Abstract
A gibberellin (GA) biosynthetic pathway was discovered operating in root tips of 7-d-old pumpkin (Cucurbita maxima) seedlings. Stepwise analysis of GA metabolism in cell-free systems revealed the conversion of GA12-aldehyde to bioactive GA4 and inactive GA34. Highest levels of endogenous GA4 and GA34 were found in hypocotyls and root tips of 3-d-old seedlings. cDNA molecules encoding two GA oxidases, CmGA20ox3 and CmGA3ox3, were isolated from root tips of 7-d-old LAB150978-treated seedlings. Recombinant CmGA20ox3 fusion protein converted GA12 to GA9, GA24 to GA9, GA14 to GA4, and, less efficiently, GA53 to GA20, and recombinant CmGA3ox3 protein oxidized GA9 to GA4. Transcript profiles were determined for four GA oxidase genes from pumpkin revealing relatively high transcript levels for CmGA7ox in shoot tips and cotyledons, for CmGA20ox3 in shoot tips and hypocotyls, and for CmGA3ox3 in hypocotyls and roots of 3-d-old seedlings. Transcripts of CmGA2ox1 were mainly found in roots of 7-d-old seedlings. In roots of 7-d-old seedlings, transcripts of CmGA7ox, CmGA20ox3, and CmGA3ox3 were localized in the cap and the rhizodermis by in situ hybridization. We conclude that hypocotyls and root tips are important sites of GA biosynthesis in the developing pumpkin seedling.
Gibberellins (GAs) are signaling molecules that regulate and integrate developmental processes during the entire life cycle of higher plants, including shoot elongation and root development (Richards et al., 2001; Olszewski et al., 2002; Fu and Harberd, 2003; Reid et al., 2004; Sun, 2004).
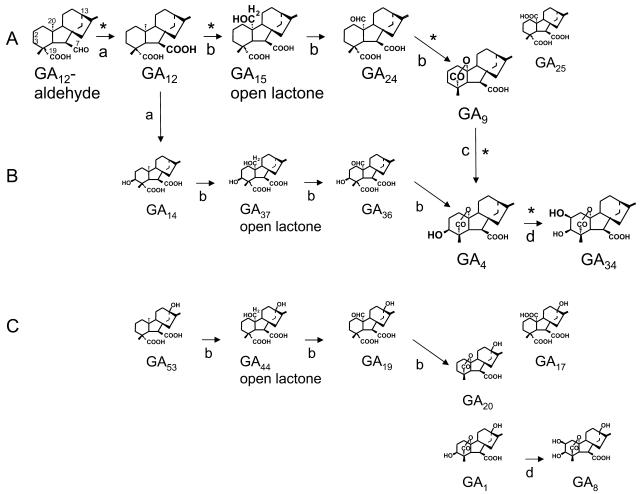
GA biosynthetic pathways are of considerable complexity (for review, see Hedden and Kamiya, 1997; Sponsel and Hedden, 2004). They are divided into nonhydroxylated, 3β-hydroxylated, and 13-hydroxylated pathways (Fig. 1, A–C, respectively; Graebe, 1987). A principal pathway to GA plant hormones can be drawn from GA12-aldehyde. First, 7-oxidation of GA12-aldehyde results in the formation of GA12. GA53 is often formed by 13-hydroxylation of GA12. The following three oxidation steps at carbon (C)-20 are catalyzed by one enzyme, the GA 20-oxidase, which, in general, leads to the formation of a C19-GA (e.g. GA9 and GA20; Fig. 1). Alternatively, C-20 oxidation leads to the formation of a carboxylic acid (e.g. GA25 and GA17). The resulting C20-GAs usually are minor products of GA 20-oxidase activity. Finally, 3-oxidation produces GA plant hormones (GA4 and GA1), which are subsequently inactivated by 2-oxidation (GA34 and GA8; Fig. 1). In many plant species, 7-oxidation and 13-hydroxylation are catalyzed by NADPH-dependent cytochrome P450 mono-oxygenases. In Arabidopsis (Arabidopsis thaliana), a multifunctional ent-kaurenoic acid oxidase with 7-oxidation catalytic properties has been characterized (Helliwell et al., 2001). In pumpkin (Cucurbita maxima), however, 7-oxidation is catalyzed by an additional multifunctional GA 7-oxidase that belongs to the class of 2-oxoglutarate-dependent dioxygenases (Lange, 1997). Recently, recombinant pumpkin GA 7-oxidase has been shown to catalyze 3-oxidation of GA12 (Frisse et al., 2003), which might initiate an early 3β-hydroxylated pathway via GA14 to GA4 (Fig. 1).
Figure 1.
GA biosynthetic pathways from GA12-aldehyde in pumpkin seedlings: nonhydroxylated pathway (A), 3β-hydroxylated pathway (B), and 13-hydroxylated pathway (C; Graebe, 1987). GA biosynthetic steps identified in cell-free systems are labeled with asterisks. Reactions are catalyzed by recombinant GA7ox (a), GA20ox2 (b), GA3ox3 (c), and GA2ox1 (d; Frisse et al., 2003). Structures and metabolic relationships are discussed in the text.
Most of the genes coding for enzymes of the GA biosynthetic pathway have been isolated and characterized and our understanding is increasing about their tissue-cell specificity during plant development (for review, see Sponsel and Hedden, 2004). Genes encoding GA oxidases from pumpkin are summarized in Table I. Localization of the transcripts of GA biosynthetic genes indicates complex tissue- and developmental-specific expression patterns in many species (Yamaguchi et al., 2001). Endogenous GAs have frequently been identified in shoots and occasionally in roots of higher plants (for review, see MacMillan, 2002). In seedlings, GA oxidase genes are mainly expressed in rapidly developing tissues, including shoot and root tips (for review, see Olszewski et al., 2002). Recently, a growing body of evidence supports the hypothesis that GAs are produced close to or at their site of action (for review, see Sponsel and Hedden, 2004), which might still include unknown transport mechanisms (Fleet and Sun, 2005). There is some indication that GAs derived from cotyledons play a role in cell division during tissue reunion in the cortex of hypocotyls, as has been shown for cucumber and tomato (Asahina et al., 2002). It has been shown that GAs promote root growth in Arabidopsis by inducing the degradation of the DELLA proteins RGA (repressor of gal-3) and GAI (GA insensitive) and that other hormones are involved in regulating this action (Achard et al., 2003; Fu and Harberd, 2003; Fleet and Sun, 2005). However, direct evidence for GA biosynthesis occurring in roots is missing.
Table I.
