Abstract
Trypsin proteinase inhibitors (TPIs) of Nicotiana attenuata are major antiherbivore defenses that increase dramatically in leaves after attack or methyl jasmonate (MeJA) elicitation. To understand the elicitation process, we characterized the proteolytic fragmentation and release of TPIs from a multidomain precursor by proteases in MeJA-elicited and unelicited plants. A set of approximately 6-kD TPI peptides was purified from leaves, and their posttranslational modifications were characterized. In MeJA-elicited plants, the diversity of TPI structures was greater than the precursor gene predicted. This elicited structural heterogeneity resulted from differential fragmentation of the linker peptide (LP) that separates the seven-domain TPI functional domains. Using an in vitro fluorescence resonance energy transfer assay and synthetic substrates derived from the LP sequence, we characterized proteases involved in both the processing of the TPI precursor and its vacuolar targeting sequence. Although both a vacuolar processing enzyme and a subtilisin-like protease were found to participate in a two-step processing of LP, only the activity of the subtilisin-like protease was significantly increased by MeJA elicitation. We propose that MeJA elicitation increases TPI precursor production and saturates the proteolytic machinery, changing the processing pattern of TPIs. To test this hypothesis, we elicited a TPI-deficient N. attenuata genotype that had been transformed with a functional NaTPI gene under control of a constitutive promoter and characterized the resulting TPIs. We found no alterations in the processing pattern predicted from the sequence: a result consistent with the saturation hypothesis.
Proteinase inhibitors (PIs) are one of the most important classes of defense proteins in plants. The accumulation of PIs is elicited by various biotic and abiotic stresses, including mechanical wounding, insect attack, pathogen attack, and UV exposure as well as signal molecules, such as systemin, methyl jasmonate (MeJA), ethylene, abscisic acid, fungal cell wall oligomers, and the fatty acid amino acid conjugates found in larval oral secretions (Ryan, 1990; O'Donnell et al., 1996; Koiwa et al., 1997; Korth and Dixon, 1997; Roda et al., 2004). Although plant PIs may play physiological roles as storage proteins or regulators of endogenous proteases (Koiwa et al., 1997), their role as antifeedants against insect herbivores is their best-documented function (Ryan, 1990; Broadway, 1996; Jongsma and Bolter, 1997; Zavala et al., 2004a). Ingested plant PIs inhibit digestive proteases of the attacking insect herbivore, imposing a physiological stress that retards growth and development and increases mortality (Jongsma and Bolter, 1997). When fed diets rich in PIs, insects counter this plant defense by producing proteases that are PI insensitive and/or PI degrading (Jongsma and Bolter, 1997; Girard et al., 1998; Giri et al., 1998). An analysis of the sequence variation revealed that the active domains of PI genes carry the signatures of an evolutionary arms race between plants and their enemies (Ryan, 1990); hence, the ability to produce a wide spectrum of structurally and functionally divergent PIs is likely important for PI defensive function.
PIs are classified according to their amino acid sequence into structural families (Rawlings et al., 2004). A plant often produces PIs of different structural families, and homologous PIs form a multigene family or multidomain PIs, which collectively can inhibit a broad range of insect gut proteases. Some examples of multidomain inhibitors include the double-headed Bowman Birk inhibitors in the Leguminosae, the cereal trypsin/α-amylase inhibitors, and the inhibitors of the potato (Solanum tuberosum) PI-II family found in Solanaceae (Atkinson et al., 1993; Lee et al., 1999; Moura and Ryan, 2001). A recent analysis of genes of the PI-II family (Barta et al., 2002) suggests that unequal cross-overs and gene conversion events led to the evolution of the structural diversity found in the Solanaceae. These mechanisms account for the structural diversity in evolutionary time; however, the possibility that plants increase the structural diversity of the PIs they produce during an ecological interaction has not been examined. Moreover, although numerous ecological interactions are known to rapidly increase the accumulation of PI transcripts and the activity of PIs (Ryan, 1990; Karban and Baldwin, 1997; van Dam et al., 2001; Glawe et al., 2003), a comparative structural analysis of PIs in control and elicited plants has not been conducted. Here, we report a novel means of producing a spectrum of different trypsin proteinase inhibitor (TPI) structures after elicitation: modulating proteolytic processing of multidomain precursor. It has become increasingly evident that proteases play a key role in complex physiological processes associated with plant defense responses at the level of perception, signaling, and execution (for review, see van der Hoorn and Jones, 2004). Although these functions are known to be important, the precise role of individual proteases and how these proteolytic pathways are regulated during defense elicitation remain unknown.
Nicotiana attenuata Torr. ex Watts (Solanaceae), a tobacco species native to the Great Basin desert of the southwest United States, has been studied for its array of induced direct and indirect defenses against herbivore attack (Baldwin, 2001). As one of its potent direct defenses, N. attenuata produces TPIs in response to caterpillar attack, mechanical wounding, and MeJA elicitation (van Dam et al., 2001; Glawe et al., 2003). Recently, we have determined the cDNA sequence of a multidomain TPI precursor of the PI-II family expressed in N. attenuata (NaTPI; Zavala et al., 2004a). That this molecule functions as an antiherbivore defense has been demonstrated by genetically silencing NaTPI expression in N. attenuata plants and by transforming a naturally occurring N. attenuata genotype that is unable to accumulate NaTPI transcripts and protein with a functional NaTPI gene to restore TPI expression (Zavala et al., 2004b; Zavala and Baldwin, 2004).
In this work, we characterize the posttranslational modifications of TPIs derived from the multidomain precursor, which were isolated as a set of isoinhibitors from N. attenuata leaves. MeJA elicitation of N. attenuata not only increased TPI transcripts and accumulation, but it also increased the structural diversity of TPIs by differentially fragmenting the linker peptide that separates the TPI functional domains. Three different classes of proteases organized the steps in the orchestrated cascade of TPI processing. We determined how MeJA elicitation affected the activity of the processing proteases and the level of their substrate, the TPI precursor that determines the maturation of the TPI phenotype after elicitation. In order to test whether the change in the processing pattern after elicitation results from saturation of the proteolytic machinery, we elicited a genotype of N. attenuata (S++) collected in Arizona, which is completely deficient in TPI production at a transcriptional level (Glawe et al., 2003) and was transformed with the full-length seven-domain TPI from the wild-type genotype expressed under a constitutive promotor (cauliflower mosaic virus 35S). We then characterized the TPI peptides that were produced after elicitation. Our results were consistent with the hypothesis that MeJA elicits irregular processing due to the saturation of the proteolytic machinery that results from overproduction of the TPI precursor. This research highlights the dynamic relationship between molecules entering the processing machinery and the diversity of the processed products that function in the induced defense response.
RESULTS
Inducibility of the TPI Precursor by MeJA
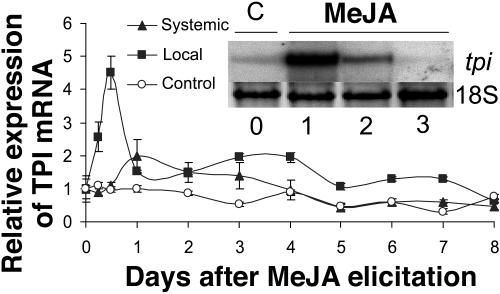
To examine the constitutive and MeJA-inducible levels of TPI mRNA, real-time PCR and northern-blot analysis were performed on total RNA from N. attenuata leaves. In comparison to unelicited plants, TPI transcripts increased 4.5-fold 12 h after elicitation in the treated leaf, with levels waning to those of control plants by day 8 (Fig. 1). Transcripts also increased in unelicited leaves that were phylotactically connected to the treated leaves (systemic leaves; one position younger than the local MeJA-treated leaf) with the largest elicitations (2-fold) observed 24 h after treatment (Fig. 1). Similarly, northern-blot analysis showed the largest amount of 1.4-kb TPI transcript levels in the systemic leaves 24 h after MeJA elicitation (Fig. 1, inset). These results demonstrate that TPI mRNA is highly induced after MeJA elicitation, attaining higher levels in the locally treated leaves than in the untreated systemic leaves, a pattern that mimics the leaves' response to Manduca sexta larvae attack (Zavala et al., 2004a).
Figure 1.
Fold induction (mean ± se) of the TPI transcripts of N. attenuata by real-time PCR in local (+1; squares) and systemic (0; triangles) leaves after MeJA (150 μg) elicitation relative to unelicited (control; circles) leaves from three replicate plants harvested separately for eight consecutive days after elicitation. Inset: Northern-blot analysis of TPI transcripts of systemic leaves for uninduced controls (C) and plants elicited with 150 μg of MeJA 1, 2, and 3 d after elicitation.
Purification and Characterization of TPIs from N. attenuata
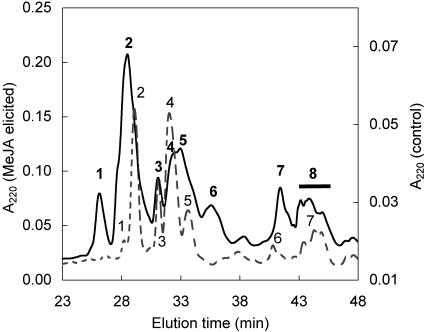
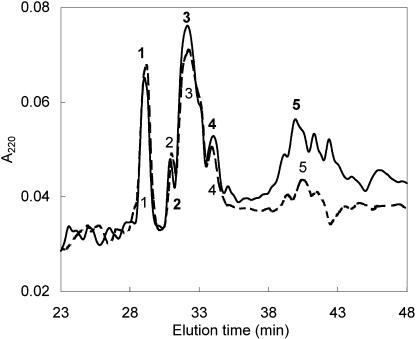
TPIs were purified from the protein fraction of unelicited (control) and MeJA-elicited leaves of the N. attenuata genotype collected in Utah (wild type). The two-step isolation scheme included separation of total TPI pool by gel permeation chromatography followed by reverse phase (RP)-HPLC fractionation. The RP-HPLC analysis revealed that MeJA elicitation increased the intensity and the number of peaks, suggesting that new TPIs appear after elicitation (Fig. 2). The total recovery of TPIs was about 6.6-fold higher in the MeJA-elicited leaves than in the unelicited leaves (Table I; ANOVA, F3,7, = 404; P < 0.0001), which demonstrates that the production of mature TPIs increases after MeJA elicitation.
