Abstract
Previously, we reported that the TAPETUM DETERMINANT1 (TPD1) gene is required for specialization of tapetal cells in the Arabidopsis (Arabidopsis thaliana) anther. The tpd1 mutant is phenotypically identical to the excess microsporocytes1 (ems1)/extra sporogenous cells (exs) mutant. The TPD1 and EMS1/EXS genes may function in the same developmental pathway in the Arabidopsis anther. Here, we further report that overexpression of TPD1 alters the cell fates in the Arabidopsis carpel and tapetum. When TPD1 was expressed ectopically in the wild-type Arabidopsis carpel, the number of cells in the carpel increased significantly, showing that the ectopic expression of TPD1 protein could activate the cell division in the carpel. Furthermore, the genetic analysis showed that the activation of cell division in the transgenic carpel by TPD1 was dependent on EMS1/EXS, as it did not happen in the ems1/exs mutant. This result further suggests that TPD1 regulates cell fates in coordination with EMS1/EXS. Moreover, overexpression of TPD1 in tapetal cells also delayed the degeneration of tapetum. The TPD1 may function not only in the specialization of tapetal cells but also in the maintenance of tapetal cell fate.
Cell division and differentiation are essential for the development of multicellular organisms. In recent years, studies have shown that cell signaling is important for the regulation of cell division and differentiation. Several genes that take roles in the cell signaling have been identified regulating the cell fates in plants. For example, the CLAVATA gene family controls the cell division and cell differentiation via regulating the interaction between different cell layers in the shoot apical meristem (Clark et al., 1996, 1997; Brand et al., 2000). ERECTA plays a role in regulating the cell growth process originating from the shoot apical meristem (Torii et al., 1996). The HAESA gene is involved in the differentiation of the abscission zone of floral organs (Jinn et al., 2000). SPOROCYTELESS/NOZZLE is required for sporogenous cell differentiation in the formation of anther primordium at an early stage of anther development (Schiefthaler et al., 1999; Yang et al., 1999; Balasubramanian and Schneitz, 2000). Although more and more genes have been identified as being involved in the regulation of cell fates in plants, little is known about the mechanism of cell fate regulation.
In the Arabidopsis (Arabidopsis thaliana) anther, the reproductive microsporocytes and somatic tapetal cells originate from the same precursor, the archesporial cells. The hypodermal archesporial cells in the anther primordium undergo a periclinal division to give rise to outer primary parietal cells and inner primary sporogenous cells. The inner primary sporogenous cells differentiate into microsporocytes, and the outer parietal cells undergo another periclinal division to give rise to two sets of secondary parietal cells. The inner secondary parietal cells differentiate into tapetal cells, while the outer secondary parietal cells further divide and develop into middle layer and endothecium cells (Sanders et al., 1999). Genetic studies have revealed that TAPETUM DETERMINANT1 (TPD1) and EXCESS MICROSPOROCYTES1 (EMS1)/EXTRA SPOROGENOUS CELLS (EXS) are required for the specialization of tapetal cells (Zhao et al., 2002; Yang et al., 2003). In the absence of TPD1 or EMS1/EXS, the inner secondary parietal cells develop into microsporocytes instead of tapetal cells. This finding indicates that the regulation pathway defined by TPD1 and EMS1/EXS plays a decisive role in the specialization of tapetal cells.
The characterization of the tpd1 mutant showed that TPD1 and EMS1/EXS may function in the same developmental pathway, as the tpd1 mutant was phenotypically identical to the ems1/exs mutant (Canales et al., 2002; Zhao et al., 2002; Yang et al., 2003). The TPD1 gene encodes a small protein of 176 amino acids (Yang et al., 2003). The EMS1/EXS encodes a putative receptor protein kinase and may define a signaling pathway that regulates the cell differentiation and development (Canales et al., 2002; Zhao et al., 2002). EMS1/EXS is expressed mostly in the developing tapetum in anthers. TPD1 is expressed mostly in the developing microsporocytes in anthers at stage 5. Comparing the expression of TPD1 to that of EMS1/EXS revealed that they have a complementary expression pattern in anthers at stage 5, the stage when the tpd1 and ems1/exs mutant phenotypes in the anther were observed. This implies that the TPD1 and EMS1/EXS genes may control the tapetal fate via signaling interaction between sporogenous cells and tapetal cells.