Pumpkin GA oxidases, coding genes, expression sites, and accession numbers
| GA Oxidases | Coding Gene | Previously Described as | Expression Sites | Accession Nos. |
|---|---|---|---|---|
| 7-Oxidase | CmGA7ox | GA 7-oxidase, Cm 7-ox | Developing seed, seedling | U61386 |
| 20-Oxidases | CmGA20ox1 | GA 20-oxidase, Cm 20-ox | Embryo of developing seed | X73314 |
| CmGA20ox2 | GA 20-oxidase, Cm 20-ox | Endosperm of developing seed | U61385 | |
| CmGA20ox3 | Cm 20-ox-RT | Developing seed, seedling | AJ308480 | |
| 3-Oxidases | CmGA3ox1 | GA 2β-3β-hydroxylase; Cm 2, 3-ox | Endosperm of developing seed | U63650 |
| CmGA3ox2 | Cm 3-ox; GA 3-oxidase | Embryo of developing seed | AJ006453 | |
| CmGA3ox3 | Cm 3-ox-RT | Seedling | AJ302040 | |
| 2-Oxidases | CmGA2ox1 | Cm 2-ox, GA 2-oxidase | Developing seed, seedling | AJ302041, AJ315663 |
| CmGA2ox2 | Truncated GA 2-oxidase | Seedling | AJ315662 |
To address this question and investigate potential sites of GA biosynthesis in pumpkin seedlings, endogenous GA levels were analyzed and a GA biosynthetic pathway in root tips of 7-d-old pumpkin seedlings was identified. Characteristic genes of this pathway were isolated, including those coding for GA 20-oxidase (CmGA20ox3) and GA 3-oxidase (CmGA3ox3; Table I). GA oxidase transcripts were quantified in different parts of developing pumpkin seedlings by competitive reverse transcription (RT)-PCR and localized in roots and, more specifically, in root tips by in situ hybridization.
RESULTS
Endogenous GAs of Developing Pumpkin Seedlings
In preliminary experiments, seedlings were treated for 7 d with inhibitor of GA biosynthesis 1-(4-trifluormethyl)-2-(1,2,4-triazolyl-(1))-3-(5-methyl-1,3-dioxan-5-yl)-propen-3-ol (LAB150978), with GA4, and with both LAB150978 and GA4 (Table II). LAB150978 serves as an inhibitor of ent-kaurene oxidase, an enzyme that catalyzes early steps of the GA biosynthetic pathway (Jung et al., 1986). Primary root elongation was inhibited significantly in the presence of the GA inhibitor, while GA4 or GA4 plus LAB150978 treatment had no significant effect. In contrast, radial growth of primary root tips was elevated in LAB150978-treated plants, while GA4 treatment resulted in significantly thinner root tips. Our results, summarized in Table II, indicate that GAs are important for normal hypocotyl and root growth of pumpkin seedlings.
Table II.
Effect of LAB150978 (10−6m), of GA4 (10−6m), and of LAB150978 plus GA4 (both 10−6m) treatment on hypocotyl and root growth of 7-d-old pumpkin seedlings
Data are shown as mean ± se (n=10).
| Treatment | Hypocotyl Length | Primary Root Length | φ of Primary Root Tip a |
|---|---|---|---|
| cm | cm | mm | |
| Control | 2.9 ± 0.3 | 15.6 ± 1.7 | 0.70 ± 0.07 |
| LAB150978 | 1.7 ± 0.2b | 11.1 ± 2.0b | 0.91 ± 0.05b |
| GA4 | 5.4 ± 0.3b | 15.7 ± 2.1 | 0.57 ± 0.04b |
| LAB150978 + GA4 | 5.0 ± 0.7b | 14.7 ± 8.5 | 0.76 ± 0.12 |
Measured at 2 mm from the tip of the primary roots.
Statistically significant differences between control plants and treated plants at the 5% level; Student's t test.
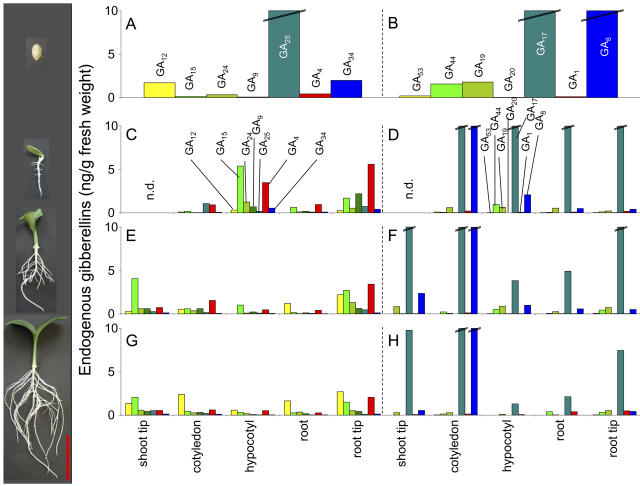
Endogenous GAs were quantified from embryos of dry pumpkin seeds and from different parts of 3-, 5-, and 7-d-old pumpkin seedlings by gas chromatography-mass spectrometry (GC-MS) single ion monitoring (Fig. 2). Levels of precursors of the early 3β-hydroxylated pathway (GA14, GA37, and GA36) were low in all tissues (<0.1 ng g−1 fresh weight; data not shown) with the exception of embryos from dry seeds where GA14 and GA36 levels were 1.9 and 0.6 ng g−1 fresh weight, respectively. Embryos of pumpkin seeds contain very high levels of the tricarboxylic acids GA25 and GA17, as well as catabolic GA8 (Fig. 2, A and B). High levels of the latter two GAs persist during the first 7 d of seedling development (Fig. 2, D, F, and H). Levels of 13-hydroxylated GAs decreased during the first 7 d of seedling development, with the exception of the plant hormone GA1, which was found at low levels in roots and root tips of 7-d-old seedlings (Fig. 2, D, F, and H).
Figure 2.
Endogenous GAs in embryos of dry seeds (A and B) and in different tissues of 3-d-old (C and D), 5-d-old (E and F), and 7-d-old (G and H) pumpkin seedlings. Images at the left illustrate pumpkin seed and seedlings at respective developmental stages (bar = 5 cm). GAs of the non- and 3β-hydroxylated pathway are depicted in A, C, E, and G, and GAs of the 13-hydroxylated pathway are depicted in B, D, F, and H. Double lines across the top of a bar indicate values >10 ng GAs/g fresh weight. Measurements have been repeated at least once with similar results. Shoot tips were analyzed together with cotyledons of 3-d-old seedlings. n.d., Not determined.
Precursors of the nonhydroxylated pathway were mainly found in shoot and root tips and in the hypocotyls, with decreasing levels in seedlings getting older (Fig. 2, C, E, and G). Highest levels of the plant hormone GA4 were identified in root tips and hypocotyls of 3-d-old seedlings, which decrease at later stages of development (Fig. 2, C, E, and G). Catabolic GA34 was found in the embryo and in early stages of seedling development, particularly in hypocotyls and root tips. These results indicate that mainly hypocotyls and root tips of developing seedlings contain relatively high concentrations of bioactive GAs compared to the other tissues. In addition, hypocotyl elongation was maximal between days 3 and 5, while the primary root length doubled approximately every second day between days 3 and 7 of seedling development (Fig. 2, left).
