Abstract
Peroxisomes perform diverse and vital functions in eukaryotes, and abnormalities in peroxisomal function lead to severe developmental disorders in humans. Peroxisomes are also involved in a wide array of physiological and metabolic functions unique to plants, yet many aspects of this important organelle are poorly understood. In yeast and mammals, various steps in peroxisome biogenesis require the function of peroxin (PEX) proteins, among which PEX12 is a RING finger peroxisomal membrane protein involved in the import of matrix proteins. To investigate the role of PEX12 in plants, we identified a T-DNA knockout allele of PEX12 and generated partial loss-of-function pex12 mutants using RNA interference. We show that pex12 null mutants are developmentally arrested during early embryogenesis, and that the embryo-lethal phenotype can be rescued by overexpression of the PEX12-cyan fluorescent protein fusion protein, which targets to the peroxisome. Using virus-induced gene-silencing techniques, we demonstrate that peroxisomal number and fluorescence of the yellow fluorescent protein-peroxisome targeting signal type 1 protein are greatly reduced when PEX12 is silenced. RNA interference plants with partial reduction of the PEX12 transcript exhibit impaired peroxisome biogenesis and function, inhibition of plant growth, and reduced fertility. Our work provides evidence that the Arabidopsis (Arabidopsis thaliana) PEX12 protein is required for peroxisome biogenesis and plays an essential role throughout plant development.
Peroxisomes perform diverse and crucial functions in eukaryotes, and peroxisomal deficiencies cause developmental disorders in humans and disrupt many physiological and developmental processes in fungi (Powers and Moser, 1998; Gould and Valle, 2000; Parsons et al., 2001; Titorenko and Rachubinski, 2004). Plant peroxisomes, which include glyoxysomes, leaf-type peroxisomes, and unspecialized peroxisomes, contain different sets of enzymes depending on the cell type and developmental stage. They are involved in a wide range of functions, including lipid metabolism, photorespiration, nitrogen metabolism, stress response, and synthesis of some plant hormones (Beevers, 1979; Olsen and Harada, 1995; Hayashi and Nishimura, 2003).
More than 30 yeast genes have been identified that encode components for various aspects of peroxisome biogenesis, including peroxisome assembly, matrix protein import, and peroxisome proliferation (Purdue and Lazarow, 2001; Charlton and Lopez-Huertas, 2002; Brown and Baker, 2003). Proteins in this category are named peroxins, or PEX proteins. The peroxisome protein import machinery is composed of a number of proteins facilitating the import process, although the architecture of this machinery is still elusive. PEX2, PEX10, and PEX12 are RING finger proteins hypothesized to function in close proximity to facilitate the import of cargo proteins and the recycling of receptors in yeast and mammals, and possibly in other eukaryotes. These three peroxins are integral membrane proteins whose N- and C-terminal domains are both predicted to be exposed to the cytosol, yet their precise biochemical function is still unclear (Purdue and Lazarow, 2001; Brown and Baker, 2003). A complicating factor is that some PEX proteins appear to play additional roles outside of the peroxisome. For instance, in the yeast Yarrowia lipolytica, PEX2 is also involved in facilitating the exit of plasma membrane- and cell wall-associated proteins from the endoplasmic reticulum before they enter the secretory pathway (Titorenko et al., 1997). In the filamentous yeast Podospora anserina, PEX2 is also required for nuclear fusion during sexual sporulation (Berteaux-Lecellier et al., 1995). Our current knowledge still cannot explain the functional complexity of these PEX proteins.
The Arabidopsis (Arabidopsis thaliana) genome is predicted to encode about 15 proteins homologous to the yeast peroxins (Mullen et al., 2001; Charlton and Lopez-Huertas, 2002); a few of the genes have been cloned and partially characterized (Lin et al., 1999, 2004; Hayashi et al., 2000, 2005; Zolman et al., 2000; Hu et al., 2002; Schumann et al., 2003; Sparkes et al., 2003; Zolman and Bartel, 2004; Woodward and Bartel, 2005). PEX2 and PEX10 encode essential peroxins because null mutations of either gene cause embryonic lethality (Hu et al., 2002; Schumann et al., 2003; Sparkes et al., 2003). PEX2 was also found to be involved in developmental pathways regulated by the DET1 protein (Hu et al., 2002). Electron microscopy of pex10 null embryos revealed defects in the formation of lipid bodies, protein bodies, and the endoplasmic reticulum, in addition to the lack of peroxisomes (Schumann et al., 2003), although it is hard to distinguish whether these deficiencies are direct or indirect results from the loss of PEX10 function. Owing to embryo lethality of the knockout alleles, clear evidence showing the involvement of these RING peroxins in Arabidopsis peroxisome biogenesis and their roles in later stages of development has yet to be presented.
Mutations in PEX12 lead to failure of matrix protein import in yeast and mammals and result in the Zellweger syndrome, a lethal neurological disorder in humans (Gould and Valle, 2000). To determine the role of PEX12 in plant cellular functions and to address its role in plant development, we characterized Arabidopsis PEX12 and identified mutants deficient in this gene. Here, we report the subcellular localization of the Arabidopsis PEX12 protein and analysis of pex12 mutants generated by T-DNA insertion and two independent strategies of RNA interference (RNAi). This work establishes an essential role for Arabidopsis PEX12 in peroxisome biogenesis and in plant growth and development.