Figure 2.
RP-HPLC separation of TPIs from N. attenuata of the wild-type genotype. Numbers refer to TPI fractions containing inhibitory activity against trypsin. RP-HPLC profile of TPIs from uninduced control leaves (dashed line, right y axis) and leaves elicited by MeJA (solid line, left y axis) are compared. Both samples were prepared from the same amount of leaf tissue. Note the scaling differences between the two y axes reflecting the different TPI levels in the constitutive and elicited material. The chromatography was performed on a C4 Vydac column equilibrated in 0.1% (v/v) trifluoroacetic acid (TFA), eluted with a 0.5%/min gradient of 60% (v/v) acetonitrile, and monitored by A220.
Table I.
Quantification of TPIs isolated from leaf tissue of N. attenuata
The recovery of RP-HPLC purified TPIs was determined by protein assays, and the relative distribution of isoinhibitors was calculated from the yield of N-terminal sequencing signals. The TPIs were compared for MeJA-elicited and control plants of the Utah genotype, which produces TPIs (wild type), and the Arizona genotype, which had been transformed with the TPI precursor gene of the wild-type genotype under control of a constitutive promotor (S++).
| Distribution of Isoinhibitors
|
|||||
|---|---|---|---|---|---|
| Genotype | Elicitation | Total TPI Yield | Irregular Processinga
|
Trimming Ratiob
|
|
| % | EEK-/EK- | DRI-/RI- | |||
| μgg−1 dry wt | |||||
| Wild type | Control | 56 ± 1 | 0 | 0/0 | 93/7 |
| MeJA | 370 ± 10 | 24 | 81/19 | 68/32 | |
| S++ | Control | 87 ± 4 | 0 | 0/0 | n.d.c |
| MeJA | 79 ± 3 | 0 | 0/0 | n.d.c | |
The relative content of irregularly processed TPIs in total the TPI pool. Irregular processing was defined by unremoved LP at the N terminus (EEK- or EK- termini).
The relative distribution of trimmed isoinhibitors. The trimming indicates one residue variability at the N terminus of irregularly (EEK- and EK- termini) and regularly processed (DRI- and RI- termini) TPIs.
n.d., Not determined.
The fractions with significant inhibitory activity against trypsin and chymotrypsin were collected and characterized by N-terminal sequencing and matrix-assisted laser-desorption ionization time of flight (MALDI-TOF) mass spectrometry (Table II). The N-terminal sequencing revealed that the fractions from unelicited leaves exclusively contained TPIs predicted from the TPI precursor coded by N. attenuata's pi gene (Fig. 3; Zavala et al., 2004a) and demonstrated that the isolated TPIs are small peptides of about 5.6 to 6.5 kD (Table II). MeJA elicitation increased the variability of the processing of termini of TPI peptides, which increased the number of TPI molecular species (isoinhibitors; Tables I and II). While the C-terminal trimming pattern that varied in one residue was independent of elicitation, the N-terminal processing of the TPI peptides resulted in two main subpopulations of TPIs in MeJA-elicited leaves. One subpopulation was processed as in control leaves (regular processing), having N termini starting DRICT- (or to a lesser degree, RICT-; Table II). The second subpopulation, specific for MeJA-elicited leaves, displayed an N-terminal sequence of EEKKNDRICT- (or to a lesser degree, EKKNDRICT-), which we referred to as irregularly processed TPIs (Table II). The N-terminal extension EEKKN corresponds to the linker peptide separating the individual TPI domains in the TPI precursor (Fig. 3; Zavala et al., 2004a). Since we did not detect this extension attached to the C terminus of any TPI, this linker peptide is removed initially at residues ↓E↓E and subsequently at residues ↓D↓R.
Table II.
Characterization of TPIs from the wild-type genotype
The assignment of the amino acid sequence of TPI peptides was based on N-terminal sequencing. The molecular mass was determined by MALDI-TOF mass spectrometry, and the predicted mass was calculated from cDNA sequence. The core sequence corresponds to [RIC…CPR] residues of the individual TPI domains (Fig. 3). The analyzed TPI fractions (see Fig. 2 for numbering) were isolated from unelicited plants (control) or plants elicited with MeJA.
| Fraction | Mass Observed | Mass Calculated | N-Terminal Sequence | TPI Domain | Assigned Structure |
|---|---|---|---|---|---|
| D | D | ||||
| Control 1 | 5758 | 5758 | DRIXT | B | D[core]S |
| 5759 | DRIXT | D | D[core] | ||
| Control 2 | 5670 | 5671 | DRIXT | B | D[core] |
| 5727 | 5729 | DRIXT | F | D[core] | |
| 5748 | 5744 | DRIXT | C or E | D[core] | |
| Control 3 | 5833 | 5831 | DRIXT | C | D[core]S |
| 5831 | DRIXT | F | D[core]T | ||
| Control 4 | 5743 | 5743 | DRIXT | C or E | D[core] |
| 5845 | 5845 | DRIXT | E | D[core]T | |
| 5847 | D | D[core]S | |||
| Control 5 | 5646 | 5644 | RIXTN | D | [core] |
| 5643 | RIXTN | B | [core]S | ||
| 5760 | 5760 | DRIXT | D | D[core] | |
| 5759 | DRIXT | B | D[core]S | ||
| Control 6 | 6142 | AXPRN | A | n.a.a | |
| 6265 | KAXPR | A | n.a. | ||
| Control 7 | 5130–6265 | AXPRN/KAXPR | A | n.a. | |
| MeJA 1 | 6231 | 6226 | EKKND | F | EEKKND[core] |
| 6332 | 6330 | EKKND | C | EKKND[core]S | |
| 6330 | F | EKKND[core]T | |||
| 6391 | 6388 | EEKKN | D | EEKKND[core] | |
| 6387 | B | EEKKND[core]S | |||
| 6460 | 6459 | EEKKN | C | EEKKND[core]S | |
| 6459 | F | EEKKND[core]T | |||
| MeJA 2 | 5557 | 5556 | RIXTN | B | [core] |
| 5617 | 5614 | RIXTN | F | [core] | |
| 5672 | 5671 | DRIXT | B | D[core] | |
| 5730 | 5729 | RIXTN | E | [core]T | |
| 5731 | RIXTN | D | [core]S | ||
| 5729 | DRIXT | F | D[core] | ||
| 5761 | 5760 | DRIXT | D | D[core] | |
| 5759 | DRIXT | B | D[core]S | ||
| 5831 | 5830 | DRIXT | C | D[core]S | |
| 5830 | F | D[core]T | |||
| 6372 | 6372 | EEKKN | C or E | EEKKND[core] | |
| 6475 | 6475 | EEKKN | D | EEKKND[core]S | |
| 6477 | EEKKN | E | EEKKND[core]T | ||
| MeJA 3 | 5834 | 5831 | DRIXT | C | D[core]S |
| 5831 | F | D[core]T | |||
| MeJA 4 | 5630 | 5628 | RIXTN | C or E | [core] |
| 5742 | 5743 | DRIXT | C or E | D[core] | |
| MeJA 5 | 5629 | 5628 | RIXTN | C or E | [core] |
| 5742 | 5743 | DRIXT | C or E | D[core] | |
| 5846 | 5845 | DRIXT | E | D[core]T | |
| 5847 | D | D[core]S | |||
| 5733 | 5729 | DRIXT | F | D[core] | |
| 5730 | RIXTN | E | [core]T | ||
| 5732 | D | [core]S | |||
| MeJA 6 | 5646 | 5644 | RIXTN | D | [core] |
| 5643 | B | [core]S | |||
| 5761 | 5760 | DRIXT | D | D[core] | |
| 5759 | B | D[core]S | |||
| MeJA 7 | 6137 | AXPRN | A | n.a. | |
| 6265 | KXPR | A | n.a. | ||
| MeJA 8 | 5130–6265 | KAXPR/AXPRN | A | n.a. |
n.a., Not assigned.
Figure 3.
Domain structure of TPI precursor from N. attenuata. The homologous domains in TPI precursor (GenBank AF 542547) are aligned, and their final processing into seven mature TPIs (TPI-A to -F) and VTS is indicated. The two-chain TPI-A is composed of N-terminal and C-terminal parts (TPI-A1 and TPI-A2, respectively); N-glycosylation signal in TPI-A2 is underlined. Bold letters signify the residues differing among the repeats and the residues specific for TPI-A. The box delineates position of the linker peptide separating TPI domains; the position of putative reactive site in the aligned domains is marked by an asterisk (Zavala et al., 2004a).
From calculations based on the total recovery of both processing subpopulations in MeJA-elicited leaves, TPIs with irregular termini represent about one-quarter of all TPIs produced (Table I). The relative production of TPIs with minor processing sites (E*EKKND*R-) compared with those with major processing sites (↓EEKKN↓DR-) was higher in MeJA-elicited leaves. MeJA-elicited leaves had TPIs with an EEK-:EK- ratio of 81:19 and TPIs with a DRI-:RI- ratio of 68:32, while these ratios for the TPIs isolated from unelicited leaves were 0:0 and 93:7, respectively (Table I).
Posttranslational Processing of the Two-Chain TPI
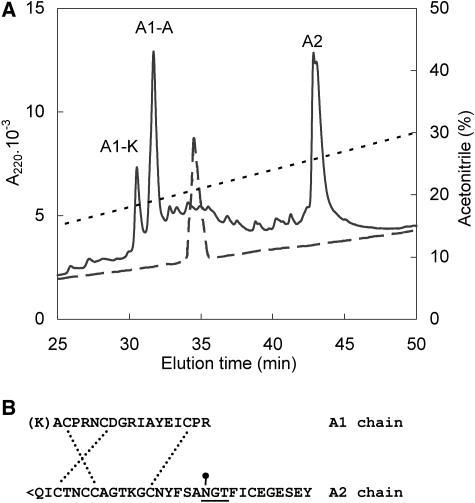
The TPI fractions with unassigned molecular masses (Table II) were positioned at the end of RP-HPLC elution profiles (Fig. 2). These fractions were analyzed by native eletrophoresis with an in-gel visualization, showing trypsin inhibitory activity and migrating in two bands. The MALDI-TOF mass spectrometry of these bands revealed a peptide of about 6.3 kD and a mixture of peptides with masses spanning 5.1 to 6.1 kD, respectively. Comparisons of the N-terminal sequences of the unassigned TPIs (Table II) with the cDNA sequence of the TPI precursor suggested that the peptides starting with KACPR- (or ACPR-) were derived from N-terminal half-domain (TPI-A1) of the precursor (Fig. 3; Zavala et al., 2004a). However, their molecular masses were higher than expected for TPI-A1, suggesting that TPI-A1 could be associated with C-terminal half-domain (TPI-A2). Similar results have been observed for chymotrypsin inhibitor from N. alata (Lee et al., 1999).