GA Metabolism in Cell-Free Systems Prepared from Root Tips of 7-d-Old Pumpkin Seedlings
GA biosynthetic pathways were investigated in cell-free systems prepared from root tips of untreated seedlings and of seedlings that have been treated with LAB150978. The identities of the products were confirmed by full-scan GC-MS (Table III). From 14C-GA12-aldehyde 14C-GA12 was formed, indicating the presence of GA 7-oxidase activity. Conversion was higher in root tips of untreated seedlings than in LAB150978-treated seedlings. 14C-GA12 was converted to 14C-GA15, and 14C-GA24 was metabolized to 14C-GA9, indicating the presence of GA 20-oxidase activity different from the one previously identified in developing pumpkin seeds (Table III; Lange et al., 1994; Lange, 1997). No difference of GA 20-oxidation was observed between untreated and LAB150978-treated seedlings. Incubation with 14C-GA9 gave two products, 14C-GA4 and 14C-GA34, indicating GA 3-oxidase and 2-oxidase activity, respectively (Table III). GA 3-oxidation was higher in LAB150978-treated than in untreated plants. Finally, 14C-GA4 was metabolized to 14C-GA34, a product of GA 2-oxidase activity (Table III). These results confirm the presence of a specific GA biosynthetic pathway in root tips distinct from the pathways operating in developing pumpkin seeds; in root tips, C19-GAs are the major end products and in developing seeds C20-GAs are mainly formed (Lange et al., 1993a, 1993b).
Table III.
Metabolism of 14C-labeled GAs by cell-free systems prepared from root tips of 7-d-old pumpkin seedlingsa
| Substratec
|
Productsb
|
||||
|---|---|---|---|---|---|
| Compounds Formed | Untreated Plants | LAB150978-Treated Plants | KRI | Characteristic Ions at m/z (% Relative of Base Peak)d | |
| pmol | pmol | ||||
| GA12-ald. | GA12 | 78.3 | 29.4 | 2422 | 368(0.01), 360(0.01), 308(100), 300(60), 291(27), 285(13), 247(37), 246(28), 241(25), 240(19) |
| GA12 | GA15 | 62.8 | 61.5 | 2746 | 352(15), 344(1), 320(8), 312(14), 290(44), 284(18), 245(100), 239(72), 197(57), 195(30) |
| GA24e | GA9 | 28.4 | 34.0 | 2418 | 332(4), 330(0), 300(96), 298 (30), 272(100), 270(9), 229(56), 228(75), 227(46), 226(35) |
| GA9f | GA4 | 24.1 | 58.8 | 2563 | 420(17), 418(4), 402(14), 400(4), 330(48), 328(17), 291(94), 289(38), 286(100), 284(83) |
| GA34 | 13.8 | 20.2 | 2672g | ||
| GA4f | GA34 | n.d.h | 40.4 | 2672 | 508(100), 506(0), 418(9), 416(2), 358(13), 356(2), 290(32), 288(22), 219(84), 217(40) |
Protein concentrations of root tips of untreated plants and of LAB150978-treated plants were 10.8 and 11.5 mg/mL, respectively.
Identification of 14C-GA metabolic products by GC-MS on the basis of mass spectra (Gaskin and MacMillan, 1992) and Kovats retention indices (KRI) of the methyl ester trimethylsilylether derivatives.
GA12-aldehyde and GA12 were (1-, 7-, 12-, 18-14C4)-labeled; GA24, GA9, and GA4 were (17-14C)-labeled.
Based on ions above a mass-to-charge ratio (m/z) of 50.
Incubation volumes 6 times the standard assay.
Incubation volumes 2 times the standard assay.
Spectrum was seriously contaminated with extraneous ions. The identification was made on the basis of co-occurence of characteristic ions (Gaskin and MacMillan, 1992) at the appropriate KRI.
Not determined.
Cloning and Expression of GA Oxidase Genes from Root Tips of 7-d-Old Pumpkin Seedlings
The GA biosynthetic pathway discovered in root tips requires 20-oxidases and 3-oxidases with different catalytic properties to the ones identified previously from developing pumpkin seeds (Lange, 1998; Frisse et al., 2003). To further characterize these enzymes, first their encoding cDNA molecules were isolated from an amplified pBluescript SK+ cDNA library derived from root-tip poly(A)+ RNA of LAB150978-treated pumpkin seedlings. The cDNA library was screened for cDNA molecules encoding GA 20-oxidases and 3-oxidases by a PCR-based strategy using respective degenerated oligonucleotide primers (Frisse et al., 2003). Two clones, designated CmGA20ox3 and CmGA3ox3, were isolated that share high homology to known sequences of other GA 20- and GA 3-oxidases, respectively.
Catalytic properties of recombinant CmGA20ox3 (GA 20-oxidase) and CmGA3ox3 (GA 3-oxidase) proteins were investigated by expression of the respective cDNA molecules in pUC18 in Escherichia coli NM522 (Table IV). Cell lysates of recombinant CmGA20ox3 protein, prepared from the full-length clone, oxidized 14C-GA12 to -GA15 and 14C-GA24 to -GA9 in standard enzyme assays (data not shown). GA 20-oxidation activity highly increased in cell lysates prepared from the predicted open reading frame (ORF) of the CmGA20ox3 clone (designated CmGA20ox3 ORF) and, therefore, was used for further characterization (Table IV). Recombinant CmGA20ox3 ORF protein is capable of oxidizing and subsequently removing the C-20 position of 14C-labeled substrates GA12, GA24, GA14, and, less efficiently, GA53. Of all GAs tested, recombinant CmGA3ox3 protein oxidizes only 14C-GA9 at the C-3β position (Table IV). No conversion of the 14C-labeled substrates GA12-aldehyde, GA12, GA15, GA24, GA25, GA13, and GA4 was observed with recombinant CmGA3ox3 protein (data not shown). No conversion of the 14C-labeled substrates GA12-aldehyde, GA12, GA15, GA24, GA25, GA13, GA9, GA4, GA14, and GA53 was obtained in standard incubation assays, with cell lysates of E. coli harboring the pUC18 plasmid without the cDNA insert (data not shown).
Table IV.
Metabolism of 14C-GAs by cell lysates from E. coli transformed with pUC18 clones Cm20ox3-ORF and CmGA3ox3
| Clone
|
Substrateb
|
Productsa
|
|||
|---|---|---|---|---|---|
| Compounds Formed | % by HPLC | KRI | Characteristic Ions at m/z (% Relative of Base Peak)c | ||
| Cm20ox3-ORF | GA12d | GA9 | 100 | 2418 | 338(12), 330(8), 306(53), 298(61), 276(58), 270(100), 251(89), 243(99), 232(78), 226(91) |
| GA24 | GA9 | 100 | 2420 | 332(0), 330(0), 300(40), 298(1), 272(89), 270(5), 243(100), 229(57), 228(77), 227(41), 226(17) | |
| GA14 | GA4 | 100 | 2561 | 426(1), 418(0), 336(4), 328(2), 292(56), 284(32), 231(99), 230(100), 225(46), 224(53) | |
| GA53 | GA44/19e | 12 | |||
| GA20 | 88 | 2544 | 426(42), 418(100), 409(1), 403(10), 381(29), 375(68), 365(9), 359(19), 307(10), 301(27) | ||
| CmGA3ox3 | GA9 | GA4 | 48 | 2563 | 420(1), 418(1), 330(7), 328(6), 286(55), 284(20), 227(99), 226(100), 225(58), 224(37) |
Identification of 14C-GA metabolic products by GC-MS on the basis of mass spectra (Gaskin and MacMillan, 1992) and Kovats retention indices (KRI) of the methyl ester trimethylsilylether derivatives.