RESULTS
Null Mutants of PEX12 Cease to Develop during Early Embryogenesis
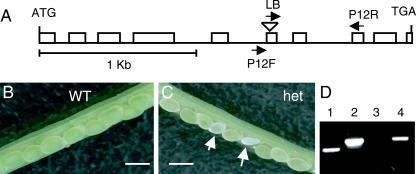
AtPEX12 (At3g04460) is a single-copy gene encoding a putative protein of 44 kD. It shares approximately 27% protein sequence identity with its yeast and mammalian orthologs and contains a C5-type RING finger motif with five conserved Cys (Fig. 1), which is different from the C3HC4-type RING found in AtPEX2 and AtPEX10. To study the function of PEX12 in plant development, we first identified from the Salk Arabidopsis T-DNA knockout collection (Alonso et al., 2003) a line containing a T-DNA insertion in the sixth exon of PEX12 (SALK_13612; Fig. 2A). We were unable to identify plants homozygous for the insertion after screening more than 200 T3 progeny from T2 heterozygous plants. Immature siliques of wild-type plants contained green seeds (Fig. 2B) that turned brown at maturity and, occasionally, one or two aborted seeds. However, immature siliques from heterozygous plants had approximately 22% white seeds (Fig. 2C), which became purplish and shriveled as the siliques matured. These data suggest that the pex12 insertion allele is segregating as a single recessive mutation, which may confer embryonic lethality in homozygotes.
Figure 1.
Amino acid sequence alignment of PEX12 from Arabidopsis and other species. Arabidopsis, AtPEX12 (Q9M841); Homo sapiens, HsPEX12 (O00623); Pichia pastoris, PpPEX12 (Q01961). Sequences were aligned using Megalign from DNASTAR. Underlined are putative transmembrane domains. Boxes indicate the conserved Cys residues in the C-terminal RING finger motif.
Figure 2.
PEX12 gene structure and knockout phenotypes. A, Schematic representation of PEX12, showing exons as white boxes and introns as single lines. The triangle indicates the position of the T-DNA insertion. Primers shown were used for genotyping. B and C, Immature siliques of wild-type and heterozygous plants. Arrows point to mutant seeds. Bars = 0.5 mm. D, PCR genotyping of embryo DNA. Lanes 1 to 2, Normal embryos; lanes 3 to 4, abnormal embryos. Primers used for 1 and 3, P12F/P12R; for 2 and 4, LB/P12R.
To confirm that the abnormal embryos indeed were homozygous for the T-DNA insertion allele, we dissected both abnormal and normal embryos (torpedo stage) from the seeds and pooled approximately 10 embryos of each type for DNA extraction. PCR analysis demonstrated that normal embryos contained both the insertion and the wild-type alleles (Fig. 2D, lanes 1 and 2), suggesting that they were composed of both wild-type and heterozygous embryos. However, only the T-DNA insertion-specific band was amplified from the abnormal embryos (Fig. 2D, lanes 3 and 4), indicating that these embryos were homozygous knockouts.
To determine which step of embryo development was disrupted by the pex12 null mutation, we analyzed developing seeds in heterozygous plants using Nomarski optics. Knockout embryos of pex12 showed retarded growth and in most cases were able to develop to the heart stage, but failed to grow further into torpedo and mature embryos (Fig. 3, C and D). Occasionally, the mutant embryos ceased to grow at globular or early torpedo stage (data not shown). Our data suggest that homozygous pex12 null embryos were delayed in growth and eventually stopped developing during early stages of embryogenesis.
Figure 3.
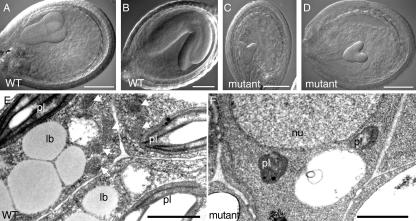
Characterization of pex12 knockout embryos. A to D, Nomarski optics of developing seeds. A and C are from the same silique, as are B and D. E and F, Electron micrographs of wild-type (E) and mutant (F) embryos. Bars in A to D = 0.1 mm; in E and F = 1 μm. lb, Lipid body; pl, plastid; nu, nucleus; WT, wild type. White arrows in E point to peroxisomes.
We performed electron microscopic analysis with the knockout embryos to determine the effect of PEX12 deficiency at the ultrastructural level. Embryos subjected to examination were from heterozygous plants 6 d after fertilization, when wild-type embryos are usually at the torpedo stage of embryogenesis. Wild-type embryos contained well-developed peroxisomes (also called glyoxysomes in seeds), lipid bodies, and plastids (Fig. 3E). The pex12 null embryos, however, lacked peroxisomes, contained small and underdeveloped plastids, and were missing characteristic lipid body structures (Fig. 3F). Based on these observations, we conclude that PEX12 is required at least for the formation of the peroxisome. The impairment of other subcellular structures, such as lipid bodies and plastids, could be an indirect consequence of the loss of PEX12. For example, lipid bodies are physically associated with peroxisomes during seed germination, providing them with fatty acid substrates. It is thus conceivable that the function and presence of lipid bodies can be affected by a feedback mechanism when no functional peroxisomes are present to carry out lipid metabolism; however, a direct role of PEX12 in lipid body formation cannot be completely ruled out.
PEX12-CFP Is Localized to the Peroxisome and Able to Rescue the pex12 Knockout Plants
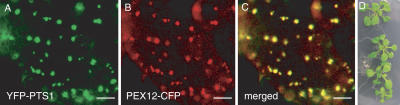
To determine the subcellular localization of AtPEX12, we analyzed wild-type Arabidopsis plants coexpressing yellow fluorescent protein (YFP)-peroxisome targeting signal type 1 (PTS1) and PEX12-cyan fluorescent protein (CFP) by fluorescence microscopy. PTS1, which is composed of Ser, Lys, and Leu, is widely used as a tag to localize proteins to the peroxisome in diverse systems. It is the targeting sequence for the majority of known peroxisomal matrix proteins and is recognized by the PTS1 receptor PEX5 (Subramani et al., 2000). PEX12-CFP displayed a punctate pattern of fluorescence, which is typical for peroxisomes, and was colocalized with YFP-PTS1 (Fig. 4, A–C), suggesting that PEX12 is also peroxisomal in Arabidopsis.
Figure 4.