To further understand the function of TPD1, we overexpressed the TPD1 in wild-type plants under the control of a constitutive cauliflower mosaic virus 35S promoter (Rogers et al., 1987). Characterization of the transgenic plants showed that ectopic expression of TPD1 activated the cell division in the transgenic carpel and delayed the degeneration of tapetum. Genetic analysis further revealed that the activation of cell division by 35S::TPD1 in carpel is dependent on the normal EMS1/EXS function. This study further demonstrates that the TPD1 product regulates the cell fates via genetic interaction with the EMS1/EXS receptor protein kinase and is involved in the regulation of tapetal cell fate.
RESULTS
Ectopic Expression of TPD1 Altered the Patterning of the Arabidopsis Siliques
The TPD1 mRNA was overexpressed in wild-type plants under the control of a constitutive cauliflower mosaic virus 35S promoter (Rogers et al., 1987). In a screen for transgenic plants, we obtained a total of 51 T1 transformants. Six plants were fully sterile, and the rest (45 plants) produced wider siliques with variable sizes compared to nontransformed plants. We then selected one fully fertile transgenic plant that produced the widest siliques for further characterization (Fig. 1A). We measured the width of siliques from the homozygous 35S::TPD1 transgenic plants in T3 generation. The average silique width was 1.91 ± 0.14 mm compared to 1.11 ± 0.08 mm of nontransformed silique (Fig. 1B). This result showed that the width of the transgenic silique was 172.2% of nontransgenic wild-type silique.
Figure 1.
Ectopic expression of TPD1 altered the pattern of siliques. A, A transgenic plant with wider siliques caused by ectopic expression of 35S::TPD1 in wild type. B, A wild-type plant. C, A transgenic plant with wider sterile siliques caused by expression of 35S::TPD1 in the tpd1 mutant. D, A tpd1 mutant plant. E, A tpd1 mutant silique. F, A transgenic sterile silique with a wider patterning, showing that ectopic expression of 35S::TPD1 altered the patterning of silique in the tpd1 mutant. G, A sterile ems1/exs-2 silique. H, A transgenic sterile silique with normal patterning, showing that the overexpression of 35S::TPD1 in ems1/exs-2 mutant did not alter the patterning of the silique. Bar = 1 cm in A to D; bar = 1 mm in E to H.
To further confirm the relationship between the wider silique and the ectopic expression of TPD1, the transgenic TPD1 gene was introduced into the tpd1 mutant background by genetic cross. In the selfing F2 population, fertile and sterile plants segregated as 3:1. The ectopic expression of TPD1 under the control of the 35S promoter cannot recover the fertility of the tpd1 mutant plant because the 35S promoter does not function in early sporogenous tissue (Jenik and Irish, 2000). As expected, the sterile transgenic tpd1 mutant plants also produced wider siliques. In the selfing F3 population, the average width of the homozygous 35S::TPD1 transgenic tpd1 siliques was 1.10 ± 0.11 mm (Fig. 1, C and F) compared to 0.72 ± 0.06 mm of nontransgenic tpd1 mutant silique (Fig. 1, D and E). This measurement showed that the width of the transgenic tpd1 silique was 152.8% of nontransgenic tpd1 silique. In the same selfing F3 population, the average width of the homozygous 35S::TPD1 transgenic fertile siliques (heterozygous TPD1/tpd1 background) was 1.90 ± 0.15 mm, an increase of 71.1%, compared to nontransgenic wild-type silique. All these results showed that the ectopic expression of TPD1 altered the patterning of the carpel.