GA12, GA14, and GA53 were (1-, 7-, 12-, 18-14C4)-labeled; GA24 and GA9 were (17-14C)-labeled.
Based on ions above a mass-to-charge ratio (m/z) of 50.
Incubation volumes 0.6 times the standard assay.
Preliminary identification according to HPLC retention time.
Transcript Analysis of GA Oxidases in Developing Pumpkin Seedlings
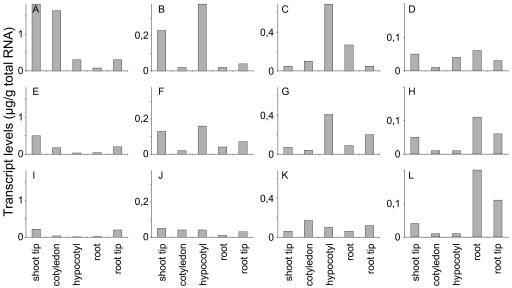
Expression levels of mRNA were quantified for GA oxidase-encoding genes in different tissues from 3-, 5-, and 7-d-old pumpkin seedlings by competitive RT-PCR (Fig. 3). The primers used were based on the sequence of CmGA7ox and CmGA2ox1 cDNAs that were originally isolated from pumpkin endosperm and embryo, respectively (Lange, 1997; Frisse et al., 2003), and on the sequence of CmGA20ox3 and CmGA3ox3 clones from root tips of the pumpkin seedling.
Figure 3.
Transcript levels of CmGA7ox (A, E, and I), CmGA20ox3 (B, F, and J), CmGA3ox3 (C, G, and K), and CmGA2ox1 (D, H, and L) genes in different parts of 3- (A–D), 5- (E –H), and 7-d-old (I–L) pumpkin seedlings as determined by quantitative RT-PCR.
Highest transcript levels of CmGA7ox, CmGA20ox3, and CmGA3ox3 were identified in 3-d-old seedlings (Fig. 3, A–C, E–G, and I–K): CmGA7ox transcripts were found mainly in shoot tips and cotyledons of 3-d-old seedlings, with moderate levels in hypocotyls and root tips (Fig. 3A), CmGA20ox3 transcript levels were highest in shoot tips and hypocotyls (Fig. 3B), and CmGA3ox3 transcripts were mostly detectable in hypocotyls (Fig. 3C). Moderate transcript levels of CmGA7ox, CmGA20ox3, and CmGA3ox3 were visible in root tips of seedlings 5 d after imbibition (Fig. 3, E–G). In contrast, CmGA2ox1 transcript levels increased in roots from day 3 to day 7 after imbibition of the seedling (Fig. 3, D, H, L). Highest CmGA2ox1 transcript levels were found in roots, together with moderate levels in shoot and root tips. Additionally, transcript levels of three previously cloned GA oxidase genes from developing pumpkin seeds (Lange, 1997; Lange et al., 1997; Frisse et al., 2003) were analyzed in untreated 5-d-old seedlings and either were not detectable (CmGA20ox1, CmGA20ox2, and CmGA3ox1) or were identified at very low levels in root tips (CmGA3ox2, 0.02 μg transcript per gram of total RNA; data not shown).
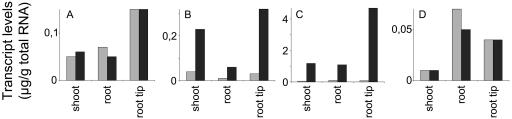
The impact of LAB150978 treatment on transcript levels was analyzed for four GA oxidase genes in 7-d-old seedlings. Transcript levels of CmGA7ox and CmGA2ox1 did not alter significantly between treated and untreated plants (Fig. 4, A and D). However, transcript levels of CmGA20ox3 and of CmGA3ox3 genes were low in general and, after LAB150978 treatment, increased dramatically in all parts of the seedling (Fig. 4, B and C).
Figure 4.
Transcript levels of CmGA7ox (A), CmGA20ox3 (B), CmGA3ox3 (C), and CmGA2ox1 (D) genes as determined by competitive RT-PCR in tissues from shoots, roots, and root tips of 7-d-old seedlings. Seedlings were treated with water (gray bars) or with LAB150978 (10−6 m; black bars).
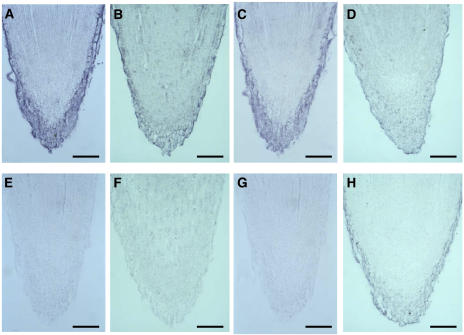
Expression patterns for transcripts of four GA oxidases were investigated by in situ hybridization in root tips of 7-d-old pumpkin seedlings. CmGA7ox, CmGA20ox3, and CmGA3ox3 transcripts were localized in the cap of the root and the rhizodermis (Fig. 5, A–C and E–G). However, only weak signals for CmGA2ox1-encoding transcripts were detectable in root tips by in situ hybridization (Fig. 5, D and H). No differences in expression pattern of these transcripts were detectable between 3- and 7-d-old plants and between untreated and LAB150978-treated plants by in situ hybridization. However, the intensity of the signals obtained for CmGA20ox3 and CmGA3ox3 were stronger in LAB150978-treated plants (data not shown).
Figure 5.
Localization of CmGA7ox (A and E), CmGA20ox3 (B and F), CmGA3ox3 (C and G), and CmGA2ox1 (D and H) mRNA in longitudinal sections from root tips of 7-d-old pumpkin seedlings by in situ hybridization. The sections were hybridized to either antisense (A–D) or sense (E–H) RNA probes of the entire respective cDNA (A, C, E, and G) or the predicted ORF (B, D, F, and H) that was labeled with digoxygenin. Bar = 0.1 mm.
DISCUSSION
In pumpkin, GA biosynthetic pathways have been elucidated mainly in cell-free systems prepared from developing seeds (Lange et al., 1993a, 1993b; for review, see Graebe, 1987). These pathways are still active in mature pumpkin seeds even 36 h after imbibition. However, they disappear rapidly in later stages of the developing seedling (Lange et al., 1997). The main products formed via those seed-specific pathways in pumpkin are GAs containing C-20 carboxylic acids (e.g. GA25 and GA17; Fig. 1). In fact, the mature embryo (as well as the testa; data not shown) contains large quantities of those tricarboxylic GAs that have no known physiological function (Fig. 2).