Subcellular localization of PEX12-CFP and its functional complementation of the pex12 null mutant. A to C, Localization of PEX12-CFP to the peroxisome. Cells shown are from leaf tissue of a plant coexpressing YFP-PTS1 and PEX12-CFP. Bars = 10 μm. D, Rescue of the pex12 knockout plant by PEX12-CFP. Top seedling, Wild type; bottom seedlings, T3 progeny from three independently rescued pex12 knockout lines.
We also transformed plants heterozygous for the knockout allele with the PEX12-CFP fusion construct driven by the 35S constitutive promoter. T2 plants were screened for homozygosity for the pex12 knockout allele and overexpression of the PEX12-CFP protein. The embryo-lethal phenotype of the knockout plants was rescued by the transgene (Fig. 4D), providing further evidence that the lack of a functional PEX12 gene was responsible for the lethal phenotype and suggesting that the PEX12-CFP fusion protein functions properly.
Virus-Induced Gene Silencing of PEX12
The embryo-lethal phenotype of the pex12 knockout plants prevented us from further elucidating the potential roles of PEX12 in peroxisome biogenesis and in later stages of development; thus, mutants with reduced levels of PEX12 were needed. To this end, two RNAi strategies were employed to knock down the expression of PEX12: (1) infecting plants with viruses containing a fragment of the PEX12 coding sequence and (2) making transgenic plants stably expressing a PEX12 double-stranded RNAi (dsRNAi) construct.
A gene-silencing system based on the bipartite geminivirus cabbage leaf curl virus (CbLCV) was recently developed that can efficiently induce diffusible, homology-based systemic silencing of endogenous genes in Arabidopsis (Turnage et al., 2002; Robertson, 2004). This system is composed of two small circular viral DNA genomes: CbLCV A and CbLCV B. To attenuate the viral symptom, the coat protein-encoding AR1 gene was deleted from the A genome and replaced by a fragment of the gene to be silenced. The B genome carries the movement protein for systemic infection (Turnage et al., 2002). To silence the PEX12 gene, a 247-bp cDNA fragment of PEX12 was cloned into the CbLCV A vector in sense or antisense orientation. Viruses containing the silencing constructs were bombarded into Arabidopsis plants in the YFP-PTS1 background. Leaf tissue from infected plants was observed under the fluorescent microscope 3 to 4 weeks after bombardment, when genes encoded by the viruses are expressed at high levels in new leaves. As a control, we also bombarded some plants with viruses containing the CHLORATA42 (CH42) gene. CH42 encodes the small subunit of the chloroplast magnesium-chelatase (Koncz et al., 1990) and confers an albino phenotype when silenced, owing to the lack of chlorophyll production (Supplemental Fig. 1). This control is used as an indicator for massive viral replication and systemic movement and therefore serves as a guide to determine the time for RNA and fluorescent microscopic analyses.
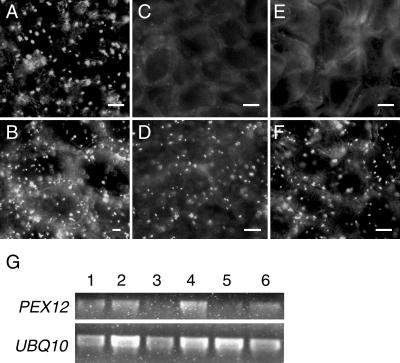
Plants infected by both the sense and antisense PEX12-silencing constructs exhibited a strong reduction in the number of peroxisomes as well as peroxisomal fluorescence of the YFP-PTS1 protein in new leaves (Fig. 5, C and E) compared to old leaves (Fig. 5, D and F), whereas plants infected by the empty vector control did not show a significant difference between old and new leaf tissue (Fig. 5, A and B). Reverse transcriptase (RT)-PCR analysis was subsequently performed to determine the expression level of PEX12 in these tissues. Figure 5G shows that, in plants bombarded with the PEX12-silencing constructs, the transcript level of PEX12 in the new tissue was significantly lower than in the old tissue, suggesting that PEX12 is required for peroxisome biogenesis in leaves. Despite the fact that the CbLCV virus used in this work was attenuated by removal of the AR1 gene, plants still displayed mild viral symptoms after infection, such as wrinkled leaves, stunted growth, and lack of inflorescence. As such, the mutant phenotypes caused by PEX12 silencing in adult plants could not be unambiguously determined by this approach.
Figure 5.
Virus-induced gene silencing of PEX12. A to F, YFP-PTS1 fluorescence in plants infected by virus containing the CbLCV empty vector (A and B), vector containing a fragment of PEX12 in the sense orientation (C and D), and vector containing the antisense fragment of PEX12 (E and F). A, C, and E, Leaves from new growth; B, D, and F, old leaves of the same plants. Bars = 10 μm. G, RT-PCR analysis of PEX12 and UBIQUITIN10 (UBQ10) transcripts. Lanes 1 to 6 are PCR products amplified from RNA from A to F.
Silencing of PEX12 by dsRNAi in Transgenic Plants
To elucidate more clearly the impact of PEX12 on plant development, a second RNAi approach was performed simultaneously. Given the lethal phenotype of the null mutants, we aimed to generate weaker and partial loss-of-function mutants of PEX12 by using a smaller fragment of the gene, rather than the entire coding region, in the RNAi construct. The same 247-bp cDNA fragment of PEX12 was cloned as inverted repeats into the dsRNAi vector pFGC5941, obtained from the Arabidopsis Biological Resource Center (ABRC). Plants containing YFP-PTS1 were transformed with the pFGC5941-derived PEX12-silencing construct under the control of the 35S promoter.