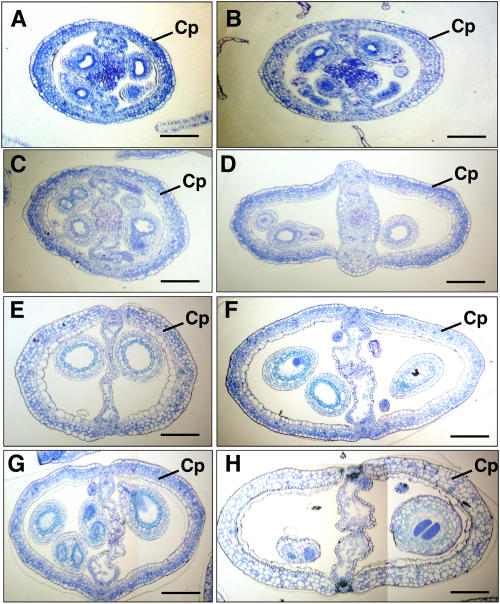
To investigate the cell mechanism of the alternation of transgenic silique, we compared the numbers of epidermal cells of the transgenic and wild-type siliques (Fig. 2). The wild-type and transgenic siliques were collected based on four time points: anthesis, and 2, 5, and 10 d after pollination. Forty middle transversal sections from each sample were counted for the average number of epidermal cells. In the transgenic siliques, the average numbers of epidermal cells per silique section at different stages were 169.2 ± 14.2, 205.5 ± 17.5, 236.8 ± 20.1, and 249.9 ± 20.1, respectively. In wild-type nontransgenic siliques, the average numbers of epidermal cells per section at different stages were 142.2 ± 10.4, 160.9 ± 11.9, 163.8 ± 10.5, 164.3 ± 12.8, respectively. These results show that the width increase of the transgenic silique results from the increase in the number of cells. These results also show that the number of cells increases continuously during the whole carpel developmental period compared to wild-type carpel, which has a relatively constant cell number after anthesis. These findings suggest that the ectopic expression of TPD1 activates the cell division in the transgenic carpel.
Figure 2.
Comparison of the wider transgenic siliques to the wild-type siliques. The transversal sections show that the wider transgenic siliques have more cells than the wild-type siliques. A, There are 142.2 ± 10.4 epidermal cells in a transversal section of a wild-type silique at anthesis stage. B, There are 169.2 ± 14.2 epidermal cells in a transversal section of a wider transgenic silique at anthesis stage. C, There are 160.9 ± 11.9 epidermal cells in a transversal section of a wild-type silique at a stage 2 d after anthesis. D, There are 205.5 ± 17.5 epidermal cells in a transversal section of a wider transgenic silique at a stage 2 d after anthesis. E, There are 163.8 ± 10.5 epidermal cells in a transversal section of a wild-type silique at a stage 5 d after anthesis. F, There are 236.8 ± 20.1 epidermal cells in a transversal section of a wider transgenic silique at a stage 5 d after anthesis. G, There are 164.3 ± 12.8 epidermal cells in a transversal section of a wild-type silique at a stage 10 d after anthesis. H, There are 249.9 ± 20.1 epidermal cells in a transversal section of a wider transgenic silique at a stage 10 d after anthesis. Cp, Carpel. Bars = 25 μm.
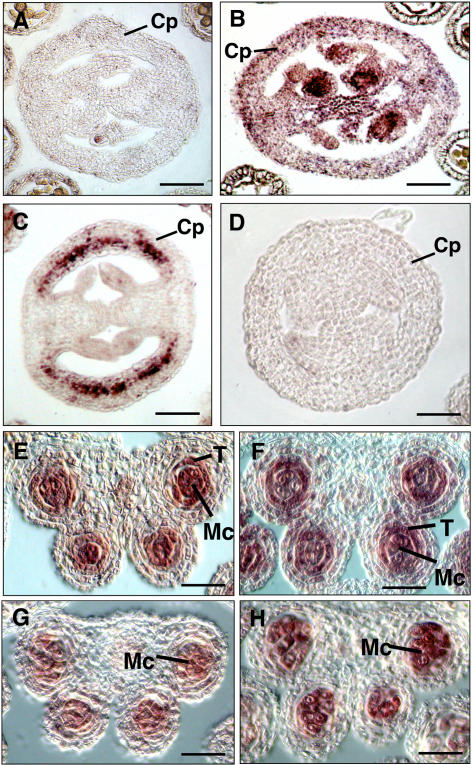
Studies have suggested that the action of the 35S promoter is not uniform in the flower and may be variable (Jefferson et al., 1987; Jack et al., 1994; Bossinger and Smyth, 1996; Wilkinson et al., 1997; Sieburth et al., 1998; Jenik and Irish, 2000). Therefore, we performed RNA in situ hybridization to investigate whether the ectopic expression of TPD1 was consistent with the developmental pattern of the transgenic carpel. RNA in situ hybridization showed that the TPD1 mRNA signal was found much more strongly in transgenic carpel (Fig. 3C) compared to that in nontransgenic wild-type carpel (Fig. 3A). These results indicate that the ectopic expression of TPD1 is consistent with the wider-carpel phenotype. RNA in situ hybridization also showed that the expression of 35S::TPD overlapped with that of EMS1/EXS in carpel (Fig. 3B).