There is growing evidence for the presence of root-based GA biosynthesis from many plant species, including pumpkin (Yamaguchi et al., 1996; Smith et al., 1998), pea (Pisum sativum; Lester et al., 1999; Martin et al., 1999; Elliott et al., 2001; Davidson et al., 2003, 2004), Arabidopsis (Chiang et al., 1995; Silverstone et al., 1997; Thomas et al., 1999), and rice (Oryza sativa; Sakamoto et al., 2001, 2004; Monna et al., 2002; Sasaki et al., 2002; Kaneko et al., 2003; Itoh et al., 2004). In this study, we present direct evidence for GA biosynthesis in root tips of young pumpkin seedlings. The pathway involves oxidations at four different positions of the GA molecule, C-7, C-20, C-3β, and C-2β (Fig. 1) and differs from the one previously identified in developing pumpkin seeds. The main products of GA 20-oxidase activity are C19-GAs, which are different from developing pumpkin seeds where the main products are C20-GAs (Lange et al., 1994; Lange, 1997). No metabolism of GA15 was observed, indicating a different GA 3-oxidase activity to the one that is present in developing seeds where GA 3-oxidases efficiently convert C20-GAs, including GA15 (Lange et al., 1997; Frisse et al., 2003). All GAs obtained in the metabolic studies were identified as endogenous compounds of the seedling. Investigations based on studies of endogenous GA levels led to the presumption that in vegetative tissues bioactive GAs are synthesized via a late 3β-hydroxylation step that occurs after C19-GAs are formed (Graebe, 1987; Hedden, 1999). Our metabolic studies confirm this supposition for GA biosynthesis in pumpkin seedlings. Moreover, none of the early 3β-hydroxylated precursors (GA14, GA37, and GA36) accumulated during seedling development. However, the presence of an early 3-oxidation step in the GA biosynthetic pathway of root tips cannot be completely excluded. Recently, we demonstrated that recombinant pumpkin GA 7-oxidase hydroxylated, at low efficiency, GA12 at the C-3β position, leading to the formation of GA14 (Frisse et al., 2003), which can be further metabolized to GA4 by recombinant CmGA20ox3 (Fig. 1; Table IV).
GA metabolic studies demonstrate that the products resulting from GA 20-oxidase and GA 3-oxidase activities are different in developing pumpkin seeds and seedlings. We isolated two cDNA molecules, CmGA20ox3 and CmGA3ox3, from root tip poly(A)+ RNA of 7-d-old LAB150978-treated pumpkin seedlings, encoding GA 20-oxidase and 3-oxidase, respectively. The broad specificity of the recombinant CmGA20ox3 protein indicates that a single enzyme is capable of catalyzing alternative GA biosynthetic pathways (Fig. 1). Unlike CmGA20ox1 and CmGA20ox2 proteins from developing seeds, recombinant CmGA20ox3 protein exhibits strong decarboxylation activities and converts GA precursors GA12, GA14, and GA53 to their respective C19-GAs. Recombinant CmGA3ox3 protein, however, reveals more stringent substrate specificity than CmGA3ox1 or CmGA3ox2 protein from developing pumpkin seeds (Lange et al., 1997, Frisse et al., 2003). It catalyzes 3-oxidation, after C19-GA is formed, but not of intermediates of the pathway, which might have some importance for its activity in situ, where huge quantities of endogenous tricarboxylic GAs might otherwise inhibit its activity. Such enzymatic properties are typical for GA 20-oxidases and GA 3-oxidases from most plant species, but up to now were not known in pumpkin (Lange, 1998). However, it is likely that the cloning of the two genes from root-tip tissues was facilitated by a negative type of feedback regulation that occurred in LAB150978-treated pumpkin plants (Fig. 4) and was observed in several other plant species (for review, see Hedden and Phillips, 2000; Fleet and Sun, 2005). A positive feed-forward regulation as reported for GA 2-oxidase transcript levels from some other plant species (Martin et al., 1999; Thomas et al., 1999; Elliott et al., 2001) was not observed for CmGA2ox1 gene expression, similar to observations obtained with rice GA 2-oxidase (Sakamoto et al., 2001).
Sites of bioactive GA synthesis seem to be tissues containing rapidly expanding cells (Silverstone et al., 1997, Yamaguchi et al., 2001; for review, see Olszewski et al., 2002). Our results show that transcript and endogenous GA profiles correlate well for most of the tissues analyzed (Figs. 2 and 3). For example, in rapidly growing hypocotyl tissues, this site of GA action was paralleled by the transcript levels of the GA oxidases as well as endogenous GA levels, including the plant hormone GA4. As hypothesized recently by Sponsel and Hedden (2004), our investigations support the view that the site of GA biosynthesis and GA action might be similar, if not identical, in hypocotyls. Additional evidence comes from expression studies on GA1, GA4, and GA5 genes, all expressed in the hypocotyl of young Arabidopsis seedlings (Silverstone et al., 1997; Bouquin et al., 2001; Meier et al., 2001).
In root tips, however, transcript levels determined for pumpkin GA 20-oxidase (CmGA20ox1, CmGA20ox2, and CmGA20ox3) and GA 3-oxidase (CmGA3ox1, CmGA3ox2, and CmGA3ox3) genes do not correlate well with endogenous GA levels. GA oxidases form multigene families (Sponsel and Hedden, 2004) and it is likely that additional GA oxidases are expressed that might account for further enzyme activities and for the high endogenous GA levels determined in this tissue. It is also possible that other GA oxidases present in this tissue do not show a negative type of feedback regulation and were, therefore, not favored in the cloning process from LAB150978-treated pumpkin seedlings. However, transcripts of CmGA7ox and CmGA20ox3, together with transcripts of CmGA3ox3, are localized in root caps and rhizodermis of the root tip by in situ hybridization, indicating the contribution of their respective GA oxidases for the GA biosynthesis in this tissue. GA synthesis in root tips might have a function for root development of the young seedling. For instance, optimal GA levels are necessary to achieve stability of principal directions of root growth (Nakielski and Barlow, 1995) and regulate root elongation as well as radial root growth (see Table II; Tanimoto, 1990; Barlow et al., 1991; Yaxley et al., 2001). Recently, DELLA proteins, components of the GA-signaling cascade, have been identified to regulate root growth in Arabidopsis seedlings and this action seems to be modulated by other hormones (Fu and Harberd, 2003).
MATERIALS AND METHODS
Plant Material, Enzyme, and Total RNA Preparations
Seeds of pumpkin (Cucurbita maxima L. cv Riesenmelone, gelb genetzt) were sown in 100-mL pots containing 10 g of vermiculite moistened with 50 mL of 0.01% methanol, 0.01% methanol containing GA4 (10−6 m), or 0.01% methanol containing the plant growth retardant 1-(4-trifluormethyl)-2-(1,2,4-triazolyl-(1))-3-(5-methyl-1,3-dioxan-5-yl)-propen-3-ol (10−6 m; LAB150978; a gift from Dr. Rademacher, BASF Agricultural Center, Limburgerhof, Germany). Water and growth regulators were replenished 10 mL/d. Germination and growth of the seedlings occurred under an 18-h photoperiod at 24°C during the day and 18°C during the night. Light was supplied by Osram Powerstar HQI-T 400W/D daylight lamps, located 1 m above the plants, giving a photon fluence rate of 200 μE m−2 s−1. At the time of sampling, the plants were rinsed, dissected into shoot tips (the upper 5 mm of the stem), cotyledons, hypocotyl, roots, and root tips (the lower 2 mm of the primary and lateral roots) and immediately frozen in liquid N2 and stored at −80°C. The plant material was ground to a fine powder in liquid nitrogen with a mortar and pestle. Total RNA was isolated from the frozen powder (25 mg) with a NucleoSpin RNA plant kit, according to the manufacturer's instructions (Machery-Nagel), and treated with DNase I (Sigma; 10 units/μg total RNA) for 20 min at 37°C, followed by phenol-chloroform extraction. Total RNA was stored at −80°C and used for quantification of specific GA oxidase transcripts as described below. Cell-free enzyme preparations were obtained from the frozen powder (1 g) to which 200 mm Tris-HCl buffer (pH 7.9, as measured at 4°C) containing 10 mm dithiothreitol was added 1:1 (w/v). After thawing, the extract was centrifuged for 30 min at 40,000g and the supernatant was stored at −80°C.
