoxide, pH 7.0). Specimens were observed and photographed under a microscope. Areas of cell surface and areas occupied by chloroplasts were measured, and the ratio of the chloroplast-covered area to the area of cell surface was calculated. For cross sections, fixed leaves were washed with fixation buffer and then post-fixed with 1% OsO4 for at least 1 h. After washing out OsO4 with water, specimens were dehydrated with a graded acetone series and embedded in Spurr's resin. Sectioning of specimens was performed with a Reichert Ultramicrotome (Ultracut; Reichert-Jung). Toluidine blue-stained sections were observed.
For microbeam experiments, 3-week-old seedlings were dark adapted for 1 d prior to use. The details of microbeam equipment and associated experimental conditions are described by Kagawa and Wada (2000).
Genetic Mapping and Rescue of jac1-1 Mutant
The jac1-1 mutant in a Col-0 background and Landsberg erecta plants were crossed. F2 mutant plants were selected, and genomic DNA from the individual plants was analyzed for cosegregation with SSLP and CAPS markers. For the CAPS and SSLP markers used for mapping, refer to http://www.arabidopsis.org/aboutcaps.html. We also developed new SSLP markers from information in the Cereon Arabidopsis polymorphism collection (Jander et al., 2002).
The T-DNA vector, pBI-HI-BSKR, was used, and the 35S-GUS-NOS terminator region (HindIII-SalI restriction fragment) of T-DNA vector pBI-HI-IG (Okamoto et al., 1997) was replaced with the multi-cloning site (PvuII fragment) from pBluescript SK (+) (Stratagene). A genomic DNA fragment containing At1g75100 was amplified from BAC F9E10 (provided from Arabidopsis Biological Resource Center) by PCR with two primers, 5′-AAGCTTTTAGAGAAAGTAGCTGTCAAT-3′ and 5′-GGTACCAGCTTTAAGTATAAGTTAAGA-3′ (underline shows HindIII site and KpnI site, respectively), and was then ligated into the HindIII-KpnI site of the vector (pBI-HI-BSKR-At1g75100). The jac1-1 mutants were transformed with pBI-HI-BSKR or pBI-HI-BSKR-At1g75100 by the floral-dipping method (Clough and Bent, 1998) using the Agrobacterium EHA101 strain. Transformants were selected with 30 mg/L hygromycin. Homozygous plants in the T2 generation were used for experiments.
RT-PCR and RACE-PCR
RNA was extracted from 3-week-old or 7-week-old seedlings. First-strand cDNA was synthesized from total RNA using oligo(dT). The amount of cDNA between each genotypes was normalized using β-tubulin 2 (TUB2) as an internal marker. The genes for JAC1 and TUB2 were amplified from the normalized cDNA by PCR with specific primers. The primers used were as follows: 5′-CTCTGACCTCCGAAAGCTTGC-3′ and 5′-TCACCTTCTTCATCCGCAGTT-3′ for TUB2; and 5′-TGCAGACATTACCAAGCTCAG-3′ and 5′-TCCGAGAGTGTTGAAG-TGGTC-3′ for JAC1. Amplifications were done for 15, 20, 25, or 30 cycles with an annealing temperature of 55°C and an extension time of 2 min for JAC1 and 1 min for TUB2. The PCR products were separated by electrophoresis on an agarose gel. PCR products amplified for 25 cycles were used for Figure 6. The amplified products covering 2 to 1,947 bp of JAC1 and 741 to 1,304 bp of TUB2 (from the start codon) were confirmed by sequence analyses (ABI PRISM 3100, PE Applied Biosystems).
RACE-PCR was performed according to the manufacturer's protocol (Gibco-BRL). For 5′-RACE, first-strand cDNA was synthesized using a gene-specific primer GSP1 (5′-TGGAATCTCTGTTGCTTTTG-3′). Next, cDNA was amplified with GSP2 (5′-ATACAGGCTTCTCGTCTTGC-3′) and reamplified with GSP3 (5′-CGGGACTACGCAACACAGGT-3′) to nest the reactions. For 3′-RACE, cDNA was amplified with GSP1 (5′-ATGCCAAGGAAACTGTAAAC-3′) and nested with GSP2 (5′-AACATTCGGTCTCTTCTATC-3′). The PCR products were subcloned into the pGEM-T Easy Vector (Promega) and sequenced.