Figure 3.
Expression patterns of TPD1 in the siliques and anthers of wild-type, tpd1, ems1/exs-2, and transgenic plants. A, A transversal section of wild-type silique probed with the TPD1 antisense RNA. B, A transversal section of wild-type silique probed with the EMS1/EXS antisense RNA. C, A transversal section of the transgenic silique probed with the TPD1 antisense RNA. D, A transversal section of wild-type silique probed with the TPD1 sense RNA as a negative control. E, A wild-type anther section probed with the TPD1 antisense RNA. F, A wild-type anther section probed with the EMS1/EXS antisense RNA. G, An ems1 anther section probed with the TPD1 antisense RNA. H, A tpd1 anther section probed with the EMS1/EXS antisense RNA. Cp, Carpel; T, tapetum; Mc, microsporocyte. Bars = 20 μm.
The Activation of Cell Division in Carpel by TPD1 Is Dependent on the Normal EMS1/EXS Function
To investigate whether the activation of cell division by TPD1 in the carpel is dependent on the normal EMS1/EXS function, we introduced the transgenic TPD1 gene into the ems1/exs-2 mutant, a new ems1/exs allele (Yang et al., 2003), by genetic cross. In the selfing F3 population, the average width of the homozygous 35S::TPD1 transgenic siliques on the heterozygous ems1/exs-2 plant was 1.36 ± 0.10 mm, an increase of 22.8% compared to that of nontransgenic wild-type silique. This rate was much lower than those of transgenic siliques from the wild-type plant (72.2%) and heterozygous tpd1 plant (71.1%). This result shows that the width of transgenic siliques might be related to the expression level of EMS1/EXS. In the same selfing F3 population, the average width of the homozygous transgenic siliques from the ems1/exs-2 mutant plant was 0.70 ± 0.07 mm (Fig. 1H), while average width of nontransgenic ems1/exs-2 siliques was similar and counted as 0.71 ± 0.05 mm (Fig. 1G). This result indicates that the ectopic expression of TPD1 has no effect on the silique width in the ems1/exs mutant. In addition, introduction of a copy of EMS1/EXS cDNA under the control of the EMS1/EXS promoter into the ems1/exs-2 plant containing the 35S::TPD1 gene recovered the phenotype of wider carpel (data not shown). It is likely that TPD1 requires the normal function of EMS1/EXS to activate the cell division in the carpel. In other words, TPD1 regulates the cell fate in the carpel via coordination with EMS1/EXS.
The Interaction between TPD1 and EMS1 Is Not at Transcription Level
To test whether the TPD1 expression is up- or down-regulated by EMS1/EXS, we investigated the expression of TPD1 in the ems1/exs-2 mutant by RNA in situ hybridization. As shown in Figure 3G, the TPD1 RNA signal in the ems1/exs-2 anther was as strong as that in the wild-type anther (Fig. 3E). The expression of TPD1 is independent from the EMS1/EXS. Furthermore, we also studied the expression of EMS1/EXS in the tpd1 mutant. The result showed that the EMS1/EXS RNA signal in the tpd1 microsporocytes (Fig. 3H) was also found to be as strong as that in wild-type microsporocytes (Fig. 3F). Therefore, we conclude that the interaction between TPD1 and EMS1/EXS is not at a transcriptional level.