Construction of a pBluescript SK+ cDNA Plasmid Library
Poly(A)+ RNA (5 μg) was extracted from root tips of 7-d-old pumpkin seedlings treated with the growth retardant LAB150978, using a mRNA isolation kit, according to the manufacturer's instructions (Fast Track 2.0; Invitrogen), and used for the preparation of an oligo(dT)-primed cDNA library in pBluescript SK+ using commercial kits (pBluescript II XR cDNA library construction kit; Stratagene). A cDNA plasmid library in Escherichia coli XL 10 GOLD of 3.4×105 independent cell-forming units was obtained and amplified, 95% of which contained inserts of a length between 800 to 2,000 bp, as shown by agarose gel electrophoresis of PCR products using pBluescript specific M13 primers.
Screening of pBluescript SK+ cDNA Library by PCR
Screening of the pBluescript SK+ cDNA library for GA 20-oxidase, 3-oxidase, and 2-oxidase genes was performed by a PCR-based cloning strategy (Frisse et al., 2003) with the following modifications: For GA 20-oxidase screening, degenerate primer pairs (sense primer 5′-TNCCNTGGAA(AG)GA(AG)AC-3′ and antisense primer 5′-GG(AG)CANA(AG)(AG)AA(AG)AANGC-3′, where n is a mixture of A, C, G, and T) were used in a PCR reaction at an annealing temperature of 50°C. One clone (designated CmGA20ox3) was isolated that gave an approximately 530-bp band, which is the expected size for a putative GA 20-oxidase. As described by Frisse et al. (2003), using degenerate GA 3-oxidase primer pairs, one clone (CmGA3ox3) was identified harboring a putative GA 3-oxidase cDNA, and using degenerate GA 2-oxidase primer pairs, one clone was isolated harboring putative GA 2-oxidase cDNAs. The pBluescript SK+ plasmids containing the cDNA inserts of the three clones, CmGA7ox, CmGA3ox3, and the putative 2-oxidase clone, were custom sequenced on both strands (AGOWA). The 2-oxidase sequence corresponded to the previously cloned CmGA2ox1 from pumpkin embryos (Frisse et al., 2003).
Heterologous Expression of Recombinant GA Oxidases
DNA sequence analysis revealed that the cDNA inserts of the three clones were not in frame to the lac-promotor of the pBluescript vector. The pBluescript SK+ vector containing CmGA20ox3 cDNA was digested with XbaI and EcoRI, filling in recessed 3′-termini with Klenow fragment and then religated. The predicted ORF of the CmGA20ox3 clone was amplified by PCR (at an annealing temperature of 60°C), using sense primer 5′-NGAATTCAAACCAAACCATGCATGTCGTGAC-3′ and antisense primer 5′-NGGATCCTTTTCCTCAGGCGAGGAAAAGTG-3′, cut by EcoRI and BamHI digest and cloned into the appropriate cloning sites of pUC18 vector (and pBluescript SK− vector for preparation of riboprobes for in situ hybridization; see below). The cDNA of clone CmGA3ox3 was excised by XhoI and XbaI and recessed 3′-termini were filled in by Klenow fragment and subcloned into the SmaI site of pUC18. The plasmid vectors containing respective inserts were used to transform E. coli NM 522 cells. Protein induction and cell lysis were carried out as described by Lange (1997).
Standard Enzyme Assay and Analysis of Incubation Products
All 14C-labeled GAs were prepared as described elsewhere (Lange and Graebe, 1993; Frisse et al., 2003). Specific radioactivity for (1, 7, 12, and 18-14C4)-labeled GA12-aldehyde, GA12, GA15, GA9, GA14, and GA53, were determined to be 5.80×10l2, 5.93×1012, 5.05×1012, 5.80×10l2, 5.80×1012, and 5.4×1012 Bq mol−1, respectively. Specific radioactivity for (17-14C)-labeled substrates was 1.95×1012 Bq/mol. Cell-free enzyme preparations from different parts of 7-d-old pumpkin seedlings (450 μL each) were incubated with 2-oxoglutarate and ascorbate (4 mm each, final concentrations), and preparations of E. coli cell lysates (350 μL) were incubated with 2-oxoglutarate and ascorbate (100 mm each, final concentrations). FeSO4 (0.5 mm), catalase (1 mg/mL), and the 14C-labeled substrate (10 μL in 50% methanol; 0.33 nmol for (1, 7, 12, and 18-14C4)-labeled GAs and 1 nmol for (17-14C)-labeled GAs) were added in a total volume of 500 μL and incubated for 16 h at 37°C. Incubation products were extracted and analyzed by reverse-phase HPLC with online radiocounting, using gradients of increasing methanol in acidic water, as described by Lange and Graebe (1993). HPLC fractions were analyzed by combined GC-MS (Xu et al., 2002). Variations in incubation conditions are specified for particular experiments.