Immunoblot Analysis
A C-terminal fragment of JAC1 (residues 301 to 550 without J-domain) was cloned into the BamHI-EcoRI site of the glutathione S-transferase gene fusion vector pGEX2T (Amersham Pharmacia). The fusion protein expressed in Escherichia coli BL21 for 2 h 37°C in the 1 mm isopropyl-β-D-thiogalactoside was purified using glutathione-Sepharose 4B (Amersham Pharmacia) according to the manufacturer's protocol. Polyclonal antisera against this fusion protein raised in a rabbit (Qiagen) were used for immunoblotting. Total proteins were extracted from 3-week-old seedlings with 2× SDS gel-loading buffer (100 mm Tris-HCl, pH 6.8, 10% 2-mercaptethanol, 4% SDS, and 20% glycerol). The protein extract was diluted and the protein concentration quantified using the Bio-Rad Protein Assay according to the manufacturer's protocol. One hundred micro grams of proteins were separated by SDS-PAGE, transferred onto polyvinylidene difluoride membranes (Immobilon P; Millipore), and probed with JAC1-antisera.
Transient Expression of GFP-Fusion Genes
A synthetic GFP (S65T) driven by the CaMV 35S promoter (35S-GFP) was used (Niwa et al., 1999). The JAC1 cDNA fragment covering the interval between the predicted start and stop codons was subcloned into the BsrGI-NotI sites of 35S-GFP (35S-GFP-JAC1). Onion (Allium cepa) epidermal strips or excised Arabidopsis (Arabidopsis thaliana) rosette leaves were placed on 0.8% agar plate containing one-third-strength Murashige and Skoog salt, and the vectors were introduced by gold particle bombardment with a Biolistic Particle Delivery System-100/He (Bio-Rad). Bombardment was done twice for each sample under the following delivery conditions: gold particle diameter, 1.6 μm; helium pressure, 1,550 p.s.i.; target distance, 9 cm; and chamber vacuum pressure, 660 mm Hg. Samples were then incubated in darkness for about 15 to approximately 18 h. GFP fluorescence was observed with an Axioskop and a 50-W halogen lamp and a filter set (BP450-490, FT510, and LP515 for an excitation filter, a dichroic mirror and a barrier filter, respectively; Zeiss). Experiments presented were repeated at least five times independently with the same result.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact the corresponding author.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AB158477.
Supplementary Material
Acknowledgments
We thank Dr. Jane Silverthorne for critical reading of the manuscript; Dr. Kazuhiro Kikuchi, Dr. Masahiro Kasahara, and Dr. Fumio Takahashi for technical advice and helpful discussion; Ms. Mineko Shimizu for assistance with mutant screening; Dr. Takeshi Kanegae for providing pBI-HI-IG binary vector; Dr. Yasuo Niwa for providing 35S-GFP vector; the Arabidopsis Biological Resource Center for providing the BAC clones.
This work was supported by the Japan Society for the Promotion of Science for Young Scientists (research fellowship grant to N.S.); by the Solution Oriented Research for Science and Technology, Japan Science and Technology Corporation (grant to T.K.); and by the Education, Sports, Science and Technology of Japan (grants for Scientific Research on Priority Areas, no. 13139203; on A, no. 13304061; and on S, no. 16107002 to M.W.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.067371.