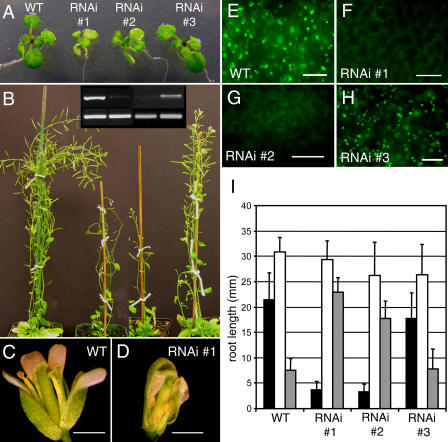
Approximately 20% of the 50 T1 transgenic plants showed fairly strong reduction of the PEX12 mRNA. The level of PEX12 suppression correlated with the severity of the mutant phenotypes in both T1 plants and their progeny. Compared with wild-type plants and RNAi plants with a subtle reduction of PEX12 expression (such as RNAi no. 3), RNAi plants with stronger reduction of PEX12 gene expression (such as RNAi nos. 1 and 2) were smaller and paler green, and developed more slowly (Fig. 6, A and B). The mutants were also less fertile with smaller gynoecia and shorter stamens in many mutant flowers (Fig. 6, C and D), and contained reduced numbers of peroxisomes and significantly weakened YFP-PTS1 fluorescence in peroxisomes (Fig. 6, E–H).
Figure 6.
Phenotypes of PEX12 RNAi plants. A and B, Two-week-old (A) and 8-week-old (B) plants. Inset in B, RT-PCR (35 cycles) of RNA from leaves of (lanes 1–4) wild-type and PEX12 RNAi number 1, number 2, and number 3 plants. Bands shown are PCR products from PEX12 (top) and UBIQUITIN10 (bottom). C and D, Flower comparison of wild-type and RNAi number 1 plants. E to H, YFP-PTS1 fluorescence in wild-type and PEX12 RNAi plant leaves. Bars in C and D = 1 mm. Bars in E to H = 20 μm. I, Root length measurements of 7-d-old light-grown RNAi seedlings on 0% (black bars) or 1% (white bars) Suc, or on plates containing 1% Suc plus 20 μm IBA (gray bars). Each error bar represents sd from approximately 25 samples.
We also tested sugar dependence and indole-3-butyric acid (IBA) response of the RNAi plants. Through β-oxidation and the glyoxylate cycle, oilseed peroxisomes participate in lipid mobilization, which provides seedlings with energy to grow. As such, peroxisomal mutant seedlings of oilseed species such as Arabidopsis usually grow poorly in the absence of exogenous sugar. Mutants defective in β-oxidation are also resistant to the inhibitory effect of IBA on root elongation because mutant peroxisomes cannot efficiently convert IBA into IAA inside this organelle (Zolman et al., 2000). T3 seeds were plated on medium with or without 1% Suc and on medium containing 20 μm IBA combined with 1% Suc; root length was measured after 7 d of growth in the light. RNAi plants numbers 1 and 2 exhibited defects in root elongation without supplemental sugar, a phenotype that was mostly rescued by Suc (Fig. 6I). In addition, these mutants were also less responsive to IBA (Fig. 6I), which is indicative of a deficiency in β-oxidation.
Taken together, our data support the role of PEX12 as a critical player in peroxisome formation and function, and in plant vegetative and reproductive growth.
Expression Profile of AtPEX12
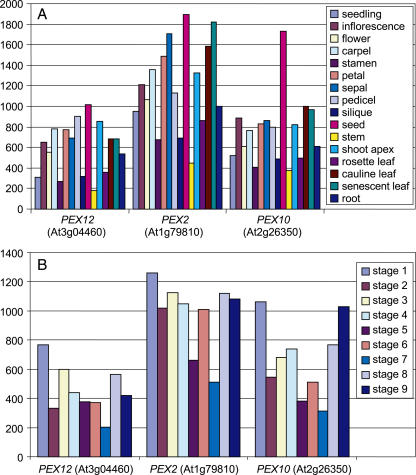
The essential role of PEX12 throughout Arabidopsis development led us to examine its expression pattern in the plant. An RT-PCR analysis of the PEX12 transcript suggested that this gene was ubiquitously expressed in young seedlings, leaves, roots, and flowers (data not shown). To assess its expression more completely, we used GENEVESTIGATOR, an Arabidopsis microarray gene expression database and analysis toolbox (Zimmermann et al., 2004; https://www.genevestigator.ethz.ch), to search for expression of the Arabidopsis PEX12 gene in various organs and at several developmental stages. The microarray data, based on experiments with the Arabidopsis full-genome chip (ATH1) arrays, showed that AtPEX12 was ubiquitously expressed and that its expression pattern correlated well with several other genes known to be required for peroxisome biogenesis, including the other two RING peroxin genes, PEX2 and PEX10 (Fig. 7). Consistent with their essential role in embryogenesis, all the three RING peroxins were most highly expressed in seeds (Fig. 7A). The transcript levels of these genes were also high during germination (Fig. 7B), when peroxisomes are needed for lipid metabolism, and in senescent plants (Fig. 7, A and B), in which leaf peroxisomes are transformed into glyoxysomes to convert membrane lipids into carbohydrates. All three PEX genes were also abundant in floral structures, namely, inflorescences, carpels, and pedicels (Fig. 7A), and at the stage when flowering is complete and siliques are formed (Fig. 7B, stage 8). These data are also in agreement with results from our study, which showed that flower formation and fertility were impaired when the expression of PEX12 was strongly reduced in Arabidopsis (Fig. 6). Interestingly, these three PEX genes were also strongly expressed in the shoot apex (Fig. 7A), where a specific function for peroxisomes has not been established, underlying their possible fundamental roles in this tissue.
Figure 7.
Expression patterns of the RING PEX genes in Arabidopsis. A, Expression in various plant organs. B, Expression at successive developmental stages, as defined by Boyes et al. (2001). Stage 1, Hypocotyls and cotyledons emerged from seed coat; stage 2, cotyledons completely open; stage 3, seedlings with two rosette leaves; stage 4, 10 rosette leaves; stage 5, first flower buds seen; stage 6, early flowering; stage 7, midflowering; stage 8, flowering completed; stage 9, senescent and ready for seed harvest. The y axis in both A and B indicates the level of gene expression. Data used for the analysis were retrieved from GENEVESTIGATOR (https://www.genevestigator.ethz.ch; Zimmermann et al., 2004).
