Abstract
Epigenetic changes accompanying plant cell dedifferentiation and differentiation are reported in 35S ribosomal DNA (rDNA) of tobacco (Nicotiana tabacum). There was a reduction of CG and CNG methylation in both intergenic and genic regions of the rDNA cistron in fully dedifferentiated callus and root compared to leaf. The rDNA hypomethylation was not random, but targeted to particular rDNA gene families at units that are clustered within the tandem array. The process of hypomethylation was initiated as early as 2 weeks after the callus induction and established epigenetic patterns were stably maintained throughout prolonged culture. However, regenerated plants and their progeny showed partial and complete remethylation of units, respectively. Nuclear run-on assays revealed a 2-fold increase of primary (unprocessed) ribosomal RNA transcripts in callus compared to leaf tissue. However, the abundance of mature transcripts in callus was elevated by only about 25%. Fluorescence in situ hybridization analysis of interphase nuclei showed high levels of rDNA chromatin condensation in both callus and leaf, with substantially less decondensed rDNA than is observed in meristematic root-tip cells. It is likely that the regions of the rDNA locus showing decondensation correspond to the clusters of hypomethylated units that occur in the tandem array at each locus. The data together indicate that the establishment of pluripotency and cell proliferation occurring with callus induction is associated with enhanced ribosomal RNA gene expression and overall rDNA hypomethylation, but is not associated with material-enhanced relaxation of chromatin structure (decondensation) at rDNA loci.
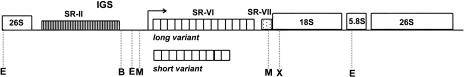
In higher eukaryotes, a large percentage of the nuclear genome is composed of ribosomal DNA (rDNA) coding for ribosomal RNA (rRNA; Hemleben and Zentgraf, 1994; Pikaard, 2000). There are variable numbers of rRNA genes in different plant species (between 1,000 to more than 50,000 genes) where they form multigene families in long tandem arrays at one or several chromosomal loci. The genic regions of the 35S rDNA-repeating unit (18S, 26S, and 5.8S subunits) are highly conserved with respect to length and nucleotide sequence; in contrast, the intragenic spacers (ITS) and the intergenic spacers (IGS) show high variability between species (Fig. 1). In all eukaryotes, including plants, the genes are transcribed using polymerase I.
Figure 1.
Restriction enzyme map of tobacco major rDNA units. Individual IGS subregions are termed as SR-II (containing C subrepeats), SR-VI (containing A1/A2 subrepeats), and SR-VII. Restriction enzymes delimiting the regions analyzed were as follows: B, BstNI; E, EcoRV; M, MboI; X, XbaI. Positions of target sites for methylation-sensitive enzymes are given in Table I. Distances are approximately to scale.
In plants, transcription is initiated from a promoter located in the IGS. There are subrepeated regions upstream and downstream of a transcription starting site that have been proposed to have regulatory function (Flavell et al., 1988; Sardana et al., 1993; Komarova et al., 2004). The primary transcript is of variable length (6–9 kb) and is processed into mature 18S, 5.8S, and 26S RNA by excision of ITS1 and ITS2 and a transcribed part of the IGS (called externally transcribed spacer [ETS]). Maturation of primary transcript, post-transcriptional modifications, and ribosome assembly occur in the nucleolus. The regulation of rRNA gene expression occurs through the suppression of whole loci (termed nucleolar dominance) and at genes within the array. Large numbers of repeats are not transcribed and are packed into transcriptionally inactive heterochromatin. Formation of rDNA heterochromatin is believed to be under epigenetic control mediated by modifications of DNA and histones. In mammals, cytosine methylation of a single site within the promoter has been shown to be important for transcription repression (Santoro and Grummt, 2001). In plant hybrids, the derepression of silent genes has been observed after treatment with drugs modulating cytosine methylation and histone acetylation at both molecular (Chen and Pikaard, 1997) and cytogenetic levels (Viera et al., 1990; Castilho et al., 1999).
Here we study rDNA methylation and activity associated with callus culture in tobacco (Nicotiana tabacum). Tobacco is an allotetraploid derived from ancestors of Nicotiana sylvestris and Nicotiana tomentosiformis. The rRNA genes show evidence of concerted evolution and have homogenized units of a tobacco-specific type (Volkov et al., 1999; Lim et al., 2000). In most tobacco varieties, there are two large gene families with closest similarity to the N. tomentosiformis parent and a single minor family of N. sylvestris origin (Kovarik et al., 2004). The tobacco-specific units with similarity to N. tomentosiformis are hypomethylated relative to the unconverted units of N. sylvestris origin (Lim et al., 2000). Compared to other repeats, including 5S rDNA, 35S rDNA is less methylated in tobacco (Kovarik et al., 2000). The copy number (Lim et al., 2000), chromosomal positions (Kenton et al., 1993; Moscone et al., 1996), and transcription starting site (Fan et al., 1995) of rRNA genes in tobacco are known. There is a single locus on chromosome T3 and three loci on chromosomes S10, S11, and S12.
rRNA gene transcription varies with demand for ribosome production and protein synthesis. The physiological changes in rRNA gene expression are reflected by changes in nucleolar volume. For example, nucleolar activity was found to increase in G2 and decrease during differentiation in onion (Cerdido and Medina, 1995). In Triticum aestivum root-tip meristematic cells, nucleolar volume declines with elevated growing temperature (and inferred metabolic activity), presumably because the nucleolus maintains a smaller granular component in cells of higher activity (Glyn et al., 1995). Exogenously applied cytokinin, but not other hormones, has been shown to stimulate polymerase I transcription of rRNA genes in Arabidopsis (Arabidopsis thaliana; Gaudino and Pikaard, 1997). Arabidopsis flower and root tissue contain twice as much 5S rRNA than leaf tissue (Mathieu et al., 2003).
Plant cell cultures contrast to those of animal cultures in that they can give rise to regenerant organisms (for review, see Grafi, 2004). Hence, plant cultures are highly suitable models for genetic studies of cell dedifferentiation and differentiation. In calli, hypomethylation and hypermethylation have been shown to influence single-copy and repeated gene loci for both endogenous and transgene sequences (Arnholdt-Schmitt et al., 1995; Smulders et al., 1995; Olhoft and Phillips, 1999; Jaligot et al., 2000; Kaeppler et al., 2000). However, few studies have demonstrated a direct correlation between methylation levels and gene expression activity (Mitsuhara et al., 2002; Fojtova et al., 2003; Avivi et al., 2004). Changes in rRNA gene methylation through cell dedifferentiation have been observed in some plant systems (Anderson et al., 1990; Vyskot et al., 1993), but not in others (Avivi et al., 2004; Komarova et al., 2004). It is unknown whether methylation increase/decrease affects the whole rDNA cistron or only specific parts, and no attempts have been made to measure transcription activity in association with methylation levels.
In this study, we have studied cytosine methylation of the tobacco 18S, 5.8S, and 26S rRNA genes in leaf material after induction and proliferative phases of callus growth and in regenerated plants and their progeny. Methylation changes were monitored across the whole rDNA cistron using a set of methylation-sensitive enzymes. Organization of rDNA chromatin at interphase was studied by fluorescence in situ hybridization (FISH). The changes in epigenetic patterns were correlated with expression measured at both primary and mature transcript levels.