Overexpression of TPD1 Delayed the Degeneration of Tapetum
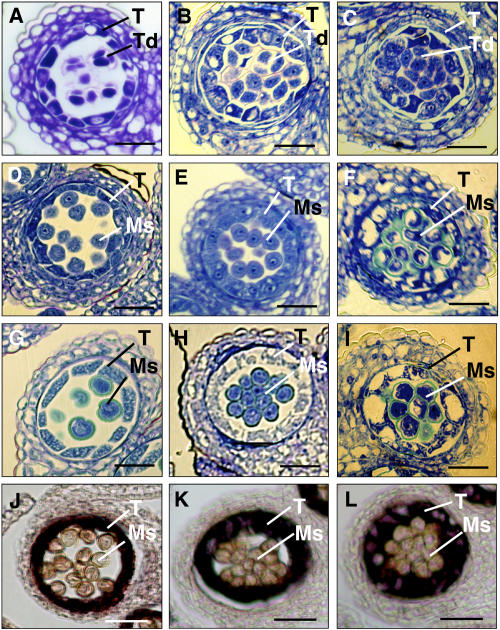
To further investigate whether overexpression of TPD1 affected development of tapetum, we examined the developmental pattern of the tapetum in transgenic plants. Among the 51 transgenic plants described previously, the six sterile transgenic plants produced enlarged tapetal cells. The tapetum was not separated from microspores (Fig. 4F) and did not degenerate when anther matured (Fig. 4I). Furthermore, the microspores were not released from the tetrad (Fig. 4F). Since the plants were male sterile, it is likely that delay of degeneration of tapetum may cause the defect of pollen development. In the other fertile transgenic plants with wider silique, the degeneration of tapetum also was delayed significantly (Fig. 4, B, E, and H) compared to nontransgenic plants (Fig. 4, A, D, and G). This result suggests that TPD1 is involved not only in the specification of tapetal cells but also in the maintenance of tapetum development. In addition, the pollen grains in the transgenic plants were stickier than those of wild type, as they still stacked together when the anther matured (Fig. 4H). However, they had been released from tetrad (Fig. 4E) and could be separated manually. The surface feature of pollen grain had likely been altered. Possibly, the delay of tapetum degeneration resulted in the pollen coat lacking the final components normally from the degenerating tapetum. The pollen still was fertile; therefore, the components might not be critical for pollen fertility.
Figure 4.
Overexpression of TPD1 delayed the degeneration of transgenic tapetal cells. A to C, Transversal sections of anthers at stage 7, showing that tetrads were formed in the wild-type (A), fertile transgenic (B), and sterile transgenic (C) anthers. D to F, The transversal sections of anthers at stage 9. D, The tapetal cells started to degenerate and microspores were separated in wild-type anther. E, The degeneration of tapetal cells was delayed and microspores were still separated in the fertile transgenic anther. F, The degeneration of tapetal cells was greatly delayed and microspores were not separated in the sterile transgenic anther. G to I, The transversal sections of anthers at stage 11. G, The tapetal cells were almost degenerated and the pollen grains were formed in wild-type anther. H, The tapetal cells were degenerated apparently more slowly in the fertile transgenic anther than in the wild-type ones. The pollen grains were formed but stacked together. I, The degeneration of tapetal cells was greatly delayed and pollen grains were not separated in the sterile transgenic anther. J to L, Overexpression of TPD1 enhanced the expression of a tapetum-specific gene, ATA7, in the transgenic tapetal cells at stage 9, showing that the ATA7 was expressed more strongly in sterile (L) and fertile (K) transgenic anthers than in wild-type anther (J). Ms, Microspores; Pg, pollen grains; T, tapetum; Td, tetrads. Bars = 20 μm.
To study the mechanism of tapetum overgrowth resulting from overexpression of TPD1, we performed RNA in situ hybridization to investigate whether overexpression of TPD1 affected the expression of tapetum-specific gene ATA7 (Rubinelli et al., 1998). As shown in Figure 4, the ATA7 RNA signal was detected more strongly in the transgenic anther than in the nontransgenic anther during anther development (Fig. 4, J, K, and L). This result indicates that overexpression of TPD1 enhances the expression of the tapetum-specific gene ATA7 in the tapetal cells. The result also shows that overexpression of TPD1 influences the development of tapetal cells, possibly via regulating the expression of tapetum-specific genes. However, it is still not clear whether TPD1 directly regulates the expression of ATA7. Increase in ATA7 expression level also might result from the overgrowth of tapetal cells.
DISCUSSION
TPD1 Regulates the Cell Fate by Genetic Interaction with EMS1/EXS
In this study, we further demonstrate that TPD1 regulates the cell fate by the interaction with EMS1/EXS. We overexpressed the TPD1 mRNA in wild-type plants under the control of the 35S promoter. Analysis of the transgenic plants overexpressing TPD1 showed that ectopic expression of TPD1 altered the development pattern of carpels. In nontransgenic wild-type carpel, the number of cells remained constant after anthesis. No cell division was observed in wild-type carpel after anthesis until the silique matured. In the transgenic carpel, the number of cells increased continuously until the silique matured. This result shows that the ectopic expression of TPD1 activates the division of the cells that are normally silent in the wild-type silique. RNA in situ hybridization showed that the TPD1 mRNA signal was found much more strongly in transgenic carpel (Fig. 3C) compared to that in nontransgenic wild-type carpel (Fig. 3A). The ectopic expression of TPD1 is consistent with the wider-carpel phenotype. RNA in situ hybridization also showed that the expression of 35S::TPD overlapped with that of EMS1/EXS in carpel (Fig. 3B). At the same time, the gained wider-silique phenotype was suppressed in the ems1/exs-2 mutant and could be recovered by expression of EMS1/EXS. The result indicates that the TPD1 function is dependent on the EMS1/EXS function.