To determine whether TPI-A1 is associated with TPI-A2, we characterized the major unassigned TPI fraction (fraction MeJA 7; Table II) in greater detail using protein chemical methods. To separate chains connected by disulfide bridges, we reductively alkylated and chromatographed this material by RP-HPLC. The peaks A1-K and A1-A contained peptides differing only in their N-terminal trimming and had KACPR- and ACPR- sequences, respectively (Fig. 4A). The MALDI-TOF data clearly demonstrated that this chain corresponded to the TPI-A1 half-domain. Since the material from the peak A2 did not produce N-terminal sequencing signal and an assignable molecular mass, it was subjected to digestion with pyroglutamate aminopeptidase, which removes N-terminal pyroglutamic acid residues, and to enzymatic N-deglycosylation in order to read the unblocked N-terminal sequence and determine the mass (Table III). These results unambiguously demonstrated that this inhibitor was a two-chain TPI derived from N- and C-terminal parts of the TPI precursor (Fig. 4B). An analogous analysis performed with the other unassigned TPI fraction (fraction MeJA 8; Table II) revealed the two-chain structures containing the A2 chain with variations in C-terminal trimming and carbohydrate moiety (data not shown).
Figure 4.
Chain composition of the two-chain TPI from N. attenuata. A, Separation of the chains. The purified two-chain TPI (5 μg) was reductively carboxymethylated, and the liberated chains were purified by RP-HPLC (solid line). The position of the original two-chain TPI (1 μg) is indicated (dashed line). The chromatography was performed on a C4 Vydac column equilibrated in 0.1% (v/v) TFA, eluted with acetonitrile gradient (dotted line), and monitored by A220. B, The deduced covalent structure of the two-chain TPI. The A1 chain is N-terminally trimmed (see A1-K and A1-A peaks in A). The Gln residue in A2 chain is modified to pyroglutamic acid (<Q). The Asn residue in the consensus glycosylation signal (underlined) contains an N-linked oligosaccharide. The interchain disulfides are connected according to those known from homologous PI-II inhibitors.
Table III.
Characterization of posttranslational modifications of the two-chain TPI
The N termini of the separated chains were determined by sequencing. The C termini were identified by assigning MALDI-TOF mass spectrometry data to cDNA sequence. The A2 chain was characterized after removal of N-terminal pyroglutamic acid (<Q) and N-linked oligosaccharide of paucimannosidic type.
| Peak | Mass Observed | Mass Calculateda | N-Terminal Sequenceb | Assigned Structure |
|---|---|---|---|---|
| D | D | |||
| 1-K | 2240 | 2239 | KACPRNCDGRIAYEICPR | KACPRNCDGRIAYEICPR |
| 1-A | 2114 | 2111 | ACPRN | ACPRNCDGRIAYEICPR |
| A2 | 4502 | (<Q)ICTNc | n.a.d | |
| 3490e | 3487 | <QICTNCCAGTKGCNYFSANGTFICEGESEY |
Mass calculated with carboxymethylated Cys residues.
Cys residues identified as carboxymethyl Cys.
Sequence determined after treatment with pyroglutamate aminopeptidase.
n.a., Not assigned.
Mass determined after treatment with N-glycosidase A.
In summary, the A2 chain of the two-chain TPIs undergoes a complex posttranslational processing and acquires the following structural determinants: (1) cyclization at the N terminus pyroglutamic acid residue from the coded Gln residue; (2) N-glycosylation at the -NGT- sequence, which is consistent with the consensus N-glycosylation signal -NXT/S- (Fig. 4B). Since the deglycosylation was effective with N-glycosidase A but not with N-glycosidase F, the N-glycans are likely to contain a core Fuc residue. The oligosaccharide structure was deduced from the decrease in molecular mass after deglycosylation (decrease of 1009 or 1171 D, respectively, for the two major populations of the A2 chain) and from the general composition of plant N-glycans (Lerouge et al., 1998). Two main N-linked oligosaccharides of paucimannosidic type were predicted with a composition of Man2-3Xyl1Fuc1GlcNAc2 (3). Proteolysis at the C terminus removes segments of the vacuolar targeting sequence (VTS) predicted from the cDNA sequence (Zavala et al., 2004a) by fragmentation in the region of -SE*Y↓V*SK- (↓, major processing; *, minor processing).
Proteases That Process the TPI Precursors
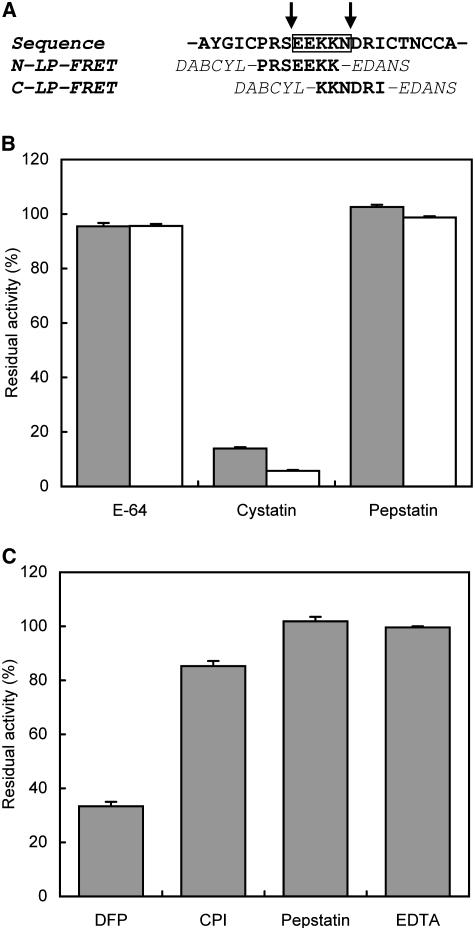
As seen from the TPI primary structure (Table I), the processing of TPI precursors involves the proteolysis at two main sites (-S↓EEKKN↓D-) to remove the linker peptides located between individual TPI domains. To characterize the processing proteases participating in this process, we designed synthetic substrates derived from the sequence of the linker peptides. The processing sites were separated in two substrates covering the N-terminal linker peptide fluorescence resonance energy transfer (N-LP-FRET) and C-terminal (C-LP-FRET) parts, which allowed the respective processing activities (Fig. 5A) to be independently monitored. The substrate structures were double labeled with an internally quenched fluorophore for use in FRET assays.
Figure 5.
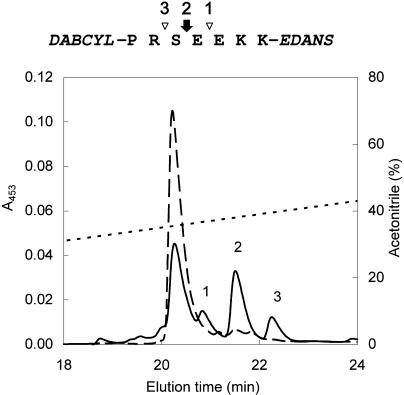
Screening of proteases involved in processing of the LP. A, The substrates were designed based on the sequence of the LP (boxed) to separate two main processing sites (arrows) where in vivo proteolysis proceeds in the TPI precursor. The synthetic substrates were double labeled with DABCYL and EDANS for use in the FRET assay. B, The activity of VPE in leaf extract was screened with C-LP-FRET substrate (open bars) and specific substrate Z-AAN-AMC (closed bars). C, The activity of subtilisin-like protease in leaf extract was screened with the N-LP-FRET substrate. For testing the inhibitory sensitivity, we treated the extract with the class-selective protease inhibitors before the assay: E-64 (500 μm), cystatin (50 μm), pepstatin (10 μm), diisopropyl fluorophosphate (DFP; 10 mm), mixture of Cys protease inhibitors (CPI; 2.05 mm), and EDTA (1 mm). The inhibition is expressed as a percentage of residual activity relative to the uninhibited control.
Because the C-terminal processing site contains an N-D bond, we tested the hypothesis that the fragmentation results from an asparaginyl endopeptidase of the Cys protease class, which prefers the Asn residue in P1 position of their substrates (Rotari et al., 2001). To characterize the putative Cys protease, the C-LP-FRET substrate was exposed to proteases in leaf extracts in the presence of class-specific inhibitors that differed in their selectivity to particular Cys proteases (Fig. 5B). Chicken cystatin significantly inhibited (about 92%) the substrate conversion, while E-64 had only a minor inhibitory effect (about 4%; ANOVA, F7,16, = 2073; P < 0.0001; Fig. 5B). This inhibitory pattern is a characteristic of the C13 family of Cys proteases (Kembhavi et al., 1993; Rotari et al., 2001), which includes plant asparaginyl endopeptidases of the vacuolar-processing enzyme (VPE) type (for review, see Muntz and Shutov, 2002). To verify the presence of a VPE in the leaf extracts of N. attenuata, we repeated the screening with the highly specific fluorescent substrate Z-AAN-AMC, which is commonly used to determine the activity of asparaginyl endopeptidases (Rotari et al., 2001). The observed inhibitory pattern was consistent with the results of the C-LP-FRET assay; both substrates were converted by specific digestion of the asparaginyl bond (Fig. 5B; P < 0.0001).
We used the N-LP-FRET substrate to examine the N-terminal processing site of the LP. This substrate was effectively digested by proteases in leaf extracts, which were strongly inhibited by the Ser protease inhibitor DFP (about 67%; ANOVA, F4,10, = 291; P < 0.0001), weakly inhibited by the cocktail of Cys protease inhibitors, and not influenced by pepstatin and EDTA, which regulate aspartic proteases and metalloproteases, respectively (Fig. 5C).
