Abstract
The copper chaperone for superoxide dismutase (CCS) has been identified as a key factor integrating copper into copper/zinc superoxide dismutase (CuZnSOD) in yeast (Saccharomyces cerevisiae) and mammals. In Arabidopsis (Arabidopsis thaliana), only one putative CCS gene (AtCCS, At1g12520) has been identified. The predicted AtCCS polypeptide contains three distinct domains: a central domain, flanked by an ATX1-like domain, and a C-terminal domain. The ATX1-like and C-terminal domains contain putative copper-binding motifs. We have investigated the function of this putative AtCCS gene and shown that a cDNA encoding the open reading frame predicted by The Arabidopsis Information Resource complemented only the cytosolic and peroxisomal CuZnSOD activities in the Atccs knockout mutant, which has lost all CuZnSOD activities. However, a longer AtCCS cDNA, as predicted by the Munich Information Centre for Protein Sequences and encoding an extra 66 amino acids at the N terminus, could restore all three, including the chloroplastic CuZnSOD activities in the Atccs mutant. The extra 66 amino acids were shown to direct the import of AtCCS into chloroplasts. Our results indicated that one AtCCS gene was responsible for the activation of all three types of CuZnSOD activity. In addition, a truncated AtCCS, containing only the central and C-terminal domains without the ATX1-like domain failed to restore any CuZnSOD activity in the Atccs mutant. This result indicates that the ATX1-like domain is essential for the copper chaperone function of AtCCS in planta.
Reactive oxygen species (ROS), including superoxide, hydrogen peroxide, and hydroxyl radicals, are generated as a by-product in cells from physiological reactions such as electron flow in chloroplasts and mitochondria and some redox reactions (Fridovich, 1978). Accumulation of ROS might cause damages to living organisms (Imlay and Linn, 1988; Bowler et al., 1992; Mehdy, 1994). Superoxide dismutase (SOD), a group of metalloenzymes, belongs to the enzymatic defense system against ROS and catalyzes the dismutation of superoxide radicals to molecular oxygen and hydrogen peroxide (Beyer et al., 1991). Copper/zinc SOD (CuZnSOD), manganese SOD (MnSOD), and iron SOD (FeSOD) are three types of SODs reported in plants.
MnSOD is localized in mitochondria, whereas FeSOD is localized in chloroplasts (Jackson et al., 1978). CuZnSOD is present in three isoforms, which are found in the cytosol, chloroplast, and peroxisome (Kanematsu and Asada, 1989; Bowler et al., 1992; Bueno et al., 1995). In Arabidopsis (Arabidopsis thaliana), three CuZnSOD genes, CSD1, CSD2, and CSD3, have been identified. The CSD1 and CSD2 activities are detected in roots, leaves, stems, and siliques, and their proteins are localized in the cytosol and chloroplast, respectively. The CSD3 is proposed to be a peroxisomal protein because its carboxyl terminus contains the Ala-Lys-Leu tripeptide, a typical peroxisomal targeting signal (Kliebenstein et al., 1998). CuZnSOD is a homodimeric copper- and zinc-containing enzyme (McCord and Fridovich, 1969). Zinc is required for the structural integrity of the protein and influences enzyme activity, whereas copper plays a catalytic role in the disproportion of superoxide (Forman and Fridovich, 1973; Beem et al., 1974). The acquirement of both metal ions was assumed to be by passive diffusion; however, Rae et al. (1999) found that the intracellular concentration of free copper is undetectable under normal physiological conditions. Therefore, cells require copper chaperones to facilitate copper transfer to specific partners (Valentine and Gralla, 1997).
Copper chaperones are a novel class of proteins involved in intracellular trafficking and delivery of copper to copper-containing proteins (Harrison et al., 1999). The proteins involved in copper trafficking in yeast (Saccharomyces cerevisiae) are Lys7 (Horecka et al., 1995; Culotta et al., 1997), ATX1 (Lin and Culotta, 1995; Lin et al., 1997), and COX17 (Glerum et al., 1996). Lys7 is the largest by molecular mass among the three; it inserts copper into CuZnSOD and was designated as copper chaperone for SOD (CCS; Culotta et al., 1997).