The previous studies showed that the expression patterns of TPD1 and EMS1/EXS in anthers overlapped with each other at early stages (Yang et al., 2003). However, no morphological defect was observed at the early stages (stage 1–3) in both tpd1 and ems1 null mutants. This finding suggests that TPD1 and EMS1/EXS are not required at the early stages of anther development. At least, the interaction between TPD1 and EMS1/EXS is not required for the formation of tapetal precursor cells.
The TPD1 is expressed in leaves, young seedlings, flower buds, and anthers (Yang et al., 2003). Similarly, EXS is also expressed in a wide range of tissues, including young buds, open flowers, siliques, and seeds (Canales et al., 2002). The further detailed analysis of the TPD1 and EMS1/EXS promoters showed that the expression of TPD1 actually does not completely overlap with EMS1/EXS in some tissues. For example, in silique, the EMS1/EXS is expressed in the developing carpel after fertilization, but the TPD1 is not expressed in the developing carpel (S.-L. Yang and D. Ye, unpublished data). Moreover, the EMS1/EXS is not required for the carpel development since the ems1 mutant has normal carpels. It is possible that EMS1/EXS does not function in the carpel development due to the lack of TPD1 expression.
TPD1 Regulates Tapetum Development
Our result showed that overexpression of TPD1 in wild-type plants delayed the degeneration of tapetum and enhanced the expression of the tapetum-specific gene ATA7 in the tapetal cells. This suggests that the TPD1 is required not only for the specialization of the tapetal cells but also for the persistence of tapetal cell fate.
In conclusion, this study further demonstrates that TPD1 regulates the cell fate via genetic interaction with EMS1/EXS. The ectopic expression of TPD1 showed that carpel expresses the downstream genes of TPD1 and EMS1/EXS out of the anther, providing a new way for further isolation of more genes involved in the TPD1 pathway.
MATERIALS AND METHODS
Plant Materials
All Arabidopsis (Arabidopsis thaliana) plants used in this study were in the Landsberg erecta background. The seeds were pregerminated on Murashige and Skoog salt agar plates with or without 50 μg mL−1 kanamycin or 25 μg mL−1 hygromycin at 22°C under a light cycle of 16 h light/8 h dark. The plants were grown in soil at 22°C under the same light cycle for germination. The tpd1-1 and ems1/exs-2 mutants were as described previously (Yang et al., 2003).
Overexpression of TPD1 in Plants
The full-length TPD1 cDNA (Yang et al., 2003) was released from pGEM-T vector and subcloned into the pCAMBIA1300 Ti-derived binary vector (CAMBIA; www.cambia.org.au) under the control of a constitutive cauliflower mosaic virus 35S promoter (Rogers et al., 1987). Then, the derived construct was introduced into Landsberg erecta by Agrobacterium-mediated infiltration. The transformants were selected using 25 μg mL−1 hygromycin. Forty-five fertile T1 plants were obtained in a screen. In T2 generation, 31 out of 45 T2 selfing families showed the segregation of three resistant:one sensitive for hygromycin. Ten seedlings from each such family were planted into soil to further produce selfing seeds. One family in which all plants produced fully fertile and wide siliques was selected for further analysis. In T3 generation, four out of the 10 plants in this family produced the seeds that all gave rise to hygromycin-resistant seedlings; the other six plants produced seedlings that segregated in three resistant:one sensitive for hygromycin. This result showed that this selected line was a single T-DNA insertion line and the four plants were homozygous 35:TPD1 transformants. The homozygous plants from this line were selected for further phenotypic and genetic characterization.
Plant Transformation
All plant transformation experiments were performed as described by Yang et al. (2003).