In order to determine which amino acid residue in the N-LP-FRET substrate is processed and confirm that the cleavage of the S-E bond is responsible for the N-LP- FRET assay result, we characterized the proteolytic digestion of N-LP-FRET by liquid chromatography-mass spectrometry (LC-MS), separated the fragments by RP-HPLC, and identified them by electrospray ionization (ESI) mass spectrometry. The analysis revealed three new peaks, and their mass data were assigned to the structure of N-LP-FRET (Fig. 6). These peaks were suppressed when the digestion was performed in the presence of DFP (data not shown), indicating that one major and two minor products are derived from the substrate. The major product was released by fragmentation of the S-E bond, which corresponds to the major processed position in this region of the TPIs. In addition, the two minor products (R-S and E-E) had their counterparts in the natively processed bonds in TPIs (Table II). These results suggest that the major protease contributing to conversion of N-LP-FRET substrate belongs to the Ser class, and its action is associated with major fragmentation directed at the S-E bond. These characteristics are similar to those of subtilisin-like proteases, which have been recently reported to be involved in proprotein processing in soybean (Glycine max; Boyd et al., 2002).
Figure 6.
Proteolysis of N-LP-FRET substrate. The substrate was incubated with leaf extract at pH 5.5, and the reaction mixture was characterized by LC-MS. The fragments were separated by RP-HPLC and identified by ESI mass spectrometry. The RP-HPLC profiles of untreated N-LP-FRET (dashed line) and the reaction mixture (solid line) are compared. The bonds digested in N-LP-FRET (arrow, major fragmentation; arrowheads, minor fragmentation) and elution of the corresponding fragments are indicated by numbers. RP-HPLC was performed on a C4 Vydac column equilibrated in 0.1% (v/v) TFA, eluted with an acetonitrile gradient (dotted line), and monitored by absorbance of DABCYL group.
We identified a subtilisin-like protease and VPE responsible for processing the TPI precursor at two major fragmentation sites of the LP (-S↓EEKKN↓DR-). The minor processing sites (-SE*EKKND*R-) identified in the TPIs can be produced by additional trimming by exopeptidases but also by action of the major processing enzymes since they are capable to cleave other selected bonds, albeit less efficiently. VPEs also weakly cut at the carboxyl site of Asp (D-R bond; Rotari et al., 2001), and subtilisin-like proteases cut S-E as well as E-E bonds (Boyd et al., 2002). The screening of processing proteases was performed with extracts of MeJA-elicited leaves. Qualitatively similar results were obtained with extracts from unelicited leaves (data not shown); however in these extracts, we observed a lower efficiency of proteolysis, indicating that the processing activity is modulated by MeJA elicitation.
Inducibility of Processing Proteases by MeJA
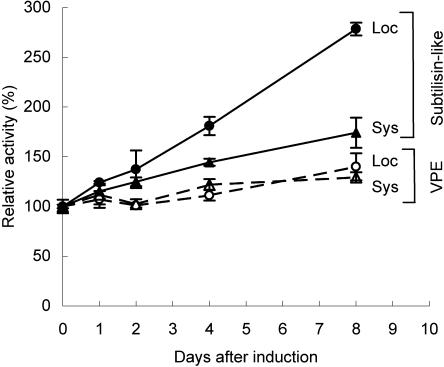
The identification of the two processing proteases and their specific substrates allowed us to examine MeJA-elicited changes in the capacity of the processing machinery. The enzymatic activity of VPE and subtilisin-like protease in MeJA-treated leaves of N. attenuata was monitored in an 8-d experiment (Fig. 7). The activity of the VPE did not change dramatically in either treated or untreated leaves on treated plants. VPE activity was weakly increased by about 15% by MeJA treatment in comparison to the unelicited control leaves (ANOVA, F4,25, L-MJ = 174; P < 0.0001; F4,25, S-MJ = 212; P < 0.0001; Fig. 7). In contrast, the activity of subtilisin-like protease gradually increased to levels 2.8-fold higher in the treated leaves (ANOVA, F4,25, L-MJ = 621; P < 0.0001) and 1.7-fold higher in the untreated leaves on treated plants 8 d after elicitation (ANOVA, F4,25, S-MJ = 103; P < 0.0001; Fig. 7). In summary, of the two proteases that participate in the TPI precursor processing, only the subtilisin-like protease strongly increases in activity after MeJA elicitation (ANOVA, F7,40, d-8 = 374; P < 0.0001; Fig. 7).
Figure 7.
Induction of processing proteases by MeJA in N. attenuata. Plants were elicited with MeJA, and the local (Loc) and systemic (Sys) leaves were harvested from three replicate plants on each of eight consecutive days after elicitation. The content (mean ± se) of VPE and subtilisin-like proteases was quantified in an activity assay with the specific substrates Z-AAN-AMC and N-LP-FRET, respectively. The relative activity levels of both VPE and subtilisin-like proteases are expressed relative to those of unelicited control leaves harvested on the same day.
Processing of the VTS
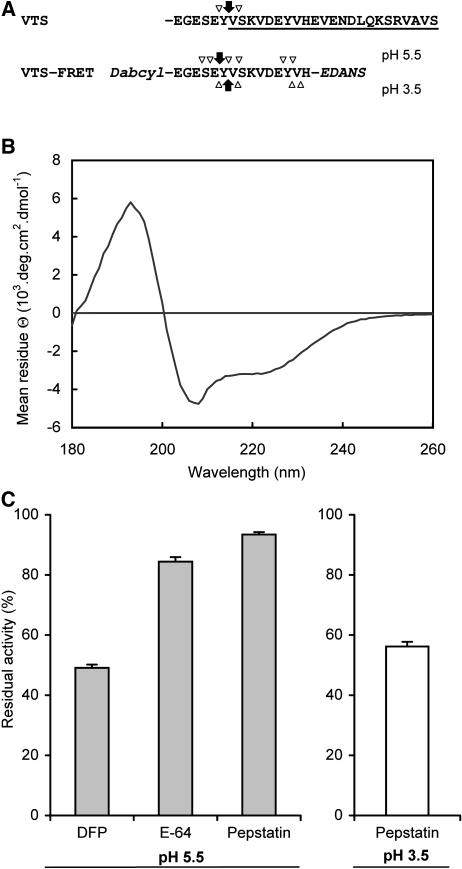
The 23-residue-long VTS is located at the C terminus of the TPI precursor and extends the sequence of the two-chain TPI (Fig. 3; Zavala et al., 2004a). An inspection of the amino acid sequence at the VTS processing site revealed a cassette with S and E residues (Fig. 8A), which resembled the N terminus of the LP processed by a subtilisin-like protease (Fig. 6). To determine which protease processed the VTS, we designed a synthetic substrate (VTS-FRET) for a FRET assay covering the sequence around the VTS processing site and an unprocessed portion of the VTS (Fig. 8A). Since a helical conformation is an important feature of VTSs in various plant proteins (Nielsen et al., 1996), we analyzed the conformation of the VTS-FRET substrate by circular dichroism spectroscopy. The obtained spectrum was typical for helical peptides, which demonstrated that the VTS-FRET substrate mimicked this important structural feature of the VTS region and hence was a suitable substrate for the processing study (Fig. 8B).
Figure 8.
Screening of proteases involved in the processing of VTS. A, The VTS-FRET substrate was designed based on the sequence of VTS (underline) surrounding the processing site where in vivo proteolysis proceeds in TPI precursor (arrows, major fragmentation; arrowheads minor fragmentation). The synthetic substrate was double labeled with DABCYL and EDANS for use in the FRET assay. The substrate was incubated with leaf extracts at pH 5.5 and 3.5, and the resulting fragments were identified by LC-MC. The major and minor bonds cleaved in VTS-FRET are labeled with arrows. B, Determination of helical conformation of VTS-FRET by circular dichroism spectroscopy. C, The proteolytic activity of leaf extracts (mean ± se) from three replicate plants was screened with VTS-FRET substrate at pH 5.5 and 3.5. The extracts were treated with the following class-selective protease inhibitors before the assay: diisopropyl fluorophosphate (DFP; 10 mm), E-64 (500 μm), and pepstatin (10 μm). The degree of inhibition was expressed in relation to the unelicited control leaves.
When we digested the VTS-FRET substrate with leaf extract proteases in the presence of class-specific protease inhibitors at pH 5.5, digestion was strongly inhibited by DFP (about 49%) and slightly inhibited by E-64 and pepstatin (ANOVA, F5,12, = 609; P < 0.0001; Fig. 8C). The fragments produced by the proteolysis of VTS-FRET were analyzed by LC-MS, which revealed the major cleavage site at the E-Y bond and several other minor cleavage bonds (Fig. 8A). In addition, the processing pattern was not identical to the native processing of the TPI-A2 chain, which was principally processed at the Y-V bond (Fig. 4B). Therefore, we examined the digestion of VTS-FRET under acidic conditions (pH 3.5), which is preferred by aspartic proteases. The acidic digestion of VTS-FRET increased the inhibitory activity of pepstatin (about 41%; P < 0.0001; Fig. 8C), and the LC-MS analysis of the resulting fragments revealed a fragment that was released by cleavage of the Y-V bond (Fig. 8, A and C). This cleavage site reflects the general preference of aspartic proteases (including those isolated from plant tissues) for hydrophobic residues around the scissible bond (Dunn, 2002; Simoes and Faro, 2004). In summary, our results suggest that the processing pattern in the region between the VTS and the TPI precursor is controlled by pH, and we conclude that an aspartic protease is likely to catalyze the removal of the VTS in vivo.