DISCUSSION
We have presented evidence that PEX12 is a peroxisomal protein in Arabidopsis and is essential for peroxisome biogenesis and plant development. First, null pex12 embryos (SALK_13612) were slow growing and eventually aborted during early embryogenesis. Second, PEX12-CFP was localized to the peroxisome and complemented the lethal phenotype of the pex12 knockout mutants. In addition, YFP-PTS1 plants infected by the CbLCV virus carrying part of the PEX12 coding sequence displayed strong reduction of the number of peroxisomes and import of PTS1-containing matrix proteins. Furthermore, transgenic plants, in which PEX12 gene expression was partially reduced by a dsRNAi construct, showed partial deficiency in peroxisome biogenesis and function, a smaller stature, and reduced fertility. Finally, a search of the GENEVESTIGATOR microarray database revealed similar expression patterns of the three RING peroxins in some tissues, supporting the essential roles of these PEX genes in seed development, germination, and flower formation.
The two classic and best-known functions for plant peroxisomes are (1) lipid mobilization through β-oxidation and glyoxylate cycle during oilseed germination and senescence, and (2) photorespiration in photosynthetic leaves (Beevers, 1979). Recent data have shown that peroxisomes also play an essential role in seed development, a complicated process that requires the coordinated action of hundreds of genes with diverse functions (Tzafrir et al., 2003). One example of the essential role for peroxisomes in embryogenesis came from the study of acyl-CoA oxidases (ACX), a family of enzymes catalyzing the first step of β-oxidation in the peroxisome with various substrate specificities. The acx3 acx4 double mutant, defective in both ACX3 (specific for medium-chain acyl-CoAs) and ACX4 (specific for short-chain acyl-CoAs), was found to abort during embryo development (Rylott et al., 2003). Here, we show that disruption of AtPEX12 caused developmental arrest of embryos mostly at the heart stage. This result is consistent with previous findings that PEX2 and PEX10 are essential for embryogenesis of Arabidopsis (Hu et al., 2002; Schumann et al., 2003; Sparkes et al., 2003). Interestingly, developmental arrest in the knockouts of PEX2 (J. Fan and J. Hu, unpublished data) and PEX10 (Schumann et al., 2003; Sparkes et al., 2003) also takes place at around the heart stage of embryogenesis. It is conceivable that peroxisome function is fundamental to post-heart stage embryo development, which requires a high level of fatty acid metabolism to provide energy for rapid embryo elongation and differentiation. Alternatively, peroxisomes may be a source of yet-to-be determined signaling molecules that promote further embryogenesis beyond the heart stage. Finally, loss of peroxisome function may also result in the accumulation of toxic levels of lipids or reactive oxygen species detrimental to the embryo because peroxisomes contain catalase, peroxisome membrane-bound ascorbate peroxidase, superoxide dismutase, and other antioxidative enzymes (del Rio et al., 2002). Irrespective of the mechanism underlying the role of peroxisomes in embryogenesis, the three RING finger peroxins appear to comprise basic components of peroxisomes, whereas the role for some other PEX genes may not be so important in Arabidopsis. For example, a null mutation of AtPEX14 caused plants to be short and pale but still fertile (Hayashi et al., 2000).
Peroxisomal matrix proteins are mislocalized in the cytoplasm in yeast and animal cells with reduced function of PEX12, indicating that this protein is particularly required for protein import into peroxisomes (Chang et al., 1997; Okumoto et al., 2000). Mammalian cells lacking PEX12 also showed accumulation of the PTS1 receptor PEX5 at the cytosolic side of the peroxisome membrane, suggesting that PEX12 may mediate recycling of PEX5 (Dodt and Gould, 1996). PEX12 was found to interact with PEX5 and PEX10 in yeast and mammals (Chang et al., 1999; Okumoto et al., 2000; Agne et al., 2003; Eckert and Johnsson, 2003). PEX10 was found to interact with PEX4, an ubiquitin-conjugating enzyme-like protein associated with the peroxisomal membrane (Eckert and Johnsson, 2003), yet the biological consequence of this interaction is unknown. Although conflicting results have been shown as to whether PEX2 is physically associated with PEX10 and PEX12 (Okumoto et al., 2000; Agne et al., 2003; Eckert and Johnsson, 2003), it has been postulated that these three RING peroxins may form a subcomplex on peroxisome membranes active in mediating protein import (Purdue and Lazarow, 2001). The similar phenotypes caused by loss of function of each of these three genes in Arabidopsis support the notion that AtPEX2, AtPEX12, and AtPEX10 act closely during peroxisome biogenesis. However, it will be necessary to test for interactions among the three RING peroxins and between these proteins and other peroxins in Arabidopsis to elucidate the biochemical mechanism underlying the action of the RING-type PEX proteins in plants.
Electron microscopic analysis of an Arabidopsis pex10 null mutant demonstrated that, in addition to the lack of characteristic peroxisomes, the lethal embryos also contained small lipid bodies with a half-unit membrane studded with ribosomes and flat lipid body discs (Schumann et al., 2003). However, these abnormal lipid body structures were not apparent in the pex12 null embryos; rather, no characteristic lipid bodies were observed in the lethal embryos examined in this study. This discrepancy may be due to the fact that there is some difference between the functions of PEX12 and PEX10. Alternatively, the mutant embryos analyzed in our study might be at a more degenerating stage. More direct evidence is needed to firmly establish a role of PEX10 or PEX12 in the formation of organelles other than the peroxisome in plants.