Phenotypic Characterization
For sectioning, the wild-type and mutant flowers at different stages were fixed overnight in FAA (50% [v/v] ethanol, 5% [v/v] acetic acid [v/v], and 3.7% [v/v] formaldehyde in water), dehydrated through an ethanol series, embedded into Historesin (Leica), and sectioned with a Leica microtome. Serial sections (2–3 μm thick) were mounted on slides and stained with 0.25% (w/v) toluidine blue O. The slides were observed under a Leica DMRB microscope and photographed with a Nikon digital camera (COOLPIX995). The selected images were compiled using the Adobe Photoshop and Microsoft PowerPoint programs.
In Situ Hybridization
In situ hybridization was performed as described previously (Yang et al., 2003). The TPD1, EMS1-specific, and ATA7 RNA probes used for in situ hybridization were also prepared as described previously (Yang et al., 2003).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AY394846 (a 935-nucleotide cloned TPD1 cDNA).
This work was supported by research grants from the National Science and Technology Board of Singapore and the Agency for Science, Technology and Research of Singapore.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.063529.
References
- Balasubramanian S, Schneitz K (2000) NOZZLE regulates proximal-distal pattern formation, cell proliferation and early sporogenesis during ovule development in Arabidopsis thaliana. Development 127: 4227–4238 [DOI] [PubMed] [Google Scholar]
- Bossinger G, Smyth D (1996) Initiation patterns of flower and floral organ development in Arabidopsis thaliana. Development 122: 1093–1102 [DOI] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Canales C, Bhatt AM, Scott R, Dickkinson H (2002) EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr Biol 12: 1718–1727 [DOI] [PubMed] [Google Scholar]
- Clark SE, Jacobsen SE, Levin JZ, Meyerowitz EM (1996) The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 122: 1567–1575 [DOI] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585 [DOI] [PubMed] [Google Scholar]
- Jack T, Brockman L, Meyerowitz EM (1994) Arabidopsis homeotic gene APETALA3 ectopic expression: transcriptional and posttranscriptional regulation determine floral organ identity. Cell 76: 703–716 [DOI] [PubMed] [Google Scholar]
- Jefferson R, Kavanagh T, Bevan M (1987) GUS fusions: glucuronidase as a sensitive and versatile gene fusion marker. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenik PD, Irish VF (2000) Regulation of cell proliferation patterns by homeotic genes during Arabidopsis floral development. Development 127: 1267–1276 [DOI] [PubMed] [Google Scholar]
- Jinn TL, Stone JM, Walker JC (2000) HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev 14: 108–117 [PMC free article] [PubMed] [Google Scholar]
- Rogers SG, Kee HJ, Horsch RB, Fraley RT (1987) Improved vectors for plant transformation: expression cassette vectors and new selectable markers. Methods Enzymol 153: 253–277 [Google Scholar]
- Rubinelli P, Hu Y, Ma H (1998) Identification, sequence analysis and expression studies of novel anther-specific genes of Arabidopsis thaliana. Plant Mol Biol 37: 607–619 [DOI] [PubMed] [Google Scholar]
- Sanders PM, Anhthu QB, Weterings K, Mclntire KN, Hsu Y, Lee PY, Trong MT, Beals TP, Goldberg RB (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11: 297–322 [Google Scholar]
- Schiefthaler U, Balasubramanian S, Sieber P, Chevalier D, Wisman E, Schneitz K (1999) Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 11664–11669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth L, Drews G, Meyerowitz EM (1998) Non-autonomy of AGAMOUS function in flower development: use of a Cre/loxP method for mosaic analysis in Arabidopsis. Development 125: 4303–4312 [DOI] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y (1996) The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson J, Twell D, Lindsey K (1997) Activities of CaMV 35S and nos promoters in pollen: implications for field release of transgenic plants. J Exp Bot 48: 265–275 [Google Scholar]
- Yang SL, Xie LF, Mao HZ, Puah CS, Yang WC, Jiang L, Sundaresan V, Ye D (2003) TAPETUM DETERMINANT 1 is required for cell specialization in the Arabidopsis anther. Plant Cell 15: 2792–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WC, Ye D, Xu J, Sundaresan V (1999) The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev 13: 2108–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao DZ, Wang GF, Speal B, Ma H (2002) The EXCESS MICROSPOROCYTE1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev 16: 2021–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]