MeJA Elicitation Does Not Change the Processing of a Constitutively Expressed TPI in the Transformed S++ Genotype
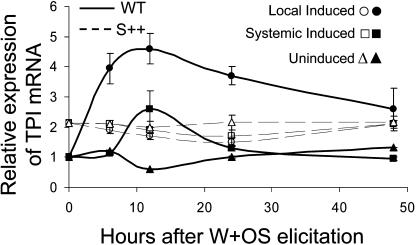
The MeJA-elicited change in the processing pattern could result from saturation of the proteolytic machinery in the wild-type genotype by the MeJA-elicited increase in TPI precursors. To test this hypothesis, we analyzed the MeJA-elicited changes in the TPIs in a natural mutant of N. attenuata collected from Arizona. This mutant does not express TPI activity or transcripts due to a mutation in the seven-domain repeat NaTPI located in the 5′ signal peptide, which results in a premature stop codon (J. Wu and I.T. Baldwin, unpublished data), and was transformed with a functional NaTPI from the Utah genotype under control of a constitutive promoter to restore the plants ability to produce TPIs (S++; Zavala et al., 2004b). To compare the constitutive and inducible levels of TPI mRNA between wild-type and S++ genotypes, real-time PCR analysis was performed on total RNA from N. attenuata leaves that had been wounded and had M. sexta oral secretion applied to their wounds (W + OS). In comparison to unelicited plants, in which TPI transcripts increased 4.5-fold 12 h after W + OS elicitation in the treated leaf and 2.5-fold in the systemic leaf of the wild-type genotype, no significant changes were found in TPI mRNA in the S++ genotype (Fig. 9). Since the S++ genotype constitutively produces TPI transcripts and activity at levels up to 70% of those found in MeJA-elicited wild-type plants and does not increase its levels of TPI transcripts or activity after elicitation (Fig. 9; Zavala et al., 2004a, 2004b), we hypothesized that if irregular processing of TPIs was also found in MeJA-elicited S++ plants, then MeJA treatment activates novel proteolytic capabilities that we have not yet identified. However, if it was not, then the MeJA-elicited irregular processing might result from saturation of the proteolytic machinery due to overproduction of the TPI precursor and the generation of semiprocessed forms. Our results were consistent with the latter hypothesis.
Figure 9.
Fold induction (mean ± se) of the TPI transcripts of N. attenuata by real-time PCR in local (+1; open and closed circles) and systemic (−1; open and closed squares) leaves of wild-type and S++ genotypes after wounded + M. sexta oral secretion (W + OS) elicitation relative to that of unelicited (uninduced; open and closed triangles) wild-type leaves from three replicate plants harvested and analyzed separately at 0, 6, 12, 24, and 48 h after elicitation. The S++ genotype was the Arizona genotype transformed with the TPI gene from the Utah genotype under control of a constitutive promoter to restore TPI production.
The RP-HPLC profiles of both unelicited and MeJA-elicited leaves from S++ plants revealed similar patterns of the separated TPI fractions (Fig. 10). The total yield of TPIs from the S++ genotype was about 41% (unelicited) and 54% (MeJA elicited) higher than that in the unelicited wild-type genotype (P = 0.04; Table I). As expected, similar amounts of TPIs (P = 0.6) with an EEK-:EK- ratio (0:0) were found in the S++ genotype either in unelicited or MeJA-elicited leaves (Table I). The TPI fractions were characterized by MALDI-TOF mass spectrometry, and the N-terminal sequencing and the primary structure of the individual TPIs were deduced (Table IV). This analysis revealed that TPIs from unelicited and MeJA-elicited leaves are processed analogously, and the fractions started with the N-terminal sequence of DRICT, or to a lesser extent, RICT. The occurrence of the two-chain TPI (fractions numbered 5 in Table IV) also suggests that the TPI precursor expressed in the transformed plants is correctly folded into a circular form. In summary, the observations that the MeJA elicitation of the S++ genotype did not change either the amount or the structure of the TPIs and that these were similar to those recovered from uninduced plants of the wild-type genotype are consistent with the hypothesis that the change in the processing pattern after MeJA elicitation results from saturation of the processing machinery.
Figure 10.
RP-HPLC separation of TPIs from the Arizona genotype S++, which was transformed with the TPI gene from Utah under control of a constitutive promoter to restore TPI production. Numbers refer to TPI fractions containing inhibitory activity against trypsin. RP-HPLC profile of TPIs from uninduced control leaves (dashed line) and MeJA-elicited leaves (solid line) are compared. Both samples were prepared from the same amount of leaf tissue. The RP-HPLC was performed under the conditions described in Figure 2.
Table IV.
Characterization of TPIs from S++ plants in which the TPI production of the Arizona genotype was restored by transformation
The assignment of the amino acid sequence of the TPI peptides is based on N-terminal sequencing. The molecular mass was determined by MALDI-TOF mass spectrometry, and the predicted mass was calculated from cDNA sequence. The core sequence corresponds to [RIC…CPR] residues of the individual TPI domains (Fig. 3). The analyzed TPI fractions (see Fig. 10 for numbering) were isolated from unelicited plants (control) or plants elicited with MeJA.
| Fraction | Mass Observed | Mass Calculated | N-Terminal Sequence | TPI Domain | Assigned Structure |
|---|---|---|---|---|---|
| D | D | ||||
| Control 1 | 5673 | 5671 | DRIXT | B | D[core] |
| 5732 | 5729 | DRIXT | F | D[core] | |
| MeJA 1 | 5673 | 5671 | DRIXT | B | D[core] |
| 5731 | 5729 | DRIXT | F | D[core] | |
| Control 2 | 5832 | 5831 | DRIXT | C | D[core]S |
| 5831 | DRIXT | F | D[core]T | ||
| MeJA 2 | 5832 | 5831 | DRIXT | C | D[core]S |
| 5831 | DRIXT | F | D[core]T | ||
| Control 3 | 5633 | 5628 | RIXT | C or E | [core] |
| 5748 | 5743 | DRIXT | C or E | D[core] | |
| 5841 | 5844 | DRIXT | E | D[core]T | |
| 5846 | D | D[core]S | |||
| MeJA 3 | 5631 | 5628 | RIXT | C or E | [core] |
| 5746 | 5743 | DRIXT | C or E | D[core] | |
| 5848 | 5844 | DRIXT | E | D[core]T | |
| 5846 | D | D[core]S | |||
| Control 4 | 5644 | 5644 | RIXTN | D | [core] |
| 5643 | RIXTN | B | [core]S | ||
| 5759 | 5760 | DRIXT | D | D[core] | |
| 5759 | DRIXT | B | D[core]S | ||
| MeJA 4 | 5646 | 5644 | RIXTN | D | [core] |
| 5643 | RIXTN | B | [core]S | ||
| 5762 | 5760 | DRIXT | D | D[core] | |
| 5759 | DRIXT | B | D[core]S | ||
| Control 5 | 6140 | AXPRN | A | n.a.a | |
| 6270 | KAXPR | A | n.a. | ||
| MeJA 5 | 6138 | AXPRN | A | n.a. | |
| 6269 | KAXPR | A | n.a. |
n.a., Not assigned.
DISCUSSION
To understand the mechanism of the TPI-based inducible defense response in N. attenuata leaves, we characterized the transcription of NaTPI and the complete set of posttranslational modifications of the TPI molecules. MeJA and W + OS treatments increased TPI mRNA 4.5-fold (in treated leaves) within 12 h of elicitation. Similar transcriptional responses were observed after herbivore attack (Zavala et al., 2004a). The translated seven-domain TPI precursor is processed into the active TPIs, comprising one two-chain and six single-chain domains. The two-chain TPIs were derived from the N and C termini of the NaTPI precursor, which suggested a circular organization and allowed us to generalize from the groundbreaking work that characterized the circular architecture of the multidomain precursors of PI-II family in N. alata stigmas (Lee et al., 1999). The yield of the purified TPIs increases about 6-fold after elicitation in the treated leaves (Table I). The highest TPI activity is attained between 3 and 5 d after elicitation by either MeJA treatment or caterpillar attack, and the elicited responses dramatically reduce M. sexta larvae performance (van Dam et al., 2001; Zavala et al., 2004a). The reduction in larval performance is a consequence of the high concentrations of TPI inhibitors, but posttranscriptional changes may also increase the defensive function of TPIs.
The regular processing pattern of constitutively produced TPIs in the wild-type genotype of N. attenuata is analogous (with regard to proteolytic events) to the processing of the TPI precursor-derived PIs from N. alata stigmas (Atkinson et al., 1993; Heath et al., 1995). However, the processing pattern of the TPI precursor in N. attenuata changes after MeJA elicitation; specifically, a new subpopulation of TPI molecules is added that contains the N-terminal EEKKN extension of the LP that is normally removed in the constitutively produced TPIs. The cleavage in this proteolysis-sensitive region is controlled by proteases with different substrate specificity acting at two major processing sites of the LP (-S↓EEKKN↓D-). Two different effector proteases were identified with specific synthetic substrates used in FRET in vitro assays. An asparaginyl endopeptidase of the VPE type (C13 protease family) was found to cleave the N-D bond of the single-chain TPI. The VPE is also responsible for cleavage of the N-Q bond, which releases a two-chain TPI and generates the capping of the newly formed N terminus. In Arabidopsis (Arabidopsis thaliana), a VPE protease with specificity for asparaginyl bonds was recently found to process vacuolar proteins in protein storage and lytic vacuoles (Shimada et al., 2003; Gruis et al., 2004). In addition, another Asn-specific protease initiates equistatin proteolysis when this PI is expressed heterologously in potato (Outchkourov et al., 2003). Here, we report that an additional protease, a subtilisin-like protease(s) of the S8 protease family with a cleavage preference similar to C1 proteases (Barnaby et al., 2004), was responsible for the cleavage of the S-E bond in the LP. These subtilisin-like proteases have substrate specificity for bonds containing Glu residues and process β-conglycinin in soybean seedlings (Boyd et al., 2002). Hence, the activity of both the subtilisin-like and the VPE proteases can influence the processing pattern of the TPI precursor after MeJA elicitation.
MeJA elicitation gradually increased the activity of the subtilisin-like protease 3-fold in treated leaves. In contrast, the activity of the VPE was not increased after MeJA elicitation. While the activity of other proteases is known to be elicited in the context of an induced defense response, the activity of the effector proteases has not been associated with the processing pattern of their products. Interestingly, the expression of γVPE in vegetative tissues of Arabidopsis is up-regulated by ethylene or salicylic acid, but not by MeJA treatment (Kinoshita et al., 1999). Of the many subtilisin-like proteases that have been described, only a few of the P69 subfamily have been screened for their defensive role, and they were found to respond to pathogen attack and salicylic acid treatment in Arabidopsis and tomato (Lycopersicon esculentum; Jorda et al., 1999; Jorda and Vera, 2000). These results suggest that saturation of the VPE, but not the subtilisin-like protease, in MeJA-elicited leaves is responsible for the irregular processing of the TPI precursor. Since the subtilisin-like protease seems to be elicited independently of the activity of the VPE, we predict that a different pattern of irregular processing would be observed if plants are induced with salicylic acid or other elicitors of the VPE.