RESULTS
Methylation Analysis in Plants, Derived Calli, and Roots
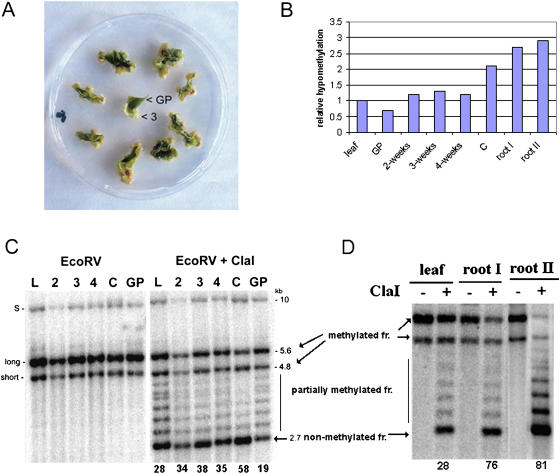
We isolated DNA from leaf tissue, different stages of callus development, green tissue that did not give rise to callus, and root tips of pot-grown seedlings (Fig. 2). Methylation was analyzed in the IGS by Southern-blot hybridization using the 18S rDNA probe. Three prominent hybridization bands after EcoRV (methylation-insensitive enzyme) digestion (at 10-, 5.6-, and 4.8-kb; Fig. 2C, left) are consistent with the existence of three rRNA gene families in allotetraploid tobacco, two major families being derived from the N. tomentosiformis parental unit and a third minor family of N. sylvestris origin that is in low abundance (Kovarik et al., 2004). Combined EcoRV and ClaI digestion revealed that units from the 5.6-kb band, and to a lesser extent the 4.8-kb band, were digested in all tissues. The 10-kb band, which is closely similar to a band in N. sylvestris, remained largely undigested due to an absence of restriction sites (Volkov et al., 1999). However, there were notable differences in the extent of ClaI digestion at the other bands between tissues. The parental leaf and the undifferentiated green tissue (GP) showed a ladder of ClaI fragments. These arise from variable levels of methylation in the regularly spaced ClaI sites (at approximately 270-bp intervals) in the A1/A2 subrepeats found in the IGS (Volkov et al., 1999). In dedifferentiating tissue, mature callus, and roots (Fig. 2, C and D), the number of ClaI-undigested units was reduced while the 2.7-kb fragment, corresponding to unmethylated ClaI sites, became stronger. Hybridization ladders of fragments were less pronounced in calli and roots compared to leaf and GP tissues, suggesting considerable hypomethylation of A1/A2 subrepeats.
Figure 2.
Hypomethylation of rDNA units in dedifferentiated callus and root tip cells. A, A single leaf cut into several pieces was laid onto callus induction medium. At 2-, 3-, and 4-week intervals, the dedifferentiated cells formed at the edges of green tissue were collected and DNA extracted. The nondedifferentiated green tissue (GP) was analyzed after 3 weeks of cultivation. “3”, 3-week callus tissue from which DNA was extracted. B, Graph showing relative hypomethylation of rDNA units in tobacco tissues. The data are taken from hybridization experiments in C and D. The hypomethylation is expressed as the ratio of signal in nonmethylated fraction (2.7 kb) to total signal (%). The value obtained for leaf was arbitrarily chosen as 1. Abbreviations are as in C. C, Methylation changes in rDNA units during callus formation. DNAs digested with EcoRV alone or in combination with methylation-sensitive ClaI are shown. Southern-blot hybridization of restricted genomic DNA was carried out with the 18S rDNA probe to reveal methylation of the IGS SR-VI subregion. The 5.6- and 4.8-kb EcoRV/ClaI hybridization bands represent molecules with fully methylated A1/A2 subrepeats; a ladder of fragments in the 3.1- to 4.8-kb region represents partially methylated molecules. The 2.7-kb band (arrow) is indicative of demethylated ClaI sites proximal to the18S gene. Note that the 2.7-kb band becomes stronger in the course of callus proliferation. DNA from the green part of nondedifferentiating tissue was slightly less digested with ClaI than the original leaf. L, Original leaf; 2, 3, 4, dedifferentiating tissue 2, 3, and 4 weeks, respectively, after passage of the leaf explants onto callus-inducing medium; C, fully dedifferentiated callus; GP, green tissue; S, rDNA family derived from N. sylvestris; long, short, rDNA families derived from N. tomentosiformis. Numbers below lanes indicate intensities of hybridization signals in nonmethylated 2.7-kb fractions as a percentage of total signal in lanes. D, Methylation analysis of root and leaf rDNA. Southern-blot hybridization was carried out as in C. DNAs of leaf, 10-mm (I), and 3-mm root tips (II) were isolated from a single plant. Quantification of signals was carried out as in C.
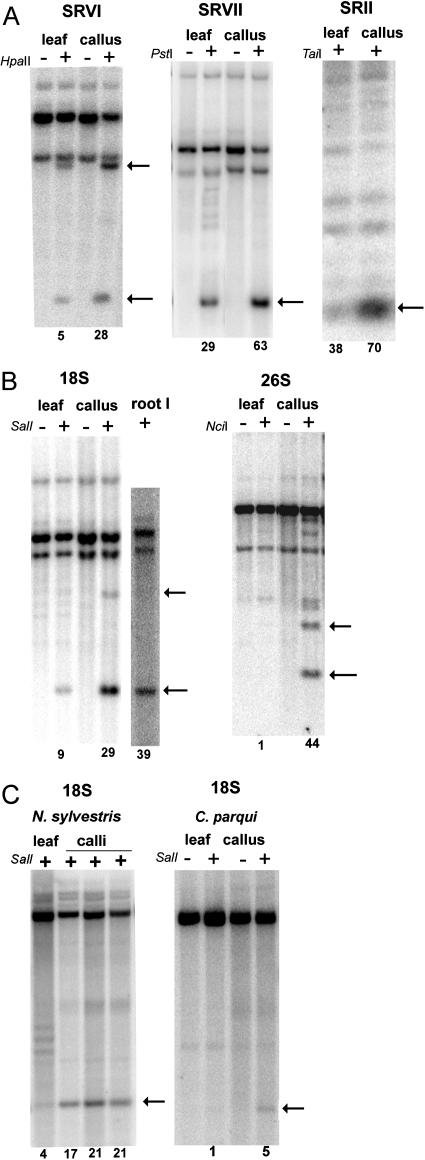
Methylation of different subregions of the rDNA cistron was analyzed with a range of restriction enzymes and probes (Table I; Fig. 3). The sites in the ETS were studied using HpaII and PstI enzymes; methylation in the nontranscribed region was assayed using the TaiI enzyme (Fig. 3A). It is evident that bands corresponding to unmethylated molecules (indicated by arrows) were stronger in lanes loaded with callus DNA compared to lanes loaded with leaf DNA. The enhanced digestibility of callus rDNA with methylation-sensitive enzymes was also observed with SalI and NciI enzymes that have recognition sites in the 18S and 26S genes, respectively (Fig. 3B). In general, the digestibility of sites in the genic region was lower than in the IGS. Nevertheless, rDNA from calli was consistently more efficiently cleaved with methylation-sensitive enzymes than rDNA from leaf.
Table I.
Enzymes and probes used for the analysis of cytosine methylation in 35S rDNA
| Region Analyzed | Enzyme Limiting the Regiona | Methylation-Sensitive Enzyme | DNA Probe | No. of Sites/Region Analyzed |
|---|---|---|---|---|
| IGS-SR VI | EcoRV | ClaI (ATmCGAT)b | 18S | 4–7c |
| IGS-SR VI | MboI | HpaII (mC mCGG) | A1/A2 | 1 |
| IGS-SR VII | EcoRV | PstI (mCTGmCAG) | 18S | 1c |
| IGS-SR II | BstNI | TaiI (AmCGT) | 26S | 17 |
| 18S | EcoRV | SalI (GTmCGAC) | 18S | 1 |
| 26S | - | XhoI (CTmCGAG) | 26S | 1 |
| 26S | BstNI | NciI (CmCSGG) | 26S | 6 |
Enzymes are not sensitive to the presence of 5-methylC (see Fig. 1).