References
- Babourina O, Newman I, Shabala S (2002) Blue light-induced kinetics of H+ and Ca2+ fluxes in etiolated wild-type and phototropin-mutant Arabidopsis seedlings. Proc Natl Acad Sci USA 99: 2433–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum G, Long JC, Jenkins GI, Trewavas AJ (1999) Stimulation of the blue light phototropic receptor NPH1 causes a transient increase in cytosolic Ca2+. Proc Natl Acad Sci USA 96: 13554–13559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett TJ, Traub LM, Fremont DH (2002) Accessory protein recruitment motifs in clathrin-mediated endocytosis. Structure 10: 797–809 [DOI] [PubMed] [Google Scholar]
- Briggs WR, Christie JM, Salomon M (2001) Phototropins: a new family of flavin-binding blue light receptors in plants. Antioxid Redox Signal 3: 775–788 [DOI] [PubMed] [Google Scholar]
- Christie JM, Reymond P, Powell G, Bernasconi P, Raibekas AA, Liscum E, Briggs WR (1998) Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science 282: 1698–1701 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Gall WE, Higginbotham MA, Chen C-Y, Ingram MF, Cyr DM, Graham TR (2000) The auxilin-like phosphoprotein Swa2p is required for clathrin function in yeast. Curr Biol 10: 1349–1358 [DOI] [PubMed] [Google Scholar]
- Greener T, Grant B, Zhang Y, Wu X, Greene LE, Hirsh D, Eisenberg E (2001) Caenorhabditis elegans auxilin: a J-domain protein essential for clathrin-mediated endocytosis in vivo. Nat Cell Biol 3: 215–219 [DOI] [PubMed] [Google Scholar]
- Harada A, Sakai T, Okada K (2003) phot1 and phot2 mediate blue light-induced transient increases in cytosolic Ca2+ differently in Arabidopsis leaves. Proc Natl Acad Sci USA 100: 8583–8588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala E, Oeller PW, Liscum E, Han I-S, Larsen E, Briggs WR (1997) Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science 278: 2120–2123 [DOI] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo JA, Gabrys H, Capel J, Alonso JM, Ecker JR, Cashmore AR (2001) Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature 410: 952–954 [DOI] [PubMed] [Google Scholar]
- Kagawa T, Kasahara M, Abe T, Yoshida S, Wada M (2004) Function analysis of phototropin2 using fern mutants deficient in blue light-induced chloroplast avoidance movement. Plant Cell Physiol 45: 416–426 [DOI] [PubMed] [Google Scholar]
- Kagawa T, Sakai T, Suetsugu N, Oikawa K, Ishiguro S, Kato T, Tabata S, Okada K, Wada M (2001) Arabidopsis NPL1: a phototropin homolog controlling the chloroplast high-light avoidance response. Science 291: 2138–2141 [DOI] [PubMed] [Google Scholar]
- Kagawa T, Wada M (1996) Phytochrome- and blue light-absorbing pigment-mediated directional movement of chloroplasts in dark-adapted prothallial cells of fern Adiantum as analyzed by microbeam irradiation. Planta 198: 488–493 [Google Scholar]
- Kagawa T, Wada M (1999) Chloroplast avoidance response induced by blue light of high fluence rate in prothallial cells of the fern Adiantum as analyzed microbeam irradiation. Plant Physiol 119: 917–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa T, Wada M (2000) Blue light-induced chloroplast relocation in Arabidopsis thaliana as analyzed by microbeam irradiation. Plant Cell Physiol 41: 84–93 [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, Meagher MB (1999) Actin-organelle interaction: association with chloroplast in Arabidopsis leaf mesophyll cells. Cell Motil Cytoskeleton 44: 110–118 [DOI] [PubMed] [Google Scholar]
- Kasahara M, Kagawa T, Oikawa K, Suetsugu N, Miyao M, Wada M (2002) Chloroplast avoidance movement reduces photodamage in plants. Nature 420: 829–832 [DOI] [PubMed] [Google Scholar]
- Kelley WL (1998) The J-domain family and the recruitment of chaperone power. Trends Biochem Sci 23: 222–227 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, et al (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301: 376–379 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M, Shimazaki K (2001) phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414: 656–660 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Shimazaki K (1999) Blue light activates the plasma membrane H+-ATPase by phosphorylation of the C-terminus in stomatal guard cells. EMBO J 18: 5548–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam BC-H, Sage TL, Bianchi F, Blumwald E (2000) Role of SH3 domain-containing proteins in clathrin-mediated vesicle trafficking in Arabidopsis. Plant Cell 12: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon SK (2001) Clathrin uncoating: auxilin comes to life. Curr Biol 11: R49–R52 [DOI] [PubMed] [Google Scholar]