To complete the analysis of all processing sites in the TPI precursor, we examined the region at which the VTS is removed from the C terminus of the precursor molecule. This region, with the major cleavage site of -ESEY↓VS-, displayed a constant proteolytic pattern regardless of MeJA elicitation. Interestingly, a sequence rich in Glu residues (-ESE-) is located upstream of the major cleavage site, which is homologous to the processing site of the subtilisin-like protease in the LP. However, with the FRET in vitro assay and the VTS-derived synthetic substrate, we demonstrated that an aspartic protease is responsible for cleavage in this Glu-rich region. This suggests that the site at which the VTS processing takes place is likely sterically hindered or separated by differential compartmentation from the subtilisin-like protease. Analysis of the sorting of the PI precursor in N. alata stigmas demonstrated that the C-terminal VTS is essential for vacuolar targeting and removed during its transport from prevacuolar to vacuolar compartment (Miller et al., 1999, 2000).
We also demonstrated that two new posttranslational modifications are produced in N. attenuata's TPIs, neither of which has been described in the multidomain members of the PI-II family: the N-linked glycosylation and the formation of pyroglutamic acid residue. The latter modification proceeds from the N terminus of the A2 chain from the two-chain TPI by a mechanism that converts the Gln to pyroglutamic acid residue after cleaving the N-Q bond. This finding is consistent with the recent identification of the same highly specific event that introduces pyroglutamic acid into pumpkin (Cucurbita maxima) seed proteins when its PV100 precursor is processed (Yamada et al., 1999). The N-terminally capped proteins are thought to be resistant against aminopeptidase degradation. The protective function of the capped proteins is demonstrated by the detection of the stabilized A2 chain only in one processing variant, in contrast to all other noncapped chains derived from TPI precursors that were trimmed of one residue during their maturation.
To summarize, we determined the structure of the processing sites in the TPI precursor and predicted their respective effector proteases. At least three proteases from different classes, two of which are synchronized for a two-step fragmentation of the LP with the primary cleavage controlled by a subtilisin-like protease, contribute to the proteolytic machinery. Such a dual model with two proteases cooperating to remove a short internal peptide segment may be a general mechanism for the processing of plant protein precursors because a dual processing of 2S albumins (by aspartic protease and VPE) was recently demonstrated in Arabidopsis (Shimada et al., 2003). In addition, we present a novel screening method for a processing protease with the potential to cleave the C-terminal type of a helical VTS of plant proproteins. A comparison of the C-terminal VTS of different proproteins reveals that these structures have little sequence homology, including the sequence of the VTS-processing site (Sticher et al., 1993; Miller et al., 1999). The lack of sequence homology of different proproteins suggests that different types of proteases are involved in the processing of individual C-terminal VTSs. However, our results suggest that an aspartic protease may be a common VTS-processing enzyme for the multidomain precursors of the PI-II family. These proproteins from several species contain a VTS and VTS cleavage bond that is homologous to that which we characterized in this study, which is also utilized in the PI precursor from N. alata (Miller et al., 1999).
MeJA elicitation in N. attenuata significantly up-regulates only one of the two proteases that control the processing of the LP in TPI precursor. Since after elicitation the processing machinery is supplied with dramatically higher levels of substrate, the MeJA-elicited change in processing pattern likely results from the accumulation of the irregular TPIs that are generated by the activity of the inducible protease. To dissect the effect of MeJA elicitation on the processing proteases from its effect on their substrate, we expressed the NaTPI precursor gene under a constitutive promoter in an N. attenuata genotype from Arizona, which is naturally deficient in TPI production. The transformation restored the ability of the Arizona genotype to produce TPIs, but neither MeJA nor W + OS elicitation increased TPI activity or transcript level (Fig. 9; Zavala et al., 2004b). Since the TPIs in control plants as well as in MeJA-elicited transgenic plants were found to be regularly processed, we conclude that the differential regulation of processing proteases changes the processing pattern of the TPI precursor only when high substrate levels saturate the less abundant protease (the VPE). Saturation of the VPE could account for the appearance of the irregular TPIs with the processing pattern of the abundant subtilisin-like protease.
We examined the 3D structure of the tomato PI-II inhibitor in complexes with proteases (Barrette-Ng et al., 2003) and found that the N terminus of the PI-II family of molecules is located in the vicinity of the reactive site, which binds to a protease. Therefore, the N-terminal extension EEKKN- in homologous TPIs from N. attenuata might modulate the interaction of irregular TPIs with the target proteases. It is tempting to speculate that this molecular mechanism functions to broaden the spectrum of inhibitory specificities of TPI isoinhibitors. The detailed mapping of inhibitory specificity of regular and irregular TPIs onto a panel of ecologically relevant target proteases will be necessary to test this hypothesis. Such an induced diversity may be particularly important for the defensive function of PIs and may enhance their defensive efficacy against herbivores and pathogens on either ecological or evolutionary time scales. The discovery of multiple regulatory pathways controlling production of defensive compounds, including those described here, will help increase our understanding of the complex biological processes that are set in motion when herbivores attack plants.
MATERIALS AND METHODS
Plant Material and Elicitation
Seeds of Nicotiana attenuata Torr. ex Watts (Solanaceae) used in these experiments originated from collections from natural populations in Utah (Baldwin, 1998) and Arizona (Glawe et al., 2003). The genotype collected from Arizona has MeJA-inducible nicotine production identical to that found in plants collected from Utah but completely lacks the ability to produce TPIs or accumulate TPI mRNA (Glawe et al., 2003). Plants of the Arizona genotype were transformed with the full-length cDNA of the seven-domain N. attenuata pi gene (S++) from the wild-type Utah genotype in the sense orientation under control of a cauliflower mosaic virus 35S promoter (Zavala et al., 2004b). Homozygous S++ plants from the T2 generation harboring one copy of the transgene were used in this experiment (Zavala et al., 2004b).
Seeds were germinated with 5 mL of 50× diluted liquid smoke (House of Herbs) and 50 μL of 0.1 m gibberellic acid (Fluka) for 1 h and the seedlings transferred to 1-liter pots filled with peat-based substrate. The plants were grown in growth chambers or in a glasshouse with a 16/8-h light/dark cycle and 1000 to 1500 μmol m−2 s−1 photosynthetic photon flux density at 28°C and 65% relative humidity. Rosette-stage plants were elicited by either wounding and applying Manduca sexta oral secretion to the puncture wounds (W + OS) or applying 150 μg of MeJA (Sigma-Aldrich) with an enantiomeric composition close to its thermodynamic equilibrium (90.1% 1R, 2R MeJA and 8.3% 1R, 2S MeJA for the two naturally occurring epimers) in 20 μL of lanolin paste to the midvein of fully expanded leaves (node +1; one position older than the source-sink transition leaf; van Dam et al., 2001) from each plant. In order to determine the effect of MeJA elicitation on TPI production and TPI transcript accumulation, leaves either treated or untreated with MeJA growing at node +1 (local) and source-sink leaves growing at node 0 (systemic; one position younger than the leaves growing at nodes +1) were harvested at 12 h and daily for 8 d after induction. To compare TPI transcript accumulation between wild-type and S++ genotypes, leaves from both genotypes either treated or untreated with W + OS growing at node +1 (local) and source-sink leaves growing at node 0 (systemic) were harvested from three replicate plants and analyzed separately at 0, 6, 12, 24, and 48 h after elicitation.
Purification of TPIs
Unelicited and MeJA-elicited leaves were harvested 3 d after elicitation and homogenized separately using a Cat X-120 homogenizer (Ingenierurburo CAT) in 50 mm sodium citrate buffer (pH 4.3), 1 m NaCl, and 0.5% (w/v) sodium hydrosulfite. The endogenous proteases in the extract were inhibited by adding a mixture of protease inhibitors (Cocktail P9599; Sigma-Aldrich). After 12 h extraction at 4°C, the insoluble material was removed by centrifugation at 12,000g for 30 min. The soluble proteins were precipitated with ammonium sulfate at 80% saturation at 4°C and centrifuged (12,000g for 30 min). The pellet was dialyzed (1 kD cutoff) against water, lyophilized, dissolved in 0.1 m Tris-HCl (pH 8.0), 0.5 m NaCl, and chromatographed on a Sephadex G-50 Superfine column (Amersham Biosciences) equilibrated with the same buffer. The part of the elution profile with inhibitory activity against bovine trypsin and chymotrypsin was pooled, dialyzed against water, and lyophilized. The total TPI pool was chromatographed on RP-HPLC performed on a Hitachi LaChrom L7100. The material was loaded on a Vydac C4 column (214TP510) equilibrated in 0.1% (v/v) TFA at a flow rate of 3 mL min−1 and eluted with a 0.5% min−1 gradient of 60% (v/v) acetonitrile solution in 0.1% (v/v) TFA. The chromatography was monitored by measuring A220. The collected fractions with inhibitory activity against bovine trypsin and chymotrypsin were dried in a Speed-Vac concentrator (Thermo Savant).
TPI Inhibition Assay
The inhibition assay was performed with 1 nm bovine trypsin or chymotrypsin in 0.1 m Tris-HCl (pH 8.0) and 10 mm CaCl2. After preincubation with TPI inhibitor (15 min), the residual enzymatic activity was determined at 30°C with 20 μm substrate Z-FR-AMC (for trypsin) or Suc-AAPF-AMC (for chymotrypsin; Bachem). The reaction was continuously monitored in microplate format using excitation and emission wavelengths of 360 and 465 nm, respectively, with fluorescence reader GENios Plus (TECAN).
In-Gel Activity of TPIs
The purified TPIs were electrophoresed on a 12% (w/v) polyacrylamide vertical slab gel with a discontinous buffer system (Davis, 1964). The TPIs were visualized using the gel-x-ray contact print technique (Giri et al., 1998). Briefly, the resolved gels were equilibrated and immersed in 0.1% (w/v) bovine trypsin and subsequently placed on undeveloped x-ray film. The TPI activity bands were visualized as unhydrolyzed gelatin against a background of gelatin hydrolyzed by trypsin.