Recognition sequences are in parentheses (m, sensitive cytosine).
Sites are present in the N. tomentosiformis-derived gene families, but not in those of N. sylvestris origin.
Figure 3.
Southern-blot hybridization analysis of cytosine methylation in fully dedifferentiated callus and parental leaf of N. tabacum (A and B), N. sylvestris, and C. parqui (C). DNAs were predigested with methylation-insensitive restriction enzymes (Table I) to delimit intergenic (A) and genic (B and C) subregions of the rDNA cistron (Fig. 1). The digestion with methylation-sensitive enzyme is indicated with a plus (+). HpaII was used in combination with MboI methylation-insensitive restriction endonuclease; PstI and SalI with EcoRV; TaiI and NciI with BstNI. Arrows indicate fragments corresponding to molecules with nonmethylated restriction sites. The signals in bands were quantified by a phosphor imager and the relative proportion of units with nonmethylated sites is shown below the lanes.
The methylation changes in the rDNA locus may relate to somaclonal variation (Kaeppler et al., 2000) arising as a consequence of culture and occurring in an unpredictable manner. To examine this hypothesis, we analyzed methylation of calli established from several plants of two tobacco varieties, two diploid (N. sylvestris and Cestrum parqui) and one tetraploid (Tragopogon mirus) species (Fig. 3C; Table II). In general, the within-species variability was low and there were no significant differences between different calli derived from plants of the same variety. However, there were differences in methylation levels between tobacco cultivars and the diploid species, possibly reflecting a variable number of rRNA genes in the cell (Flavell et al., 1988). Generally, species and varieties with less genes had higher proportions of hypomethylated fraction. For example, tobacco cv SR-1 in which one gene family was lost (Skalická et al., 2003) had significantly less methylation of rDNA than cv Vielblattriger.
Table II.
rDNA cytosine methylation in leaf and derived calli
The calli were established from different plants and cells were cultivated for at least 6 weeks. In all experiments, the growth media contained the same concentration of auxin and cytokinin. N, Number of independent experiments; nd, not determined.
| Species | Methylation of the 18S Genea | Methylation of the IGSb | |||
|---|---|---|---|---|---|
| N | Leaf | Callus | Leaf | Callus | |
| N. tabacum cv Vielblattriger | 5 | 91 ± 5.0 | 71 ± 4.7 | 72 ± 8.2 | 42 ± 10.0 |
| N. tabacum cv SR-1 | 4 | 76 ± 3.0 | 63 ± 3.0 | 45 ± 2.4 | 33 ± 5.7 |
| N. sylvestris | 3 | 96 ± 0.6 | 80 ± 2.4 | 96 ± 0.3 | 90 ± 0.6 |
| C. parqui | 2 | 98 ± 0.3 | 94 ± 0.3 | Nd | nd |
| T. mirus | 1 | 98 | 95 | Nd | nd |
The data are based on methylation of a conserved SalI site close to the 5′-end of the 18S gene.
The data are based on methylation of ClaI and MspI sites in the ETS of N. tabacum and N. sylvestris, respectively.
Hypomethylated Units Tend to Cluster within the Tandem Array
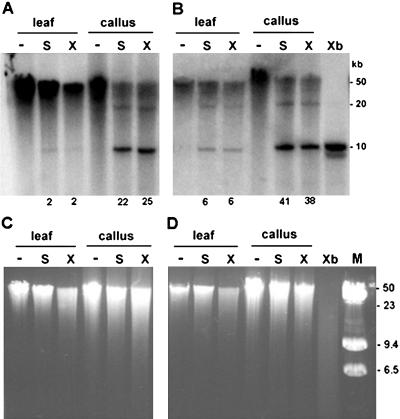
The rRNA genes are organized as long tandem arrays of homogeneous units. We were interested in determining whether the hypomethylated units are randomly distributed across the arrays or whether they are clustered. We digested DNA of high molecular mass (approximately 50 kb) with methylation-sensitive SalI and XhoI, both having a single recognition site per unit, and hybridized the blots with the 18S genic probe (Fig. 4). Liberation of an approximately 10-kb unit monomer would suggest existence of consecutive units with demethylated restriction sites in the genome, while higher molecular mass fragments corresponding to dimers (20 kb) and trimers (30 kb) would indicate interspersion of methylated units with nonmethylated units. Signal in the high molecular mass region (>50 kb) represents at least five methylated units in tandem. In both cv Vielblattriger (Fig. 4A) and SR-1 (Fig. 4B) calli, about 30% of high molecular mass material was digested with both SalI and XhoI into the approximately 10-kb monomer. The faint 20-kb band, together with near absence of 30- and 40-kb bands, shows that unmethylated units are not frequently interspersed with methylated units. In leaf, unmethylated monomers were also visible, but their relative abundance was reduced compared to calli. In conclusion, the hypomethylated units appear to be concentrated within the tandem array and probably locate to distinct chromosomal loci. Different chromatin domains possess differential methylation in tobacco (Kovarik et al., 2000) and Arabidopsis (Mathieu et al., 2002). The extent of digestion of total genomic DNA with both methylation-sensitive enzymes was comparable for leaf and callus as deduced from fluorescence signals in ethidium bromide-stained gels (Fig. 4, C and D).
Figure 4.
Large-scale organization of hypomethylated units. The DNA of approximately 50 kb was digested with methylation-sensitive SalI (S lines) and XhoI (X lines) enzymes, separated in 0.6% agarose gel, and hybridized with the 18S rDNA probe. Digestion with CG methylation-insensitive XbaI enzyme was used to reveal a repeat monomer of approximately 10 kb (Xb line). Note that callus DNAs from both Vielblattriger (A) and SR-1 (B) varieties contain significant hybridization signal in a monomeric fraction. Minus (−) indicates nondigested DNA. Numbers below lanes indicate intensities of hybridization signals in nonmethylated, approximately 10-kb fractions as a percentage of total signal in lanes (6–50 kb). Ethidium bromide gels prior to DNA transfer are shown in C and D to reveal global genome methylation at respective restriction sites. C and D correspond to blots A and B, respectively. M, Size markers (lambda + lambda/HindIII).
Methylation Analysis in Regenerated Plants and Progeny
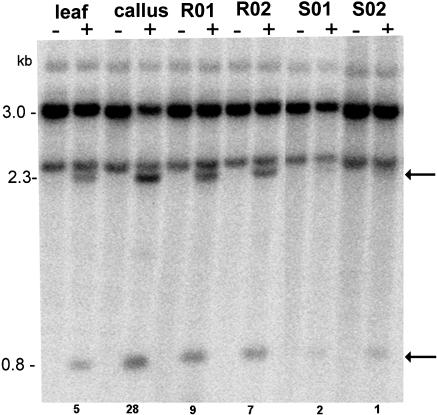
To study the maintenance of methylation patterns, we regenerated plants from two calli established from different parental plants. The regenerated plants (R0) were selfed to obtain S1 progeny. The methylation in the SR-VI region was analyzed in leaf DNA using MboI and HpaII enzymes, and representative profiles are shown in Figure 5. The unmethylated diagnostic bands were at 0.8 and 2.3 kb. As expected, the unmethylated fragments appeared to be strongest in lanes loaded with callus DNA. However, in regenerated plants (R01 and R02), the 0.8- and 2.3-kb HpaII bands became weaker, while the nondigested hybridizing fraction appeared stronger. In S1 plants, the hybridization profiles were similar to those of parental leaf. A similar trend was also obtained with other enzymes (data not shown). Thus, it seems that the epigenetic change induced in callus appeared to be reversed in regenerated plants and subsequent generations.
Figure 5.