The mechanism for peroxisome biogenesis in various organisms shares a significant degree of conservation. All of the Arabidopsis PEX genes that have been cloned and characterized show some level of functional similarity with their yeast counterparts. For example, the PEX2, PEX10, PEX14, and PEX16 proteins all target to peroxisomes (Hayashi et al., 2000; Hu et al., 2002; Schumann et al., 2003; Sparkes et al., 2003; Lin et al., 2004). PEX16 could partially complement a yeast pex16 mutant (Lin et al., 1999), and mutants of PEX5, PEX6, PEX7, and PEX14 exhibit defects in peroxisomal functions (Hayashi et al., 2000; Zolman et al., 2000; Zolman and Bartel, 2004; Woodward and Bartel, 2005). On the other hand, some level of divergence in peroxisome biogenesis has also been observed in different species. For example, the number of import pathways for matrix proteins and the capacity of the receptors seem to vary among species (Otera et al., 1998; Motley et al., 2000; Hayashi et al., 2005). In addition, a number of the yeast and mammalian PEX genes do not have sequence homologs in Arabidopsis; thus, some aspects of peroxisome biogenesis in plants may be unique. It is possible that, in addition to the orthologs of yeast and animal PEX proteins, plants also carry unique peroxins that function in plant-specific aspects of peroxisome biogenesis. Further genetic, bioinformatic, and proteomic analyses are needed to identify novel PEX proteins from plants.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants used in this study are of the Columbia background. Seeds were germinated on 1× Murashige and Skoog medium (Gibco), with or without 1% Suc, 20 μm IBA, and appropriate antibiotics, when necessary. Plants used for most experiments were grown with 16/8-h photoperiod under 80 to 100 μmol m−2 s−1 light conditions at 22°C. Plants used for bombardment in virus-induced gene silencing were grown in the same light intensity and temperature in short-day conditions with an 8/16-h photoperiod.
Genotyping of Arabidopsis Plants Containing a T-DNA Insertion in PEX12
PCR was performed with genomic DNA using Taq DNA polymerase (Promega) and conditions suggested by the manufacturer. The primers were as follows: left border (LB) from the T-DNA vector, 5′-GCGTGGACCGCTTGCTGCAACT-3′; P12F from intron 5 and exon 6, 5′-ATGCCAAGATAGATGGATACATCCTCAAGG-3′; and P12R from exon 8, 5′-GGAGGGTACACTGTTGGAGCTGATAATCTC-3′. P12F and P12R amplify a 620-bp product from the wild-type allele. LB and P12R amplify an insertion-specific product of 730 bp. An approximately 110-bp piece of DNA of unknown origin was also found at the insertion site, between the T-DNA and the PEX12 genomic DNA.
Light and Electron Microscopy
A Leica MZ125 dissecting microscope (W. Nuhsbaum) was used to observe siliques and floral structures, and a Zeiss Axiophot microscope (Carl Zeiss) was used for Nomarski optics of seeds. For Nomarski optics, fresh siliques harvested 6 to 9 d after pollination were dissected. Developing seeds were treated with clearing solution containing chloral hydrate:water:glycerol (8:2:1, v/v/v) and cleared for 1 to 4 h at room temperature or overnight at 4°C.
A Zeiss Axiophot microscope was used to visualize fluorescent proteins. For in vivo detection of YFP and CFP, leaf tissue was mounted in water and viewed with a YFP filter (excitation 500 ± 12.5 nm, emission 540 ± 20 nm) or a CFP filter (excitation 440 ± 10 nm, emission 480 ± 15 nm).
Abnormal and normal seeds from the same silique (6–7 d after flowering) were fixed separately in 2.5% glutaraldehyde, 0.1 m phosphate buffer, pH 7.2, at room temperature for 2 h, followed by a secondary fixation in 1% (w/v) OsO4 in the same buffer. Samples were dehydrated in a graded series of acetone and embedded in Spurr's epoxy resin. Ultrathin sections (70–90 nm) were cut by a MT-X ultramicrotome, stained with 2% uranyl acetate and lead citrate, and observed under a JEM-100CX II transmission electron microscope (JEOL).
Generating YFP-PTS1, PEX12-CFP, and PEX12 RNAi Plants
To make YFP-PTS1, a YFP-Ser-Lys-Leu fragment was amplified by PCR from the vector pEYFP-Peroxi (CLONTECH) and cloned into a vector derived from pPZP212 (Hajdukiewicz et al., 1994) and containing the 35S promoter. To clone PEX12-CFP, the coding region of AtPEX12 (At3g04460) was first amplified by RT-PCR from first-strand cDNA made from wild-type Col seedling mRNA, using primers At3g04460Fw (5′-AAGGATCCATCACTAACTAGAAGAAGAGA-3′) and At3g04460Rv (5′-ATGTCGACAGTGTCCTGAAACAACCTCC-3′). The resulting PCR fragment was then cloned into BamHI and SalI sites at the amino terminus of CFP (CLONTECH) in a binary vector derived from pPZP211 (Hajdukiewicz et al., 1994) and containing the 35S promoter.
To clone the PEX12 RNAi construct, the Arabidopsis vector pFGC5941 for dsRNA production was obtained from ABRC (stock no. CD3-447). A 247-bp fragment of PEX12 cDNA (position 364–611) was amplified by PCR using primers hpsipex12L (5′-GCTCTAGAGGCGCGCCGTTGTGTTACCGTATTTC-3′) and hpsipex12R (5′-CGGGATCCATTTAAATTCACTTGATGCATGTATC-3′). The PCR product was digested with AscI and SwaI, and ligated into pFGC5941 to generate plasmid pFGC5941-PEX12a. The same PCR product was digested again with BamHI and XbaI, and ligated into pFGC5941-PEX12a to generate plasmid pFGC5941-PEX12. As a result, the pFGC5941-PEX12 plasmid has a copy of the 247-bp fragment inserted into AscI to SwaI sites, and an inverted repeat of the fragment inserted into BamHI to XbaI sites.