Synthesis of FRET Substrates
The DABCYL and EDANS form a pair; the absorbance of the DABCYL acceptor overlaps with the fluorescence of the EDANS donor, thus ensuring that the fluorescence is quenched through FRET (De Angelis, 1999). Proteolytic cleavage of the FRET substrate releases a fragment containing EDANS and restores its fluorescence. The FRET substrates were synthesized by Fmoc solid phase peptide chemistry on an ABI 433A peptide synthesizer (Applied Biosystems; Atherton and Sheppard, 1989). The DABCYL and EDANS tags were incorporated at the peptide termini using Fmoc-Lys(Dabcyl)-OH and Fmoc-Glu(EDANS)-OH precursors. The peptides were prepared in the form of peptidyl amides containing the free N terminus. They were purified by RP-HPLC on a Vydac C18 column (218TP510) equilibrated in 0.1% (v/v) TFA at a flow rate of 3 mL min−1 and eluted with a 1% min−1 gradient of 90% (v/v) acetonitrile solution in 0.1% (v/v) TFA. The peptides were characterized by ESI mass spectrometry on an LCQ Classic Finnigan Mat (Thermo Finnigan).
Screening of Plant Protease Activity
The Activity Assay with FRET Substrates
The substrate (20 μm) was incubated at 30°C in 50 mm sodium citrate (pH 3.5 or 5.5), 2.5 mm dithiothreitol, and 0.1% (v/v) Brij 35 with 5 to 25 μL of leaf extract. The kinetics of the released product was continuously monitored in microplate format using excitation and emission wavelengths of 330 and 485 nm, respectively, with the fluorescence reader GENios Plus (TECAN).
The Activity Assay with Z-AAN-AMC Substrate (Bachem)
The substrate (25 μm) was incubated at 30°C in 50 mm sodium citrate (pH 5.5), 2.5 mm dithiothreitol, 2 mm EDTA, and 0.1% (v/v) Brij 35 with 5 to 25 μL of leaf extract. The kinetics of the released product was continuously monitored in microplate format using excitation and emission wavelengths of 360 and 465 nm, respectively, with the fluorescence reader GENios Plus (TECAN).
The inhibition activity of protease was measured using the activity assay by adding the specific substrate after preincubation (10 min) of the reaction mixture with a protease inhibitor. The following inhibitors were used: E-64 (500 μm), chicken cystatin (50 μm), diisopropyl fluorophosphate (10 mm), cocktail of Cys protease inhibitors (1 mm N-ethylmaleimide, 1 mm iodoacetic acid, and 50 μm E-64), pepstatin (10 μm), and EDTA (1 mm).
Preparation of the Leaf Extract
Fresh leaf tissue was homogenized using the Cat X-120 homogenizer (Ingenierurburo CAT) in 50 mm sodium citrate (pH 5.5), 2.5 mm dithiothreitol, and 1% (w/v) CHAPS. After 2 h extraction on ice, the homogenate was clarified by centrifugation (12,000g for 15 min), and the final extract was set to a concentration of 0.2 mg of protein per milliliter.
Analysis of FRET Substrate Fragments
A reaction mixture containing 150 μm of the substrate and 20 μL of the leaf extract was incubated in 100 μL of 50 mm sodium citrate (pH 3.5 or 5.5) and 2.5 mm dithiothreitol at 30°C for 4 to 12 h. The reaction was stopped by adding 10 μL of TFA, and the mixture was filtrated through Micropure-0.22 Separator (Millipore). The reaction mixture was analyzed by LC-MS (Finnigan Mat) using (1) chromatography by RP-HPLC with a Vydac C18 column (218TP54) equilibrated in 0.1% (v/v) TFA at a flow rate of 0.1 mL min−1 and eluted with a 1% min−1 gradient of 90% (v/v) acetonitrile solution in 0.1% (v/v) TFA, and (2) ESI mass spectrometry on an LCQ Classic (Finnigan Mat).
Protein Chemistry Methods
The purified two-chain TPI (25 μg) was reduced under denaturing conditions in 100 μL of 0.25 m Tris-HCl (pH 8.3), 8 m urea, 10 mm dithiothreitol, and 2 mm EDTA for 2 h at 37°C in N2 atmosphere and subsequently carboxymethylated with 22 mm iodoacetic acid for 30 min at 37°C in the dark and N2 atmosphere (Horn et al., 2002). The reaction mixture was treated with 0.1% TFA and chromatographed on a Vydac C4 column (214TP54). The RP-HPLC system was equilibrated in 0.1% (v/v) TFA at a flow rate of 1 mL min−1 and eluted with a 1% min−1 gradient of 90% (v/v) acetonitrile solution in 0.1% (v/v) TFA. To remove pyroglutamate from the N terminus, 2.5 μg of the carboxymethylated peptide was digested with 1 mU of pyroglutamate aminopeptidase (Sigma-Aldrich) in 20 μL of 50 mm sodium phosphate (pH 7.8), 10 mm dithiothreitol, and 2 mm EDTA for 6 h at 55°C. The digests were directly subjected to automatic Edman degradation. To remove N-glycans, 2.5 μg of the carboxymethylated peptide was deglycosylated at 37°C for 24 h with 0.5 mU of N-glycosidase F or with 1 mU of N-glycosidase A (Roche Diagnostics) in 30 μL of 20 mm sodium phosphate (pH 7.8), 1% mercaptoethanol, and 1 mm EDTA and in 50 μL of 20 mm sodium acetate (pH 5.0), 10 mm dithiothreitol, and 2 mm EDTA, respectively. The samples were lyophilized and subjected to MALDI-TOF mass spectrometry.
Analytical Methods
MALDI-TOF analysis was carried out using a TofSpec 2E (Waters Micromass). N-terminal sequencing was done by automated Edman degradation on an ABI 494A protein sequencer (Applied Biosystems). The circular dichroism spectra were measured with 0.4 mm peptide in 10 mm sodium phosphate (pH 6.5) and 50% (v/v) trifluoroethanol (Lazo and Downing, 1997) at 26°C using CD6 Dichrograph (Jobin Yvon, Instruments S.A.).
RNA Expression Analysis
Leaves growing at node 0 either uninduced or elicited with MeJA from four plants were harvested 1, 2, and 3 d after elicitation and pooled by harvest day for RNA isolation. Total RNA was extracted using the acid guanidine thiocyanate-phenol-chloroform method (Winz and Baldwin, 2001). RNA was quantified spectrophotometrically at 260 nm. RNA (10 μg) was size-fractionated on 1.2% (w/v) agarose-formaldehyde gel and blotted onto nylon membrane (NEN Life Science Products). Ethidium bromide staining of the gel prior to blotting revealed rRNA bands that served as loading controls. RNA gel blots were hybridized using radiolabeled probes generated by PCR using the plasmid pUCPI2/14 (Glawe et al., 2003) containing the repeat domain. Hybridization was detected using an FLA 3000 phosphor imager and quantified with Aida Image Analyzer v. 3.11 (Fuji Photo Film). For the real-time PCR analysis, leaves growing at nodes +1 and 0 of plants either uninduced or elicited with MeJA were harvested from 0 to 8 d after elicitation from four replicate plants per harvest day and used for RNA isolation. The isolated RNA was quantified spectrophotometrically and diluted to 300 ng μL−1. The diluted RNA was reverse transcribed (Applied Biosystems), and 10 ng of the reverse-transcribed template was used in a 25-μL PCR reaction containing 1× universal mix (Eurogentec), 300 nm forward (5′-TCAGGAGATAGTAAATATGGCTGTTCA-3′) and reverse primers (5′-ATCTGCATGTTCCACATTGCTTA-3′), and 300 nm of FAM-labeled Taqman probe (5′-TCCTTGCTCTCCTCCTCTTATTTGGAATGTCT-3′) with 18 s RNA (Eurogentec) as internal standard. Thermal cycling and detection were performed on an ABI Prism 7700 sequence detector.
Statistical Analysis
Data were analyzed with STATVIEW (SAS Institute). The differences in proteolytic activity were calculated as the relative activity with respect to the control (all proportions were arcsin square root transformed before statistical analysis to correct non-normality) and analyzed by ANOVA followed by Fisher's protected lsd post hoc comparison in all experiments.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AF542547.
Acknowledgments
We thank Dr. B. Schlott (Institute for Molecular Biotechnology, Jena, Germany) and Dr. Z. Voburka (Institute of Organic Chemistry and Biochemistry) for N-terminal sequencing, Dr. A. Svatoš (Max Planck Institute for Chemical Ecology) and Dr. J. Cvačka (Institute of Organic Chemistry and Biochemistry) for mass spectrometry, Dr. P. Maloň and H. Dlouhá (Institute of Organic Chemistry and Biochemistry) for circular dichroism spectroscopy, and E. Wheeler (Max Planck Institute for Chemical Ecology) for editorial assistance.
This work was supported by the Max Planck Society. M.H., L.D.-M., M.V., and M.M. were supported by the Grant Agency of the Czech Republic (grant no. 522/04/1286), the Grant Agency of the Czech Academy of Sciences (grant no. IAA4055303), and by the research project Z40550506.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.064006.