Southern-blot hybridization showing remethylation of cytosines in the SR-VI subregion in regenerated plants and their progeny. Results for two out of 15 regenerated plants (R01 and R02) and two out of 10 progeny plants (S01 and S02) are presented. Genomic DNAs digested with MboI (−) and MboI plus HpaII (+) enzymes were probed with the IGS probe. Arrows indicate positions of hypomethylated fragments. The degree of hypomethylation was quantified as described in Figure 3.
Primary and Steady-State RNA Levels
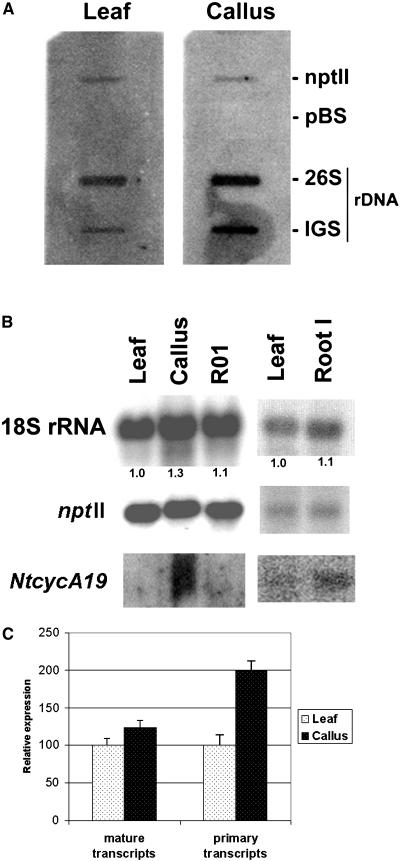
Since hypomethylation is frequently correlated with gene expression, we wished to investigate rRNA gene expression in dedifferentiated callus cells and regenerated plants. Since the rRNA genes are ubiquitously expressed, quantitative rather than qualitative differences were expected. To normalize rRNA expression patterns, we used a transgenic tobacco line carrying the nptII gene under the control of a strong 35S promoter. The reporter gene in the transgenic locus 2 is not methylated and its expression is not influenced by developmental changes (Fojtova et al., 2003). The primary transcripts were analyzed by nuclear run-on assays using isolated nuclei from leaf and callus (Fig. 6A). The slot blots with respective inserts were hybridized against the radioactively labeled nascent RNA probes from leaf and callus. While the ntpII hybridization signals were comparable for both probes, the callus RNA probe hybridized most strongly to the IGS target. The relative intensities of IGS signals between leaf and callus RNA (measured with a phosphor imager), an indication of the amount of primary, unprocessed rRNA transcript, are shown in Figure 6C (primary transcript column).
Figure 6.
Expression analysis in parental leaves, derived calli, and regenerated plants. A, Representative slot-blot hybridization with nascent RNA synthesized by run-on transcription using nuclei from the parental leaf and derived callus. The slots contained plasmids carrying the 26S gene and the A1/A2 subrepeat (IGS) as inserts. The expression of nonsilenced and nonmethylated nptII transgene served as a positive control. The plasmid vector (pBluescript; Stratagene) hybridization was used as a negative control (pBS). B, Representative northern-blot hybridization of steady-state transcripts. Total RNAs were isolated from different tissues and were subsequently hybridized on blots with the nptII, 18S rRNA, and NtcycA19 gene probes. Numbers below the lanes indicate the multiple increase in 18S/nptII ratio. For leaf, the ratio was arbitrarily chosen as 1. R01, Tissue culture regenerant; root I, RNA sample from an approximately 10-mm root tip. C, Quantification of the primary and steady-state (mature transcript) levels. The signals in A and B were quantified using a phosphor imager. The data from two run-on assays and six northern-blot experiments, independently conducted, were collected, normalized to the nptII transgene expression, and expressed as an average.
The steady-state 18S rDNA levels were determined by northern-blot hybridization (Fig. 6B). Total RNAs were isolated from leaf, roots, callus, and a regenerated plant and hybridized on blots with the nptII and 18S rRNA gene probes. Both probes were hybridized subsequently on the same blot to avoid variation caused by differences in blot preparations. The signals in each band were quantified by a phosphor imager. The data were summarized from five independent leaf and calli isolates and RNA levels were expressed as the 18S rRNA/nptII ratio (Fig. 6C, mature transcript column).
The tobacco NtcycA19 gene belonging to the A-type cyclins specifically expressed in G2 (Reichheld et al., 1996) is used here as a marker of cell cycle activity. To determine the proliferation activity of cells in different tissues, we hybridized the blots with the NtcycA19 gene probe (Fig. 6B). Calli showed the strongest hybridization signal, while in leaf the signal was almost undetectable (even with long exposures that give background noise). Signal in roots was faint and intermediate in strength between calli and leaf.
FISH
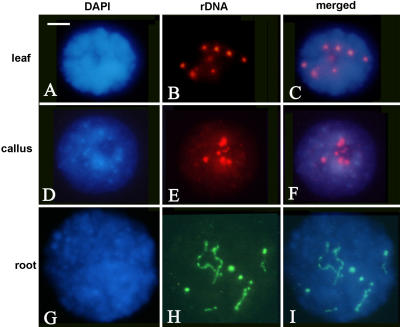
Increased chromatin decondensation is often considered a hallmark of gene activity. To determine rDNA condensation levels in interphase nuclei, we carried out FISH with the rDNA probe pTa71 (Fig. 7). In leaf, the rDNA probe produced eight hybridization signals that correspond to four pairs of rDNA loci. The leaf has the most condensed nuclei (typical nuclei are as in Fig. 7, A–C). Here, in the majority of nuclei, the heterochromatin and euchromatin are quite highly condensed and the nuclei are small and compact. Because everything is condensed, heterochromatin from rDNA is not apparent. The nucleoli are very small. The root-tips (Fig. 7, G–I) have much more decondensed nuclei, they are bigger, and the heterochromatin blocks from repeats and rDNA heterochromatin are clearly visible by DAPI staining. The nucleoli are quite large. The callus forms an intermediate morphology between leaf and root patterns (Fig. 7, D–F), but there is variability and some nuclei can be quite highly condensed as in leaf.
Figure 7.
FISH using biotin (red fluorescence)- or digoxigenin (green fluorescence)-labeled pTa71 against rDNA to interphase nuclei from mature leaf (A–C), fully dedifferentiated calli (D–F), and root-tip meristematic cells (G–I). Scale bar = 10 μm.
DISCUSSION
Hypomethylation of rDNA Units in Cell Culture
In this study, we describe changes in methylation and expression of rRNA genes in the course of culture-induced cell dedifferentiation and plant regeneration. We show that rRNA genes undergo specific hypomethylation in calli of different species. The process of hypomethylation starts as early as 2 weeks after the transfer of leaf segments onto callus-induction medium and is completed within 8 weeks. Nonproliferating green tissue did not show hypomethylation, suggesting that epigenetic change could be a direct consequence of the dedifferentiated state. Interestingly, these tissues show higher than average methylation levels compared to normal leaf (Fig. 2). This raises another possibility that the callus is derived from the least methylated fraction of cells in the complex tissue that makes up the leaf. Perhaps there might be a selection for cells with a hypomethylated rDNA phenotype in callus.
The relative hypomethylation of rDNA units was remarkably stable in calli and was maintained for at least 2 years of in vitro cultivation. In regenerated plants, the hypomethylated state was partially lost and their progeny displayed methylation levels comparable to parental leaf tissue. rRNA gene hypomethylation in callus culture was also observed in cell cultures of N. sylvestris (a diploid progenitor of tobacco) and species outside of Nicotiana, indicating that hypomethylation is not linked to allopolyploidy or to a particular species (Table II). Contrasting results were previously obtained in a post-transcriptionally silenced tobacco transgene under the control of a 35S promoter. Here the transgene acquired de novo methylation after prolonged cultivation of tobacco cells in cell culture, with hypermethylation appearing gradually over 12 months (Fojtova et al., 2003). Thus, different loci (and promoters) may respond differentially to developmental changes. The fact that root-tip cells (which include the apical meristem) showed a similar level of unit methylation as calli suggests that hypomethylation is associated with dividing cells.