- Lin C, Shalitin D (2003) Cryptochrome structure and signal transduction. Annu Rev Plant Biol 54: 469–496 [DOI] [PubMed] [Google Scholar]
- Liscum E, Briggs WR (1995) Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell 7: 473–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motchoulski A, Liscum E (1999) Arabidopsis NPH3: a NPH1 photoreceptor-interacting protein essential for phototropsim. Science 286: 961–964 [DOI] [PubMed] [Google Scholar]
- Niwa Y, Hirano T, Yoshimoto K, Shimizu M, Kobayashi H (1999) Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J 18: 455–463 [DOI] [PubMed] [Google Scholar]
- Oikawa K, Kasahara M, Kiyosue T, Kagawa T, Suetsugu N, Takahashi F, Kanegae T, Niwa Y, Kadota A, Wada M (2003) CHLOROPLAST UNUSUAL POSITIONING1 is essential for proper chloroplast positioning. Plant Cell 15: 2805–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H, Sakamoto K, Tomizawa K, Nagatani A, Wada M (1997) Photoresponses of transgenic Arabidopsis overexpressing the fern Adiantum capillus-veneris PHY1. Plant Physiol 115: 79–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DJ, Vallis Y, Noble MEM, Hunter JB, Dafforn TR, Evans PR, McMahon HT (1999) A structural explanation for the binding of multiple ligands by the α-adaptin appendage domain. Cell 97: 805–815 [DOI] [PubMed] [Google Scholar]
- Pishvaee B, Costaguta G, Yeung BG, Ryazantsev S, Greener T, Greene LE, Eisenberg E, McCaffery JM, Payne GS (2000) A yeast DNA J protein required for uncoating of clathrin-coated vesicles in vivo. Nat Cell Biol 2: 958–963 [DOI] [PubMed] [Google Scholar]
- Sakai T, Kagawa T, Kasahara M, Swartz TE, Christie JM, Briggs WR, Wada M, Okada K (2001) Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc Natl Acad Sci USA 98: 6969–6974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Briggs WR (2002) Cellular and subcellular localization of phototropin 1. Plant Cell 14: 1723–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Wada M, Kadota A (2001) External Ca2+ is essential for chloroplast movement induced by mechanical stimulation but not by light stimulation. Plant Physiol 127: 497–504 [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Wada M, Kadota A (2003) Accumulation response of chloroplasts induced by mechanical stimulation in bryophyte cells. Planta 216: 772–777 [DOI] [PubMed] [Google Scholar]
- Schmid SL (1997) Clathrin-coated vesicle formation and protein sorting: an integrated process. Annu Rev Biochem 66: 511–548 [DOI] [PubMed] [Google Scholar]
- Stoelzle S, Kagawa T, Wada M, Hedrich R, Dietrich P (2003) Blue light activates calcium-permeable channels in Arabidopsis mesophyll cells via the phototropin signaling pathway. Proc Natl Acad Sci USA 100: 1456–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tlalka M, Fricker M (1999) The role of calcium in blue-light-dependent chloroplast movement in Lemna trisulca. Plant J 20: 461–473 [DOI] [PubMed] [Google Scholar]
- Traub LM, Downs MA, Westrich JL, Fremont DH (1999) Crystal structure of the alpha appendage of AP-2 reveals a recruitment platform for clathrin-coat assembly. Proc Natl Acad Sci USA 96: 8907–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungewickell E, Ungewickell H, Holstein SEH, Lindner R, Prasad K, Barouch W, Martin B, Greene LE, Eisenberg E (1995) Role of auxilin in uncoating clathrin-coated vesicles. Nature 378: 632–635 [DOI] [PubMed] [Google Scholar]
- Wada M (1988) Chloroplast photorelocation in enucleatd fern protonemata. Plant Cell Physiol 29: 1227–1232 [Google Scholar]
- Wada M, Grolig F, Haupt W (1993) Light-oriented chloroplast positioning: contribution to progress in photobiology. J Photochem Photobiol B Biol 17: 3–25 [Google Scholar]
- Wada M, Kagawa T (2001) Light-controlled chloroplast movement. In DP Häder, M Lebert, eds, Photomovement. Elsevier, Amsterdam, pp 897–924
- Wada M, Kagawa T, Sato Y (2003) Chloroplast movement. Annu Rev Plant Biol 54: 455–468 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.