Quantitative Analysis of GA
For quantitative determination of endogenous GAs, 2 g fresh weight of frozen plant tissue were spiked with 17, 17-d2-GA standards (2 ng each; from Professor L. Mander, Canberra, Australia) and pulverized under liquid nitrogen. Eighty percent methanol-water (8 mL) was added and the extract was stirred for 1 h at 4°C. After centrifugation, the pellet was re-extracted with methanol (4 mL) for 30 min and recentrifuged. The re-extraction procedure was repeated two times. The combined methanol extracts were evaporated to dryness and resuspended in water (3 mL), adjusted to pH 8.0 (1 m KOH). Solvent partition was performed using ethyl acetate (four times, 1 mL). The aqueous phase was adjusted to pH 3 (acetic acid) followed by solvent partition with ethyl acetate (four times, 1 mL). The combined ethyl acetate fractions were evaporated to dryness, redissolved in methanol (100 μL), and methylated with ethereal diazomethane (two times, 200 μL). The methyated samples were dried and redissolved in methanol (100 μL) and water (5 mL), adjusted to pH 3.2 (acetic acid). After loading onto a C18 cartridge (Waters), the cartridge was washed with water (10 mL), pH 3.2. Methylated GAs were eluted with methanol (6 mL), which was then dried. For purification by HPLC, the residues were redissolved in 200 μL methanol-water, pH 3.2 (1:1), and applied to a C18 reverse-phase column (10 cm long, 8 mm i.d., 4 μm particle size, Novapack liquid chromatography cartridge in a RCM100 radial compression system; Waters), which was eluted with a gradient from 25% methanol in water to 100% methanol in 40 min delivered by a two-pump HPLC system (models 501 and 510; Waters) at a flow rate of 1 mL min−1. Starting from 13.5 mL, 18 fractions were collected per run, each containing 2 mL eluate and dried. Dried HPLC fractions were redissolved in 2 μL N-methyl-N-trimethylsilyltrifluoracetamide (Macherey-Nagel). The derivatized samples were analyzed using a Turbo-Mass MS system (Perkin-Elmer) equipped with a Perkin-Elmer AutoSystem XL gas chromatograph. Samples (1–2 μL) injected into a SGE BPX5 capillary column (30 m long, 0.25 mm i.d., 0.25-μm film thickness; SGE) at an oven temperature of 60°C. The split value (30:1) was open after 1 min, after which the temperature was increased by 45°C min−1 to 220°C and then with 4°C min−1 to 300°C. The He inlet was pneumatic pressure controlled at a constant flow rate of 1.5 mL min−1 and the injector, transfer line, and source temperatures were 220°C, 280°C, and 240°C, respectively. Data were acquired in the selected ion monitoring mode after 5 min. The ions monitored for quantification of endogenous GAs were 270 and 272 (GA12-aldehyde), 300 and 302 (GA12), 239 and 241 (GA15), 314 and 316 (GA24), 270 and 272 (GA9), 284 and 286 (GA25), 284 and 286 (GA4), 506 and 508 (GA34), 207 and 209 (GA53), 207 and 209 (GA44), 374 and 376 (GA19), 418 and 420 (GA20), 492 and 494 (GA17), 506 and 508 (GA1), 594 and 596 (GA8), 416 and 418 (GA14), 432 and 434 (GA37), and 284 and 286 (GA36). Identification was confirmed on the basis of retention time and the co-occurrence of additional ions. Endogenous levels were calculated on the basis of peak areas, after corrections were made for the contribution of naturally occurring isotopes and for the presence of unlabeled GAs in the internal standards, where necessary.
Quantitative RT-PCR
Quantification of CmGA7ox transcripts was performed as described by Lange et al. (1997). For each of the CmGA20ox3, CmGA3ox3, and CmGA2ox1 (Frisse et al., 2003) genes, three specific oligonucleotides were synthesized based on their cDNA sequence: clone CmGA20ox3 F (5′-AAA CCA AAC CAT GCA TGT CGT GAC-3′), R (5′-TTT TCC TCA GGC GAG GAA AAG TG-3′), and RT (5′-TTT AAA TGG AAG GGT TC-3′); clone CmGA3ox3 F (5′-CAA CAT GGC CAC CAC AAT AGC-3′), R (5′-GGG ATT AGC CTA CTT TGA CCC GGC-3′), and RT (5′-TAG GGT GGG ATT AG-3′); and CmGA2ox1 F (5′-AAT GAG AAG CTC CAC GTC CAT G-3′), R (5′-GAT GTT CGA ATC CTG TCA CCT C-3′), and RT (5′-AGA TGT TCG AAT CC-3). For the preparation of internal RNA standards, pBluescript SK plasmids containing the CmGA20ox3 clone were digested with SalI and HindIII, the CmGA3ox3 clone was digested with HincII, and the ORF of CmGA2ox1 was digested with HindIII that released a 299-, 379-, and 448-bp fragment, respectively. The vectors containing the remaining cDNAs were religated and used for standard RNA synthesis. For stability reasons, dilution of standard RNA was performed in the presence of lambda RNA (100 ng/μL). Quantification of CmGA20ox3, CmGA3ox3, and CmGA2ox1 transcripts were performed as described by Lange et al. (1997), except that total RNA (50–100 ng) was reverse transcribed using the RevertAid H Minus first-strand cDNA synthesis kit (MBI Fermentas), and PCR was performed at an annealing temperature of 60°C for CmGA20ox3 and CmGA3ox3 genes and 65°C for the CmGA2ox1 gene.
In Situ Hybridization
Sense and antisense riboprobes of full-length cDNAs of CmGA7ox (Lange, 1997) and CmGA3ox3 and of the predicted ORF encoding CmGA2ox1 (Frisse et al., 2003) and CmGA20ox3 were synthesized using the dioxygenin nucleic acid labeling kit, according to the manufacturer's protocol with T7 and T3 RNA polymerases (Roche Molecular Biochemicals) and used for in situ hybridization as described by Frisse et al. (2003).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AJ315663 and AJ302040.
Acknowledgments
We thank Anja Liebrandt for technical assistance and Julia Niemeyer for help with RT-PCR.
This work was supported by the Deutsche Forschungsgemeinschaft (grant nos. La880/4–2 and La880/4–3).
This paper is dedicated to Jan E. Graebe on the occasion of his 75th birthday.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.064162.
References
- Achard P, Vriezen WH, van der Straeten D, Harberd NP (2003) Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15: 2816–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahina M, Iwai H, Kikuchi A, Yamaguchi S, Kamiya Y, Kamada H, Satoh S (2002) Gibberellin produced in the cotyledon is required for cell division during tissue reunion in the cortex of cut cucumber and tomato hypocotyls. Plant Physiol 129: 201–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow PW, Brain P, Parker JS (1991) Cellular growth in roots of a gibberellin-deficient mutant of tomato (Lycopersicon esculentum Mill.) and its wild type. J Exp Bot 42: 339–351 [Google Scholar]
- Bouquin T, Meier C, Foster R, Nielsen ME, Mundy J (2001) Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol 127: 450–458 [PMC free article] [PubMed] [Google Scholar]
- Chiang H, Hwang I, Goodman H (1995) Isolation of the Arabidopsis GA4 locus. Plant Cell 7: 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SE, Elliott RC, Helliwell CA, Poole AT, Reid JB (2003) The pea gene NA encodes ent-kaurenoic acid oxidase. Plant Physiol 131: 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SE, Smith JJ, Helliwell CA, Poole AT, Reid JB (2004) The pea gene LH encodes ent-kaurene oxidase. Plant Physiol 134: 1123–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RC, Ross JJ, Smith JL, Lester DR, Reid JB (2001) Feed-forward regulation of gibberellin deactivation in pea. J Plant Growth Regul 20: 87–94 [Google Scholar]
- Fleet CM, Sun TP (2005) A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr Opin Plant Biol 8: 77–85 [DOI] [PubMed] [Google Scholar]