References
- Atherton E, Sheppard RC (1989) Solid Phase Peptide Synthesis: A Practical Approach. IRL Press at Oxford University Press, Oxford
- Atkinson AH, Heath RL, Simpson RJ, Clarke AE, Anderson MA (1993) Proteinase inhibitors in Nicotiana alata stigmas are derived from a precursor protein which is processed into five homologous inhibitors. Plant Cell 5: 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin I (2001) An ecologically motivated analysis of plant-herbivore interactions in native tobacco. Plant Physiol 127: 1449–1458 [PMC free article] [PubMed] [Google Scholar]
- Baldwin IT (1998) Jasmonate-induced responses are costly but benefit plants under attack in native populations. Proc Natl Acad Sci USA 95: 8113–8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnaby NG, He F, Liu X, Wilson KA, Wilson KA, Tan-Wilson A (2004) Light-responsive subtilisin-related protease in soybean seedling leaves. Plant Physiol Biochem 42: 125–134 [DOI] [PubMed] [Google Scholar]
- Barrette-Ng IH, Ng KK, Cherney MM, Pearce G, Ryan CA, James MN (2003) Structural basis of inhibition revealed by a 1:2 complex of the two-headed tomato inhibitor-II and subtilisin Carlsberg. J Biol Chem 278: 24062–24071 [DOI] [PubMed] [Google Scholar]
- Barta E, Pintar A, Pongor S (2002) Repeats with variations: accelerated evolution of the Pin2 family of proteinase inhibitors. Trends Genet 18: 600–603 [DOI] [PubMed] [Google Scholar]
- Boyd PM, Barnaby N, Tan-Wilson A, Wilson KA (2002) Cleavage specificity of the subtilisin-like protease C1 from soybean. Biochim Biophys Acta 1596: 269–282 [DOI] [PubMed] [Google Scholar]
- Broadway RM (1996) Dietary proteinase inhibitors alter complement of midgut proteases. Arch Insect Biochem Physiol 32: 39–53 [Google Scholar]
- Davis BJ (1964) Disc electrophoresis II: methods and application to human serum. Ann N Y Acad Sci 121: 404–429 [DOI] [PubMed] [Google Scholar]
- De Angelis DA (1999) Why FRET over genomics? Physiol Genomics 1: 93–99 [DOI] [PubMed] [Google Scholar]
- Dunn BM (2002) Structure and mechanism of the pepsin-like family of aspartic peptidases. Chem Rev 102: 4431–4458 [DOI] [PubMed] [Google Scholar]
- Girard C, Le Metayer M, Bonade Bottino M, Pham Delegue MH, Jouanin L (1998) High level of resistance to proteinase inhibitors may by conferred by proteolytic cleavage in beetle larvae. Insect Biochem Mol Biol 28: 229–237 [DOI] [PubMed] [Google Scholar]
- Giri AP, Harsulkar AM, Deshpande VV, Sainani MN, Gupta VS, Ranjekar PK (1998) Chickpea defensive proteinase inhibitors can be inactivated by podborer gut proteinases. Plant Physiol 116: 393–401 [PMC free article] [Google Scholar]
- Glawe GA, Zavala JA, Kessler A, Van Dam NM, Baldwin IT (2003) Ecological costs and benefits correlated with trypsin protease inhibitor production in Nicotiana attenuata. Ecology 84: 79–90 [Google Scholar]
- Gruis D, Schulze J, Jung R (2004) Storage protein accumulation in the absence of the vacuolar processing enzyme family of cysteine proteases. Plant Cell 16: 270–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath RL, Barton PA, Simpson RJ, Reid GE, Lim G, Anderson MA (1995) Characterization of the protease processing sites in a multidomain proteinase inhibitor precursor from Nicotiana alata. Eur J Biochem 230: 250–257 [DOI] [PubMed] [Google Scholar]
- Horn M, Baudys M, Voburka Z, Kluh I, Vondrasek J, Mares M (2002) Free-thiol Cys331 exposed during activation process is critical for native tetramer structure of cathepsin C (dipeptidyl peptidase I). Protein Sci 11: 933–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongsma MA, Bolter C (1997) The adaptation of insects to plant protease inhibitors. J Insect Physiol 43: 885–895 [DOI] [PubMed] [Google Scholar]
- Jorda L, Coego A, Conejero V, Vera P (1999) A genomic cluster containing four differentially regulated subtilisin-like processing protease genes is in tomato plants. J Biol Chem 274: 2360–2365 [DOI] [PubMed] [Google Scholar]
- Jorda L, Vera P (2000) Local and systemic induction of two defense-related subtilisin-like protease promoters in transgenic Arabidopsis plants. Luciferin induction of PR gene expression. Plant Physiol 124: 1049–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karban R, Baldwin IT (1997) Induced Responses to Herbivory. Chicago University Press, Chicago
- Kembhavi AA, Buttle DJ, Knight CG, Barrett AJ (1993) The two cysteine endopeptidases of legume seeds: purification and characterization by use of specific fluorometric assays. Arch Biochem Biophys 303: 208–213 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Yamada K, Hiraiwa N, Kondo M, Nishimura M, Hara-Nishimura I (1999) Vacuolar processing enzyme is up-regulated in the lytic vacuoles of vegetative tissues during senescence and under various stressed conditions. Plant J 19: 43–53 [DOI] [PubMed] [Google Scholar]
- Koiwa H, Bressan RA, Hasegawa PM (1997) Regulation of protease inhibitors and plant defense. Trends Plant Sci 2: 379–384 [Google Scholar]
- Korth KL, Dixon RA (1997) Evidence for chewing insect-specific molecular events distinct from a general wound response in leaves. Plant Physiol 115: 1299–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo ND, Downing DT (1997) Circular dichroism of model peptides emulating the amphipathic alpha-helical regions of intermediate filaments. Biochemistry 36: 2559–2565 [DOI] [PubMed] [Google Scholar]
- Lee MCS, Scanlon MJ, Craik DJ, Anderson MA (1999) A novel two-chain proteinase inhibitor generated by circularization of a multidomain precursor protein. Nat Struct Biol 6: 526–530 [DOI] [PubMed] [Google Scholar]
- Lerouge P, Cabanes-Macheteau M, Rayon C, Fischette-Laine AC, Gomord V, Faye L (1998) N-glycoprotein biosynthesis in plants: recent developments and future trends. Plant Mol Biol 38: 31–48 [PubMed] [Google Scholar]
- Miller EA, Lee MCS, Anderson MA (1999) Identification and characterization of a prevacuolar compartment in stigmas of Nicotiana alata. Plant Cell 11: 1499–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EA, Lee MCS, Atkinson AHO, Anderson MA (2000) Identification of a novel four-domain member of the proteinase inhibitor II family from the stigmas of Nicotiana alata. Plant Mol Biol 42: 329–333 [DOI] [PubMed] [Google Scholar]
- Moura DS, Ryan C (2001) Wound-inducible proteinase inhibitors in pepper. Differential regulation upon wounding, systemin, and methyl jasmonate. Plant Physiol 126: 289–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntz K, Shutov AD (2002) Legumains and their functions in plants. Trends Plant Sci 7: 340–344 [DOI] [PubMed] [Google Scholar]
- Nielsen KJ, Hill JM, Anderson MA, Craik DJ (1996) Synthesis and structure determination by NMR of a putative vacuolar targeting peptide and model of a proteinase inhibitor from Nicotiana alata. Biochemistry 35: 369–378 [DOI] [PubMed] [Google Scholar]
- O'Donnell PJ, Calvert C, Atzorn R, Wasternack C, Leyser HMO, Bowles DJ (1996) Ethylene as a signal mediating the wound response of tomato plants. Science 274: 1914–1917 [DOI] [PubMed] [Google Scholar]
- Outchkourov NS, Rogelj B, Strukelj B, Jongsma MA (2003) Expression of sea anemone equistatin in potato. Effects of plant proteases on heterologous protein production. Plant Physiol 133: 379–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Tolle DP, Barrett AJ (2004) Evolutionary families of peptidase inhibitors. Biochem J 378: 705–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roda A, Halitschke R, Steppuhn A, Baldwin IT (2004) Individual variability in herbivore-specific elicitors from the plant's perspective. Mol Ecol 13: 2421–2433 [DOI] [PubMed] [Google Scholar]
- Rotari VI, Dando PM, Barrett AJ (2001) Legumain forms from plants and animals differ in their specificity. Biol Chem 382: 953–959 [DOI] [PubMed] [Google Scholar]
- Ryan CA (1990) Protease inhibitors in plants: genes for improving defenses against insects and pathogens. Annu Rev Phytopathol 28: 425–449 [Google Scholar]
- Shimada T, Yamada K, Kataoka M, Nakaune S, Koumoto Y, Kuroyanagi M, Tabata S, Kato T, Shinozaki K, Seki M, et al (2003) Vacuolar processing enzymes are essential for proper processing of seed storage proteins in Arabidopsis thaliana. J Biol Chem 278: 32292–32299 [DOI] [PubMed] [Google Scholar]
- Simoes I, Faro C (2004) Structure and function of plant aspartic proteinases. Eur J Biochem 271: 2067–2075 [DOI] [PubMed] [Google Scholar]
- Sticher L, Hofsteenge J, Neuhaus JM, Boller T, Meins F Jr (1993) Posttranslational processing of a new class of hydroxyproline-containing proteins. Prolyl hydroxylation and C-terminal cleavage of tobacco (Nicotiana tabacum) vacuolar chitinase. Plant Physiol 101: 1239–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam NM, Horn M, Mares M, Baldwin IT (2001) Ontogeny constrains systemic protease inhibitor response in Nicotiana attenuata. J Chem Ecol 27: 547–568 [DOI] [PubMed] [Google Scholar]
- van der Hoorn RA, Jones JD (2004) The plant proteolytic machinery and its role in defence. Curr Opin Plant Biol 7: 400–407 [DOI] [PubMed] [Google Scholar]
- Winz RA, Baldwin IT (2001) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. IV. Insect-induced ethylene reduces jasmonate-induced nicotine accumulation by regulating putrescine N-methyltransferase transcripts. Plant Physiol 125: 2189–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Shimada T, Kondo M, Nishimura M, Hara-Nishimura I (1999) Multiple functional proteins are produced by cleaving Asn-Gln bonds of a single precursor by vacuolar processing enzyme. J Biol Chem 274: 2563–2570 [DOI] [PubMed] [Google Scholar]
- Zavala JA, Baldwin IT (2004) Fitness benefits of trypsin proteinase inhibitor expression in Nicotiana attenuata are greater than their costs when plants are attacked. BMC Ecol 10: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala JA, Patankar AG, Gase K, Hui D, Baldwin IT (2004. a) Manipulation of endogenous trypsin proteinase inhibitor production in Nicotiana attenuata demonstrates their function as antiherbivore defenses. Plant Physiol 134: 1181–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala JA, Patankar AG, Gase K, Baldwin IT (2004. b) Constitutive and inducible trypsin proteinase inhibitor production incurs large fitness costs in Nicotiana attenuata. Proc Natl Acad Sci USA 101: 1607–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]