In both plants and mammals, passive and active models of DNA hypomethylation have been proposed. The classic methylation maintenance model explains the loss of 5-methylcytosine residues by a passive means arising from DNA replication in the absence of maintenance methylation. In contrast, active processes involve excision of modified nucleotides without DNA replication. The following data argue for a passive demethylation of rDNA clusters during dedifferentiation: (1) The demethylation required at least two callus replication cycles (4 weeks) to complete; and (2) nondividing green tissue exposed to callus-inducing medium (left on agar) have increased methylation levels compared to the original leaf. In plants, CG and CNG motifs are probably methylated by distinct enzyme activities encoded by MET and CMT gene families, respectively (Finnegan and Kovac, 2000). In calli, both CG and CNG motifs showed reduced methylation, indicating that hypomethylation is not limited to a particular sequence motif. It is unlikely that there is a general down-regulation of DNA methyltransferase activities in calli since other repeated loci did not undergo methylation changes (Fig. 4, C and D; Fulnecek et al., 1998; Kovarik et al., 2000). Loss of methylation could be related to other epigenetic changes occurring at the level of histone modification and/or transcription factor binding. Recently, a nucleolar specific HDT1 histone deacetyltransferase gene was shown to be required for nucleolar dominance and maintenance of rRNA gene silencing in Arabidopsis polyploids (Lawrence et al., 2004). There is evidence for alterations in global histone acetylation patterns during plant development (Hodurkova and Vyskot, 2003). It will be interesting to see whether callus-associated hypomethylation is accompanied by changes in histone modification at rDNA chromatin.
Developmental Changes in rDNA Chromatin Condensation Patterns
FISH analysis was used to reveal potential differences in condensation patterns of rRNA genes at interphase. Several cytological observations in, for example, Arabidopsis and Triticum (Soppe et al., 2002) suggest that DNA methylation might control heterochromatin assembly. Most rDNA chromatin in leaf and callus tissues is highly condensed at interphase, suggesting that the observed hypomethylation in rDNA of callus does not manifest itself as global relaxation of rDNA chromatin. Perhaps partial methylation maintained mainly at the genic regions (Fig. 3) is sufficient to drive chromatin condensation. Alternatively, loss of methylated cytosines might not necessarily lead to chromatin decondensation. In this context, assembly of chromocenters appeared to be unaffected by hypomethylation of tandem arrays in Arabidopsis mutants (Tariq et al., 2003). Treatment of wheat cells with hypomethylation drugs showed variable condensation patterns depending on chromosomal positions (Glyn et al., 1995) and the differences in condensation patterns of rRNA genes were also noted in primary and adventitious roots of onion (Hasterok and Maluszynska, 2000). Despite a lack of overall decondensation, both callus and root material also show sectors in the rDNA loci of decondensed chromatin. Perhaps the decondensed rDNA sectors contain clustered hypomethylated units (Fig. 4). However, it is also possible that hypomethylated units correspond to facultative heterochromatin whose condensation levels may change in response to endogenous and exogenous stimuli. Indeed, there was variability between the root-tip interphases with condensed FISH signal ranging from as little as 5% up to 50% of total rDNA signal, a range that may reflect various stages of cell differentiation, active and quiescent cells. We favor the hypothesis that hypomethylated rDNA may not only indicate actively transcribing genes, but also may include silenced units that had been active in previous replication cycles, perhaps constituting facultative heterochromatin with the potential to be active. In some calli interphases, condensed rDNA chromatin occurred both outside and inside the nucleolus. Similar data have been observed in wheat suspension cultures where nucleoli are observed to contain some condensed rDNA heterochromatin (Leitch et al., 1993).
In tobacco protoplasts, increased condensation of rDNA chromatin was observed without material changes in methylation patterns (Williams et al., 2003). However, there are notable differences between this system and the one we describe here. First, in this study, actively dividing callus cells were analyzed, while in the Williams et al. (2003) study, nondividing protoplasts were used. Perhaps decondensation occurs only after the cell cycle is resumed. Second, conditions during protoplast treatment could elicit cell stress and this is known to alter epigenetic patterns (Kovarik et al., 1997; Labra et al., 2002; for review, see Arnholdt-Schmitt, 2004).
Hypomethylation Is Accompanied by Moderate Increase of rRNA Gene Expression
In many plants, the IGS shows lower levels of methylation than the genic regions, as shown here in tobacco (Fig. 3) and as previously reported (Flavell et al., 1988; Jupe and Zimmer, 1990; Torres-Ruiz and Hemleben, 1994). In mammals, ribosomal genes are highly methylated in the intergenic region while the genic regions are mostly methylation-free (Brock and Bird, 1997). Thus, the distribution of methylation along the rRNA gene unit appears to differ significantly between plants and mammals. A direct relationship has been established between methylation and 35S rDNA expression in plants. In Brassica allopolyploids (Chen and Pikaard, 1997) and in Triticale cereals (Viera et al., 1990), hypomethylation with the drug 5-azacytidine activated silent underdominant loci. To study the relation between rDNA expression and methylation changes accompanying dedifferentiation, we have determined levels of both primary and mature transcripts by nuclear run-on and northern-blot hybridization, respectively. As an internal control, we used a nonmethylated transgenic locus that constitutively expresses the nptII reporter gene and whose expression is not influenced by callus induction. There is about a 2-fold increase in primary transcript levels observed in calli compared to parental leaves. At the steady-state level, such difference was not so pronounced and the content of 18S rRNA in callus and root was increased by a maximum of 25% and 10%, respectively. The difference between primary and steady-state levels could be explained by increased turnover of mature rRNA in calli, regulation at the post-transcriptional level, or a combination of both. Arabidopsis tissues with lower heterochromatin content showed an elevated proportion of minor 5S rRNA species (Mathieu et al., 2003). It remains to be tested whether similar heterogeneity is displayed among the 35S rRNA, although distinguishing between individual RNA species might be difficult due to a high level of interlocus unit homogenization in tobacco (Volkov et al., 1999; Lim et al., 2000). The fact that both 5S (Mathieu et al., 2003) and 35S rRNA transcripts appear to be elevated in root suggests that expression of both loci might be subjected to orchestrated regulation, perhaps ensuring stoichiometric ratios of these structural molecules during ribosome assembly.
Increased rRNA gene expression could reflect increased demand for protein synthesis in rapidly dividing cells and could be regulated by the hormones. Indeed, cytokinin has been shown to up-regulate transcription initiation from the polymerase I promoter in Arabidopsis (Gaudino and Pikaard, 1997). It is known that root-tip cells are tissues (places) of primary cytokinin biosynthesis and callus-induction media also contain relatively high levels of cytokinin (for concentration, see “Materials and Methods”). Perhaps rRNA gene expression is stimulated by this hormone in these cells. In regenerated plants and their progeny, the rDNA expression (and methylation) appeared to return to normal (Figs. 5 and 6). In contrast, the hypomethylation drug, 5-azacytidine, induced mostly heritable changes in NOR activity (Amado et al., 1997) and maintenance of the hypomethylation state (Vyskot et al., 1995).