All PCR amplifications were carried out using Pfu DNA polymerase (Stratagene) and protocols suggested by the manufacturer. Agrobacterium-mediated transformation of Arabidopsis plants was performed using the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on Murashige and Skoog plates containing 50 ng/μL kanamycin for YFP-PTS1, 90 ng/μL gentamycin for PEX12-CFP, or 10 ng/μL BASTA for PEX12 RNAi.
Virus-Induced Gene Silencing
A 247-bp fragment from PEX12 cDNA (position 364–611) was amplified by PCR using primers husi2L (5′-GCTCTAGAGTTGTGTTACCGTATTTC-3′) and husi2R (5′-GAAGATCTTCACTTGATGCATGTATC-3′). The PCR product was digested with XbaI and BglII and cloned into the pCPCbLCV.007 vector (Turnage et al., 2002) to generate plasmid p007-husi2.
A reverse copy of the 247-bp fragment was also generated by PCR using primers husi2aL (5′-GAAGATCTGTTGTGTTACCGTATTTC-3′) and husi2aR (5′-GCTCTAGACTTCACTTGATGCATGTATC-3′). The PCR product was digested with XbaI and BglII and cloned into the pCPCbLCV.007 vector (Turnage et al., 2002) to generate plasmid p007-husi2a.
Arabidopsis plants in the YFP-PTS1 background were grown in individual pots in short-day conditions and bombarded with an equal amount of each silencing construct DNA (in CbLCV A) and the pCPCbLCV.008 DNA (CbLCV B) as described in a previous study (Turnage et al., 2002). Each plant was bombarded at the age of 3 to 4 weeks old, according to the protocol provided by Turnage et al. (2002). Two to three plants were bombarded with each construct. The experiment was repeated three times. “Old” and “new” leaf tissue was collected separately from infected plants approximately 4 weeks after bombardment. “New leaves” were those from around the center of the rosette that emerged after bombardment, and the “old leaves” were older rosette leaves that were present at the time of the bombardment. Because of the distinct colors of silenced and nonsilenced leaves, the CH42-infected plants served as a guide to distinguish “new” from “old” tissue for microscopic and RT-PCR characterizations.
RT-PCR Analysis of PEX12 Transcripts in PEX12-Silenced Plants
Total RNA was extracted with TRIzol reagent (Invitrogen) and subjected to Reverse Transcription reaction (Gibco). The PEX12-specific primers PEX12F2 (5′-GCGAGATTGAGATTGAGGAAAGACAGTGCC-3′) from exon 3 and PEX12R (5′-GGAGGGTACACTGTTGGAGCTGATAATCTC-3′) from exon 8 amplify a 684-bp product from PEX12 cDNA. The ubiquitin-specific primers UBQ10-1 (5′-TCAATTCTCTCTACCGTGATCAAGATGCA-3′) and UBQ10-2 (5′-GGTGTCAGAACTCTCCACCTCAAGAGTA-3′) from the UBIQUITIN10 gene (At4g05320) amplify a cDNA product of approximately 320 bp. PCR conditions were as follows: 94°C for 3 min, followed by cycles (27 for Fig. 5 and 35 for Fig. 6) of 94°C for 45 s, 57°C for 45 s, 72°C for 1 min, and a final extension of 72°C for 7 min.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers Q9M841, O00623, and Q01961.
Supplementary Material
Acknowledgments
We thank Drs. Andreas Weber and Gregg Howe for their comments on the manuscript, and Ms. Karen Bird for editing. We are also grateful to Renato Perez and Deb Lin for technical assistance, Dr. Dominique Robertson for the CbLCV vectors, Dr. Pablo Cerdan for the UBQ10 primers, and the Salk Institute and ABRC for the PEX12 T-DNA insertion lines and the dsRNA vector pFGC5941.
This work was supported by the U.S. Department of Energy and Michigan State University start-up funds to J.H., and by the National Science Foundation and the Howard Hughes Medical Institute (funding to J.C.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.066811.
References
- Agne B, Meindl NM, Niederhoff K, Einwachter H, Rehling P, Sickmann A, Meyer HE, Girzalsky W, Kunau WH (2003) Pex8p: an intraperoxisomal organizer of the peroxisomal import machinery. Mol Cell 11: 635–646 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Beevers H (1979) Microbodies in higher plants. Annu Rev Plant Physiol 30: 159–193 [Google Scholar]
- Berteaux-Lecellier V, Picard M, Thompson-Coffe C, Zickler D, Panvier-Adoutte A, Simonet JM (1995) A nonmammalian homolog of the PAF1 gene (Zellweger syndrome) discovered as a gene involved in caryogamy in the fungus Podospora anserina. Cell 81: 1043–1051 [DOI] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Gorlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown LA, Baker A (2003) Peroxisome biogenesis and the role of protein import. J Cell Mol Med 7: 388–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Lee WH, Moser H, Valle D, Gould SJ (1997) Isolation of the human PEX12 gene, mutated in group 3 of the peroxisome biogenesis disorders. Nat Genet 15: 385–388 [DOI] [PubMed] [Google Scholar]
- Chang CC, South S, Warren D, Jones J, Moser AB, Moser HW, Gould SJ (1999) Metabolic control of peroxisome abundance. J Cell Sci 112: 1579–1590 [DOI] [PubMed] [Google Scholar]
- Charlton W, Lopez-Huertas E (2002) PEX genes in plants and other organisms. In A Baker, IA Graham, eds, Plant Peroxisomes. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 385–426
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- del Rio LA, Corpas FJ, Sandalio LM, Palma JM, Gomez M, Barroso JB (2002) Reactive oxygen species, antioxidant systems and nitric oxide in peroxisomes. J Exp Bot 53: 1255–1272 [PubMed] [Google Scholar]
- Dodt G, Gould SJ (1996) Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J Cell Biol 135: 1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert JH, Johnsson N (2003) Pex10p links the ubiquitin conjugating enzyme Pex4p to the protein import machinery of the peroxisome. J Cell Sci 116: 3623–3634 [DOI] [PubMed] [Google Scholar]