- Frisse A, Pimenta MJ, Lange T (2003) Expression studies of gibberellin oxidases in developing pumpkin seeds. Plant Physiol 131: 1220–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Harberd NP (2003) Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421: 740–743 [DOI] [PubMed] [Google Scholar]
- Gaskin P, MacMillan J (1992) GC-MS of Gibberellins and Related Compounds. Cantock's Enterprise, Bristol, UK
- Graebe JE (1987) Gibberellin biosynthesis and control. Annu Rev Plant Physiol 38: 419–465 [Google Scholar]
- Hedden P (1999) Regulation of gibberellin biosynthesis. In PJJ Hooykaas, MA Hell, KR Libbenga, eds, Biochemistry and Molecular Biology of Plant Hormones. Elsevier Press, Amsterdam, pp 161–188
- Hedden P, Kamiya Y (1997) Gibberellin biosynthesis: enzymes, genes and their regulation. Annu Rev Plant Physiol Plant Mol Biol 48: 431–460 [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Pharmacol Sci 5: 523–530 [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Chandler PM, Poole A, Dennis ES, Peacock WJ (2001) The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc Natl Acad Sci USA 98: 2065–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Tatsumi T, Sakamoto T, Otomo K, Toyomasu T, Kitano H, Ashikari M, Ichihara S, Matsuoka M (2004) A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol Biol 54: 533–547 [DOI] [PubMed] [Google Scholar]
- Jung J, Rentzea C, Rademacher W (1986) Plant growth regulation with triazoles of the dioxanyl type. J Plant Growth Regul 4: 181–188 [Google Scholar]
- Kaneko M, Itoh H, Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Ashikari M, Matsuoka M (2003) Where do gibberellin biosynthesis and gibberellin signaling occur in rice plants? Plant J 35: 104–115 [DOI] [PubMed] [Google Scholar]
- Lange T (1997) Cloning gibberellin dioxygenase genes from pumpkin endosperm by heterologous expression of enzyme activities in Escherichia coli. Proc Natl Acad Sci USA 94: 6553–6558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T (1998) Molecular biology of gibberellin synthesis. Planta 204: 409–419 [DOI] [PubMed] [Google Scholar]
- Lange T, Graebe JE (1993) Enzymes of gibberellin synthesis. In PJ Lea, ed, Methods in Plant Biochemistry, Vol 9. Academic Press, London, pp 403–430
- Lange T, Hedden P, Graebe JE (1993. a) Biosynthesis of 12α- and 13-hydroxylated gibberellins in a cell-free system from Cucurbita maxima endosperm and the identification of new endogenous gibberellins. Planta 189: 340–349 [DOI] [PubMed] [Google Scholar]
- Lange T, Hedden P, Graebe JE (1993. b) Gibberellin biosynthesis in cell-free extracts from developing Cucurbita maxima embryos and the identification of new endogenous gibberellins. Planta 189: 350–358 [DOI] [PubMed] [Google Scholar]
- Lange T, Hedden P, Graebe JE (1994) Expression cloning of a gibberellin 20-oxidase, a multifunctional enzyme involved in gibberellin biosynthesis. Proc Natl Acad Sci USA 91: 8552–8556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T, Robatzek S, Frisse A (1997) Cloning and expression of a gibberellin 2β,3β-hydroxylase cDNA from pumpkin endosperm. Plant Cell 9: 1459–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester DR, MacKenzie-Hose AK, Davies PJ, Ross JJ, Reid JB (1999) The influence of the null le-2 mutation on gibberellin levels in developing pea seeds. Plant Growth Regul 27: 83–89 [Google Scholar]
- MacMillan J (2002) Occurrence of gibberellins in vascular plants, fungi, and bacteria. J Plant Growth Regul 20: 387–442 [DOI] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Hedden P (1999) The SLENDER gene of pea encodes a gibberellin 2-oxidase. Plant Physiol 121: 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier C, Bouquin T, Nielsen ME, Raventos D, Mattsson O, Rocher A, Schomburg F, Amasino RM, Mundy J (2001) Gibberellin response mutants identified by luciferase imaging. Plant J 25: 509–519 [DOI] [PubMed] [Google Scholar]
- Monna L, Kitazawa N, Yoshino R, Suzuki J, Masuda H, Maehara Y, Tanji M, Sato M, Nasu S, Minobe Y (2002) Positional cloning of rice semidwarfing gene, sd-1: rice “green revolution gene” encodes a mutant enzyme involved in gibberellin synthesis. DNA Res 9: 11–17 [DOI] [PubMed] [Google Scholar]
- Nakielski J, Barlow PW (1995) Principal directions of growth and the generation of cell patterns in wild-type and gib-1 mutant roots of tomato (Lycopersicon esculentum Mill.) grown in vitro. Planta 196: 30–39 [Google Scholar]
- Olszewski N, Sun TP, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell (Suppl) 14: S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JB, Symons GM, Ross JJ (2004) Regulation of gibberellin and brassinosteroid biosynthesis by genetic, environmental and hormonal factors. In PJ Davies, ed, Plant Hormones: Biosynthesis, Signal Transduction, Action! Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 179–203
- Richards DE, King KE, Ait-Ali T, Harberd NP (2001) How gibberellin regulates plant growth and development: a molecular genetic analysis of gibberellin signaling. Annu Rev Plant Physiol Plant Mol Biol 52: 67–88 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Kobayashi M, Itoh H, Tagiri A, Kayano T, Tanaka H, Iwahori S, Matsuoka M (2001) Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol 125: 1508–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, et al (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134: 1642–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, et al (2002) Green revolution: a mutant gibberellin-synthesis gene in rice. Nature 416: 701–702 [DOI] [PubMed] [Google Scholar]
- Silverstone A, Chang C, Krol E, Sun TP (1997) Developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. Plant J 12: 9–19 [DOI] [PubMed] [Google Scholar]
- Smith MW, Yamaguchi S, Ait-Ali T, Kamiya Y (1998) The first step of gibberellin biosynthesis in pumpkin is catalyzed by at least two copalyl diphosphate synthases encoded by differentially regulated genes. Plant Physiol 118: 1411–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sponsel VM, Hedden P (2004) Gibberellin biosynthesis and inactivation. In PJ Davies, ed, Plant Hormones: Biosynthesis, Signal Transduction, Action! Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 63–94
- Sun TP (2004) Gibberellin signal transduction in stem elongation and leaf growth. In PJ Davies, ed, Plant Hormones: Biosynthesis, Signal Transduction, Action! Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 304–320
- Tanimoto E (1990) Gibberellin requirement for the normal growth of roots. In N Takahashi, BO Phinney, J MacMillan, eds, Gibberellins. Springer-Verlag, New York, pp 229–240
- Thomas SG, Phillips AL, Hedden P (1999) Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA 96: 4698–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Lange T, Altpeter F (2002) Cloning and characterization of a cDNA encoding a multifunctional gibberellin 20-oxidase from perennial ryegrass (Lolium perenne L.). Plant Sci 163: 147–155 [Google Scholar]
- Yamaguchi S, Kamiya Y, Sun TP (2001) Distinct cell-specific expression patterns of early and late gibberellin biosynthetic genes during Arabidopsis seed germination. Plant J 28: 443–453 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Saito T, Abe H, Yamane H, Murofushi N, Kamiya Y (1996) Molecular cloning and characterization of a cDNA encoding the gibberellin biosynthetic enzyme ent-kaurene synthase B from pumpkin (Cucurbita maxima L). Plant J 10: 203–213 [DOI] [PubMed] [Google Scholar]
- Yaxley JR, Ross JJ, Sherriff LJ, Reid JB (2001) Gibberellin biosynthesis mutations and root development in pea. Plant Physiol 125: 627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]