Despite a relatively good correlation between hypomethylation and rDNA expression, a direct involvement of DNA methylation on control of rRNA gene transcription remains in doubt. For example, the proximal ClaI site (with respect to the 18S gene) is only approximately 40% methylated in calli. Assuming that there are approximately 6,000 units in tobacco diploid cells (Lim et al., 2000), it follows that almost 4,000 units do not contain methylation at this site. However, it is unlikely that all hypomethylated units are active since large amounts of chromatin remain highly condensed and probably inactive in the callus interphase (Fig. 7). Second, the callus-induced hypomethylation does not seem to be targeted to promoter regions, although the sites immediately upstream and downstream were less methylated than the sites in the genic regions. It is likely that both chromatin-mediated epigenetic silencing and specific transcription factors contribute to complex regulation of rRNA gene expression during plant development.
MATERIALS AND METHODS
Plant Material
Tobacco (Nicotiana tabacum L. var. Vielblattriger, SR-1), Nicotiana sylvestris (Goodspeed, 1954), and the transgenic tobacco HeLo2 line (Van Houdt et al., 2000) were used. Tragopogon mirus 2603 was from collections at the University of Florida (Kovarik et al., 2005) and Cestrum parqui was from The Royal Botanic Gardens, Kew, UK. Calli were established from leaf explants by hormonal treatment and grown in 0.7% agar containing B5 salts supplemented with Suc (30 g L−1), α-napthalene acetic acid (2.0 mg L−1), and 6-benzylaminopurine (0.2 mg L−1). In tobacco, the process of callus formation starts as early as 1 week after the initial transfer of leaf cuttings to callus induction medium and is complete within 4 to 6 weeks. Callus was transferred onto fresh agar medium every 3 to 4 weeks, when its weight had increased 4-fold over the inoculum. Cells of calli after two subcultures were considered to be fully dedifferentiated. To obtain regenerated plants, calli were transferred onto shoot-inducing medium containing α-napthalene acetic acid (0.2 mg L−1) and 6-benzylaminopurine (2.0 mg L−1). About eight to 10 plantlets were obtained after two subcultures (1 month each). After the rooting phase on growth medium without hormones, the plantlets were transferred into greenhouse conditions. The S1 plants were obtained by selfing regenerated plants. Leaf material for harvesting was obtained from fully developed plants of approximately 20-cm height. Leaves were collected in the morning to reduce potential epigenetic variability caused by environmental factors (Watson et al., 1987).
DNA Isolation and Southern-Blot Hybridization
Total genomic DNA was isolated from approximately 10 g of leaves, lyophilized calli, or approximately 100 root tips of healthy pot-grown plants by a cetyltrimethylammonium bromide method as described previously (Kovarik et al., 2000). DNA methylation was analyzed with methylation-sensitive restriction endonucleases. Genomic DNA (2 μg) was digested with an excess of the enzymes (5 units μg−1 DNA) and subjected to electrophoresis on 0.8% agarose gel. After electrophoresis, the gels were alkali blotted onto Hybond-XL membranes (Amersham Biosciences) and hybridized with 32P-labeled DNA probes (DekaLabel kit; MBI Fermentas). After washing under high-stringency conditions, the hybridization bands were visualized with a PhosphorImager (Storm; Molecular Dynamics) and the data were processed by ImageQuant software (Molecular Dynamics). Completeness of digestion was monitored by rehybridization of the blots with a chloroplast probe. Relative methylation levels of particular sites, or groups of sites, were determined by phosphor imager quantification and comparison of hybridization signals of bands corresponding to methylated and nonmethylated DNA molecules. Rectangle (Figs. 2, 3, and 5) or line (Fig. 4) integration methods were used for signal counting.
DNA Probes
In tobacco, both genic and intergenic regions of rDNA were sequenced (Volkov et al., 1999), enabling us to design probes and restriction enzymes for methylation analysis using Southern-blot hybridization (Fig. 1; Table I). The 18S probe contained a 1.7-kb EcoRI fragment of the 18S rRNA gene from Solanum lycopersicum (Kiss et al., 1989). The 26S rDNA probe was a 220-bp fragment of the 26S rRNA gene subunit from Nicotiana tomentosiformis obtained by PCR amplification of the 3′-end region using 5′-GAATTCACCCAAGTGTTGGGAT-3′ (forward) and 5′-AGAGGCGTTCAGTCATAATC-3′ (reverse) primers (Kiss et al., 1989; accession no. AF479172). For details, see Lim et al. (2000). The IGS probe was a cloned 350-bp insert containing A1/A2 subrepeats from N. tabacum (Lim et al., 2004). The nptII (neomycin phosphotransferase II) coding sequence probe was an approximately 830-bp insert of the pGEM nptII plasmid (Van Houdt et al., 2000). The 1.9-kb fragment containing tobacco cyclin A cDNA (clone NtcycA19) was used as a probe (Reichheld et al., 1996). Quantification of hybridization signals was carried out using the Storm PhosphorImager and ImageQuant software.
Run-on Transcription
Run-on transcription in isolated nuclei was performed as described by van Blokland et al. (1994) with some modifications (Fojtova et al., 2003). Young plant leaves (10–12 g) were ground in liquid nitrogen and resuspended in extraction buffer (10 mm NaCl, 10 mm MES, pH 6.0, 5 mm EDTA, 0.15 m spermidine, 20 mm β-mercaptoethanol, 0.6% Triton X-100, and 0.25 m Suc). After purification on a Percoll gradient, nuclei were resuspended in 1.2 mL of storage buffer [10 mm Tris-HCl, 100 mm (NH4)2SO4, 10 mm MgCl2, 5 mm β-mercaptoethanol], and stored with an equal amount of 99% glycerol at −80°C. Since we were unable to homogenize wet calli in liquid nitrogen due to the high water content, we lyophilized about 10 g of frozen callus tissue prior to homogenization.
For the nuclear run-on transcription assays, 800 μL of nuclei were thawed and washed in storage buffer to remove glycerol. The reaction was started by adding nucleotide mix (final concentration 0.5 mm for each C, G, and ATP) and 100 μCi of [α-32P]UTP (6,000 Ci mmol−1; ICN Biomedicals) and stopped by SDS (final concentration 3%) after incubation at 27°C for 30 min. Labeled RNA was purified by DNaseI treatment and phenol extraction and finally recovered by ethanol precipitation. The pellet was resuspended in 100 μL sterile water and used as a probe for hybridization (approximately 2×106 dpm).
Plasmids carrying relevant inserts were linearized, denatured, and about 1 to 2 μg were bound to the nylon membrane (Hybond XL; Amersham Biosciences). Hybridization was performed in ULTRAhyb buffer (Ambion) for 48 h at 42°C. Membranes were washed in standard saline phosphate/EDTA + 0.1% SDS at room temperature for 15 min and in the same solution at 60°C for 30 min. After overnight exposure to the screen, the radioactive signals were visualized using the Storm PhosphorImager.
Northern-Blot Hybridization
Total RNA was isolated using the RNeasy kit (Qiagen) from leaves, calli, and roots of the tobacco HeLo2 line. Formaldehyde agarose gel electrophoresis was carried out according to the supplier's protocol. After electrophoresis, the ethidium bromide-stained gels were photographed, blotted onto membranes (Hybond XL; Amersham Biosciences), and hybridized to [α-32P]-labeled DNA probes (>108 dmp/μg DNA; DekaLabel kit; MBI Fermentas). Hybridizations were carried out under high-stringent conditions, as described above. The membranes were exposed to the screen and radioactive signals were visualized using the Storm PhosphorImager.