- Gould SJ, Valle D (2000) Peroxisome biogenesis disorders: genetics and cell biology. Trends Genet 16: 340–345 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Nishimura M (2003) Entering a new era of research on plant peroxisomes. Curr Opin Plant Biol 6: 577–582 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Nito K, Toriyama-Kato K, Kondo M, Yamaya T, Nishimura M (2000) AtPex14p maintains peroxisomal functions by determining protein targeting to three kinds of plant peroxisomes. EMBO J 19: 5701–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Yagi M, Nito K, Kamada T, Nishimura M (2005) Differential contribution of two peroxisomal protein receptors to the maintenance of peroxisomal functions in Arabidopsis. J Biol Chem 280: 14829–14835 [DOI] [PubMed] [Google Scholar]
- Hu J, Aguirre M, Peto C, Alonso J, Ecker J, Chory J (2002) A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science 297: 405–409 [DOI] [PubMed] [Google Scholar]
- Koncz C, Mayerhofer R, Koncz-Kalman Z, Nawrath C, Reiss B, Redei GP, Schell J (1990) Isolation of a gene encoding a novel chloroplast protein by T-DNA tagging in Arabidopsis thaliana. EMBO J 9: 1337–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Cluette-Brown JE, Goodman HM (2004) The peroxisome deficient Arabidopsis mutant sse1 exhibits impaired fatty acid synthesis. Plant Physiol 135: 814–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Sun L, Nguyen LV, Rachubinski RA, Goodman HM (1999) The Pex16p homolog SSE1 and storage organelle formation in Arabidopsis seeds. Science 284: 328–330 [DOI] [PubMed] [Google Scholar]
- Motley AM, Hettema EH, Ketting R, Plasterk R, Tabak HF (2000) Caenorhabditis elegans has a single pathway to target matrix proteins to peroxisomes. EMBO Rep 1: 40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen RT, Flynn CR, Trelease RN (2001) How are peroxisomes formed? The role of the endoplasmic reticulum and peroxins. Trends Plant Sci 6: 256–261 [DOI] [PubMed] [Google Scholar]
- Okumoto K, Abe I, Fujiki Y (2000) Molecular anatomy of the peroxin Pex12p: Ring finger domain is essential for Pex12p function and interacts with the peroxisome-targeting signal type 1-receptor Pex5p and a ring peroxin, Pex10p. J Biol Chem 275: 25700–25710 [DOI] [PubMed] [Google Scholar]
- Olsen LJ, Harada JJ (1995) Peroxisomes and their assembly in higher plants. Annu Rev Plant Physiol Plant Mol Biol 46: 123–146 [Google Scholar]
- Otera H, Okumoto K, Tateishi K, Ikoma Y, Matsuda E, Nishimura M, Tsukamoto T, Osumi T, Ohashi K, Higuchi O, et al (1998) Peroxisome targeting signal type 1 (PTS1) receptor is involved in import of both PTS1 and PTS2: studies with PEX5-defective CHO cell mutants. Mol Cell Biol 18: 388–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons M, Furuya T, Pal S, Kessler P (2001) Biogenesis and function of peroxisomes and glycosomes. Mol Biochem Parasitol 115: 19–28 [DOI] [PubMed] [Google Scholar]
- Powers JM, Moser HW (1998) Peroxisomal disorders: genotype, phenotype, major neuropathologic lesions, and pathogenesis. Brain Pathol 8: 101–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdue PE, Lazarow PB (2001) Peroxisome biogenesis. Annu Rev Cell Dev Biol 17: 701–752 [DOI] [PubMed] [Google Scholar]
- Robertson D (2004) VIGS vectors for gene silencing: many targets, many tools. Annu Rev Plant Biol 55: 495–519 [DOI] [PubMed] [Google Scholar]
- Rylott EL, Rogers CA, Gilday AD, Edgell T, Larson TR, Graham IA (2003) Arabidopsis mutants in short- and medium-chain acyl-CoA oxidase activities accumulate acyl-CoAs and reveal that fatty acid beta-oxidation is essential for embryo development. J Biol Chem 278: 21370–21377 [DOI] [PubMed] [Google Scholar]
- Schumann U, Wanner G, Veenhuis M, Schmid M, Gietl C (2003) AthPEX10, a nuclear gene essential for peroxisome and storage organelle formation during Arabidopsis embryogenesis. Proc Natl Acad Sci USA 100: 9626–9631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Brandizzi F, Slocombe SP, El-Shami M, Hawes C, Baker A (2003) An Arabidopsis pex10 null mutant is embryo lethal, implicating peroxisomes in an essential role during plant embryogenesis. Plant Physiol 133: 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S, Koller A, Snyder WB (2000) Import of peroxisomal matrix and membrane proteins. Annu Rev Biochem 69: 399–418 [DOI] [PubMed] [Google Scholar]
- Titorenko VI, Ogrydziak DM, Rachubinski RA (1997) Four distinct secretory pathways serve protein secretion, cell surface growth, and peroxisome biogenesis in the yeast Yarrowia lipolytica. Mol Cell Biol 17: 5210–5226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko VI, Rachubinski RA (2004) The peroxisome: orchestrating important developmental decisions from inside the cell. J Cell Biol 164: 641–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnage MA, Muangsan N, Peele CG, Robertson D (2002) Geminivirus-based vectors for gene silencing in Arabidopsis. Plant J 30: 107–114 [DOI] [PubMed] [Google Scholar]
- Tzafrir I, Dickerman A, Brazhnik O, Nguyen Q, McElver J, Frye C, Patton D, Meinke D (2003) The Arabidopsis SeedGenes Project. Nucleic Acids Res 31: 90–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B (2005) The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol Biol Cell 16: 573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Bartel B (2004) An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proc Natl Acad Sci USA 101: 1786–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Yoder A, Bartel B (2000) Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156: 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.