FISH
FISH was carried out on leaf from healthy plants or calli as in Lim et al. (2000). Briefly, slides were pretreated with 100 μg mL−1 RNaseA for 1 h and 0.25 μg mL−1 pepsin for 5 min followed by denaturation in 70% formamide in 2× SSC (0.3 m sodium chloride and 0.03 m sodium citrate) at 70°C for 2 min. The hybridization mix contained 8 μg mL−1 biotin-labeled or digoxigenin-labeled pTa71 (contains 18–26S rDNA and IGS from Triticum aestivum) in 2× SSC supplemented with 50% (v/v) formamide, 10% (w/v) dextran sulfate, and 0.1% (w/v) SDS. After overnight hybridization at 37°C, the slides were washed in 20% (v/v) formamide in 0.1× SSC at 42°C at an estimated hybridization stringency of 80% to 85%. Sites of probe hybridization were detected for biotinylated probes with Cy3-conjugated avidin (Amersham-Pharmacia Biotech) and for digoxigenin-labeled probes with fluorescent isothiocyanate-conjugated antidigoxigenin (Roche Biochemicals). Chromosomes were counterstained with 2 μg mL−1 DAPI in 4× SSC, mounted in Vectashield (Vector Laboratories) medium and examined using a Leica DM RA2. Images were captured using Openlab (Improvision) and processed using Adobe Photoshop (Adobe Systems) by treating all images for color contrast and brightness uniformly.
Supplementary Material
Acknowledgments
We thank Drs. Kamila Skalická and Roman Matyasek for comments and helpful discussions, and Prof. Heberle-Bors (University of Vienna, Austria) for the cyclin A probe. We greatly appreciate the technical assistance of Mrs. Libuse Jedlickova.
This work was supported by the Grant Agency of the Czech Republic, Academy of Sciences (grant nos. 521–01–0775 and A50040507 to A.K. and 204–03–P104 to J.F.) and the National Environmental Research Council (grant to A.R.L.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.061788.
References
- Amado L, Abranches R, Neves N, Viegas W (1997) Development-dependent inheritance of 5-azacytidine-induced epimutations in triticale: analysis of rDNA expression patterns. Chromosome Res 5: 445–450 [DOI] [PubMed] [Google Scholar]
- Anderson S, Lewis-Smith AC, Smith SM (1990) Methylation of ribosomal RNA genes in Petunia hybrida plants, callus cultures and regenerated shoots. Plant Cell Rep 8: 554–557 [DOI] [PubMed] [Google Scholar]
- Arnholdt-Schmitt B (2004) Stress-induced reprogramming. A role for global genome regulation? Plant Physiol 136: 2579–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnholdt-Schmitt B, Hesterich S, Neumann K-H (1995) Physiological aspects of genome variability in tissue culture, I. Growth phase-dependent differential DNA methylation of the carrot genome (Daucus carrota L.) during primary culture. Theor Appl Genet 91: 809–815 [DOI] [PubMed] [Google Scholar]
- Avivi Y, Morad V, Ben-Meir H, Zhao J, Kashkush K, Tzfira T, Citovsky V, Grafi G (2004) Reorganization of specific chromosomal domains and activation of silent genes in plant cells acquiring pluripotentiality. Dev Dyn 230: 12–22 [DOI] [PubMed] [Google Scholar]
- Brock GJR, Bird A (1997) Mosaic methylation of the repeat units of the human ribosomal RNA genes. Hum Mol Genet 6: 451–456 [DOI] [PubMed] [Google Scholar]
- Castilho A, Neves N, Rufini-Castiglione M, Viegas W, Heslop-Harrison JS (1999) 5-Methylcytosine distribution and genome organization in triticale before and after treatment with 5-azacytidine. J Cell Sci 112: 4397–4404 [DOI] [PubMed] [Google Scholar]
- Cerdido A, Medina FJ (1995) Subnucleolar location of fibrillarin and variation in its levels during the cell cycle and during differentiation of plant cells. Chromosoma 103: 625–634 [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Pikaard CS (1997) Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Genes Dev 11: 2124–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Yakura K, Miyanishi M, Sugita M, Sugiura M (1995) In vitro transcription of plant RNA polymerase I-dependent rRNA genes is species-specific. Plant J 8: 295–298 [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Kovac KA (2000) Plant DNA methyltransferases. Plant Mol Biol 43: 189–201 [DOI] [PubMed] [Google Scholar]
- Flavell RB, O'Dell M, Thompson WF (1988) Regulation of cytosine methylation in ribosomal DNA and nucleolus organizer expression in wheat. J Mol Biol 204: 523–534 [DOI] [PubMed] [Google Scholar]
- Fojtova M, Van Houdt H, Depicker A, Kovarik A (2003) Epigenetic switch from posttranscriptional to transcriptional silencing is correlated with promoter hypermethylation. Plant Physiol 133: 1240–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulnecek J, Matyasek R, Kovarik A, Bezdek M (1998) Mapping of 5-methylcytosine residues in Nicotiana tabacum 5S rRNA genes by genomic sequencing. Mol Gen Genet 259: 133–141 [DOI] [PubMed] [Google Scholar]
- Gaudino RJ, Pikaard CS (1997) Cytokinin induction of RNA polymerase I transcription in Arabidopsis thaliana. J Biol Chem 272: 6799–6804 [DOI] [PubMed] [Google Scholar]
- Glyn MC, Egertova M, Gazdova B, Kovarik A, Bezdek M, Leitch AR (1995) The influence of 5-azacytidine on the condensation of the short arm of rye chromosome 1R in Triticum aestivum L. root tip meristematic nuclei. Chromosoma 106: 485–492 [DOI] [PubMed] [Google Scholar]
- Goodspeed TH (1954) The Genus Nicotiana, Vol 16. Chronica Botanica, Waltham, MA
- Grafi G (2004) How cells differentiate: lessons from plants. Dev Biol 268: 1–6 [DOI] [PubMed] [Google Scholar]
- Hasterok R, Maluszynska J (2000) Different rRNA gene expression in primary and adventitious roots of Allium cepa L. Folia Histochem Cytobiol 38: 181–184 [PubMed] [Google Scholar]
- Hemleben V, Zentgraf U (1994) Structural organisation and regulation of transcription by RNA polymerase I of plant nuclear ribosomal genes. In L Nover, ed, Results and Problems in Cell Differentiation 20: Plant Promoters and Transcription Factors. Springer-Verlag, Berlin/Heidelberg, pp 3–24 [DOI] [PubMed]
- Hodurkova J, Vyskot B (2003) Histone H4 acetylation patterns during seed germination and early plant development. Biol Plant (Prague) 46: 23–28 [Google Scholar]
- Jaligot E, Rival A, Beule T, Dussert S, Verdeil JL (2000) Somaclonal variation in oil palm (Elaesis guineensis Jacg.): the DNA methylation hypothesis. Plant Cell Rep 19: 684–690 [DOI] [PubMed] [Google Scholar]
- Jupe ER, Zimmer EA (1990) Unmethylated regions in the intergenic spacer of maize and teosinte ribosomal RNA genes. Plant Mol Biol 14: 333–347 [DOI] [PubMed] [Google Scholar]
- Kaeppler SM, Kaeppler HF, Rhee Y (2000) Epigenetic aspects of somaclonal variation in plants. Plant Mol Biol 43: 179–188 [DOI] [PubMed] [Google Scholar]
- Kenton A, Parokonny AS, Gleba YY, Bennett MD (1993) Characterization of the Nicotiana tabacum L. genome by molecular cytogenetics. Mol Gen Genet 240: 159–169 [DOI] [PubMed] [Google Scholar]
- Kiss T, Szkukalek A, Solymosy F (1989) Nucleotide sequence of a 17S (18S) rRNA gene from tomato. Nucleic Acids Res 17: 2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova NY, Grabe T, Huigen DJ, Hemleben V, Volkov RA (2004) Organization, differential expression and methylation of rDNA in artificial Solanum allopolyploids. Plant Mol Biol 56: 439–463 [DOI] [PubMed] [Google Scholar]
- Kovarik A, Koukalova B, Bezdek M, Opatrny Z (1997) Hypermethylation of tobacco heterochromatic loci in response to osmotic stress. Theor Appl Genet 95: 301–306 [Google Scholar]
- Kovarik A, Koukalova B, Lim KY, Matyasek R, Lichtenstein CP, Leitch AR, Bezdek M (2000) Comparative analysis of DNA methylation in tobacco heterochromatic sequences. Chromosome Res 8: 527–541 [DOI] [PubMed] [Google Scholar]
- Kovarik A, Matyasek R, Lim KY, Skalická K, Koukalova B, Knapp S, Chase M, Leitch AR (2004) Concerted evolution of 18-5.8-26S rDNA repeats in Nicotiana allotetraploids. Biol J Linn Soc 82: 615–625 [Google Scholar]
- Kovarik A, Pires JC, Leitch AR, Lim KY, Sherwood A, Matyasek R, Rocca J, Soltis DE, Soltis PS (2005) Rapid concerted evolution of nuclear ribosomal DNA in two allopolyploids of recent and recurrent origin. Genetics 169: 931–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labra M, Ghiani A, Citterio S, Sgorbati S, Sala F, Vannini C, Ruffini-Castiglione M, Bracale M (2002) Analysis of cytosine methylation pattern in response to water deficit in pea root tips. Plant Biol 4: 694–699 [Google Scholar]
- Lawrence RJ, Earley K, Pontes O, Silva M, Chen ZJ, Neves N, Viegas W, Pikaard CS (2004) A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell 13: 599–609 [DOI] [PubMed] [Google Scholar]
- Leitch AR, Schwarzacher T, Wang ML, Leitch IJ, Surlan-Momirovich G, Moore G, Heslop-Harrison JS (1993) Molecular cytogenetic analysis of repeated sequences in a long term wheat suspension culture. Plant Cell Tissue Organ Cult 33: 287–296 [Google Scholar]
- Lim KY, Kovarik A, Matyasek R, Bezdek M, Lichtenstein CP, Leitch AR (2000) Gene conversion of ribosomal DNA in Nicotiana tabacum is associated with undermethylated, decondensed and probably active gene units. Chromosoma 109: 161–172 [DOI] [PubMed] [Google Scholar]
- Lim KY, Skalická K, Koukalova B, Volkov RA, Matyasek R, Hemleben V, Leitch AR, Kovarik A (2004) Dynamic changes in the distribution of a satellite homologous to intergenic 26-18S rDNA spacer in the evolution of Nicotiana. Genetics 166: 1935–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O, Jasencakova Z, Vaillant I, Gendrel A-V, Colot V, Schubert I, Tourmente S (2003) Changes in 5S rDNA chromatin organization and transcription during heterochromatin establishment in Arabidopsis. Plant Cell 15: 2929–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O, Picard G, Tourmente S (2002) Methylation of an euchromatin-heterochromatin transition region in Arabidopsis thaliana chromosome 5 left arm. Chromosome Res 10: 455–466 [DOI] [PubMed] [Google Scholar]
- Mitsuhara I, Shirasawa-Seo N, Iwai T, Nakamura S, Honkura R, Ohashi Y (2002) Release from post-transcriptional gene silencing by cell proliferation in transgenic tobacco plants: possible mechanism for noninheritance of the silencing. Genetics 160: 343–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscone EA, Matzke MA, Matzke AJ (1996) The use of combined FISH/GISH in conjunction with DAPI counterstaining to identify chromosomes containing transgene inserts in amphidiploid tobacco. Chromosoma 105: 231–236 [PubMed] [Google Scholar]
- Olhoft PM, Phillips RL (1999) Genetic and epigenetic instability in tissue culture and regenerated progenies. In HR Lerner, ed, Plant Responses to Environmental Stresses: From Phytohormones to Genome Reorganization. Marcel Dekker, New York, pp 111–148
- Pikaard CS (2000) The epigenetics of nucleolar dominance. Trends Genet 16: 495–500 [DOI] [PubMed] [Google Scholar]
- Reichheld JP, Chaubet N, Shen WH, Renaudin JP, Gigot C (1996) Multiple A-type cyclins express sequentially during the cell cycle in Nicotiana tabacum BY2 cells. Proc Natl Acad Sci USA 93: 13819–13824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R, Grummt I (2001) Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol Cell 8: 719–725 [DOI] [PubMed] [Google Scholar]
- Sardana R, O'Dell M, Flavell R (1993) Correlation between the size of the intergenic regulatory region, the status of cytosine methylation of rRNA genes and nucleolar expression in wheat. Mol Gen Genet 236: 155–162 [DOI] [PubMed] [Google Scholar]
- Skalická K, Lim KY, Matyásek R, Koukalová B, Leitch AR, Kovarík A (2003) Rapid evolution of parental rDNA in a synthetic tobacco allotetraploid line. Am J Bot 90: 988–996 [DOI] [PubMed] [Google Scholar]
- Smulders MJM, Rus-Kortekaas W, Vosman B (1995) Tissue culture-induced DNA methylation polymorphisms in repetitive DNA of tomato calli and regenerated plants. Theor Appl Genet 91: 1257–1264 [DOI] [PubMed] [Google Scholar]
- Soppe WJ, Jasencakova Z, Houben A, Kakutani T, Meister A, Huang MS, Jacobsen SE, Schubert I, Fransz PF (2002) DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J 21: 6549–6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq M, Saze H, Probst AV, Lichota J, Habu Y, Paszkowski J (2003) Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc Natl Acad Sci USA 100: 8823–8827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Ruiz RA, Hemleben V (1994) Pattern and degree of methylation in ribosomal RNA genes of Cucurbita pepo L. Plant Mol Biol 26: 1167–1179 [DOI] [PubMed] [Google Scholar]
- van Blokland R, van der Geest N, Mol JN, Kooter JM (1994) Transgene-mediated suppression of chalcone synthase expression in Petunia hybrida results from an increase in RNA turnover. Plant J 6: 861–877 [Google Scholar]
- Van Houdt H, Kovarik A, Van Montagu M, Depicker A (2000) Cross-talk between posttranscriptionally silenced neomycin phosphotransferase II transgenes. FEBS Lett 467: 41–46 [DOI] [PubMed] [Google Scholar]
- Viera R, Queiroz A, Morais L, Barao A, Mello-Sampavo T, Viegas WS (1990) 1R chromosome nucleolus organiser region activation by 5-azacytidine in wheat×rye hybrids. Genome 33: 707–712 [Google Scholar]
- Volkov RA, Borisjuk NV, Panchuk II, Schweizer D, Hemleben V (1999) Elimination and rearrangement of parental rDNA in the allotetraploid Nicotiana tabacum. Mol Biol Evol 16: 311–320 [DOI] [PubMed] [Google Scholar]
- Vyskot B, Gazdova B, Siroky J (1993) Methylation patterns of two repetitive DNA sequences in tobacco tissue cultures and their regenerants. Biol Plant (Prague) 35: 321–327 [Google Scholar]
- Vyskot B, Koukalova B, Kovarik A, Sachambula L, Reynolds D, Bezdek M (1995) Meiotic transmission of a hypomethylated repetitive DNA family in tobacco. Theor Appl Genet 91: 659–664 [DOI] [PubMed] [Google Scholar]
- Watson JC, Kaufman LS, Thompson WF (1987) Developmental regulation of cytosine methylation in the nuclear ribosomal RNA genes of Pisum sativum. J Mol Biol 193: 15–26 [DOI] [PubMed] [Google Scholar]
- Williams L, Zhao J, Morozova N, Li Y, Avivi Y, Grafi G (2003) Chromatin reorganization accompanying cellular dedifferentiation is associated with modifications of histone H3, redistribution of HP1, and activation of E2F-target genes. Dev Dyn 228: 113–120 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.