CCS possesses three functionally distinct protein domains. The N-terminal ATX1-like domain, bearing striking homology to ATX1 and containing the MXCXXC copper-binding site, is required for CCS function under strict copper limitation conditions in vivo (Schmidt et al., 1999). The central domain displays sequence homology to its target protein, CuZnSOD, and physically interacts with SOD1 in human (Homo sapiens) and yeast (Casareno et al., 1998; Lamb et al., 2000, 2001; Schmidt et al., 2000). The C-terminal domain carries a conserved Cys (CXC) motif and may play a crucial role in copper transfer to CuZnSOD (Schmidt et al., 1999). Besides, the interaction with SOD1 also requires the C-terminal domain (Schmidt et al., 2000). Although the C-terminal domain is relatively short (30–40 residues), it is unique to CCS proteins from diverse species among identified copper chaperone proteins (Schmidt et al., 1999). Genes that complement yeast lys7, atx1, and cox17 mutants have been identified in tomato (Lycopersicon esculentum) and Arabidopsis. Two of the cDNA clones, LeCCS (AF117707, tomato) and AtCCS (AF179371, Arabidopsis), are predicted to encode chloroplast-localized proteins but with an incomplete chloroplast-targeting transit peptide (Zhu et al., 2000). Wintz and Vulpe (2002) searched the Arabidopsis genome and concluded that AtCCS (AF179371.1, At1g12520) is the only yeast CCS homolog in Arabidopsis. The open reading frame of AtCCS contains two in-frame ATGs at the 5′-end of the predicted transcript. Because no other gene encodes for a potential cytosolic AtCCS-like chaperone in Arabidopsis, Wintz and Vulpe (2002) hypothesized that two transcription initiation sites may be involved in the production of different forms of AtCCS from the single AtCCS gene. However, The Arabidopsis Information Resource (TAIR) database predicts the initiation codon of AtCCS gene to be the second ATG, resulting in a polypeptide of 254 amino acids, while the Munich Information Centre for Protein Sequences (MIPS) predicts the initiation codon of AtCCS gene to be the first ATG, resulting in a polypeptide of 320 amino acids. Trindade et al. (2003) cloned a StCCS gene from potato (Solanum tuberosum) that also carried a plastidic transit peptide and was expressed only in stem-like tissues; unlike Arabidopsis, potato has another copy of the StCCS homolog that is expressed in leaves.
In yeast, a truncated yCCS without the ATX1-like domain complemented the lys7 null mutant weakly, compared with the intact yCCS. This result indicated that the ATX1-like domain is important for the yCCS activity (Schmidt et al., 1999). Furthermore, in vitro studies of LeCCS suggested that the ATX1-like domain is indispensable for copper and/or zinc ion binding during the metal delivery process (Zhu et al., 2000).
To further study the function of AtCCS, we have obtained an AtCCS knockout mutant. We show here that the mutant has lost all three forms of CuZnSOD activity. Complementation of the mutant with the cDNA encoding the 320-amino acid polypeptide, as annotated by MIPS, restored all three activities. The results suggest that one AtCCS gene encodes both the cytosolic and the chloroplastic forms of AtCCS and activates CuZnSOD activities at different subcellular locations. In addition, we show that the ATX1-like domain of AtCCS is essential for the copper chaperone function in planta.
RESULTS
AtCCS Gene in Arabidopsis
In Arabidopsis, the AtCCS gene (At1g12520) is composed of six exons and five introns. Analysis of the genomic sequence relevant to this gene revealed the existence of an additional ATG located upstream of the annotated initiation codon, and this additional ATG is in frame with the open reading frame annotated in TAIR database (Fig. 1A). This additional ATG is the start codon predicted in the MIPS Arabidopsis database (http://mips.gsf.de/proj/thal/db/index.html). In TAIR database, the AtCCS gene is predicted to encode a 254-amino acid polypeptide containing no signal peptide for organelle targeting (designated hereinafter as AtCCScytosolic [AtCCScyt]). Whereas, the AtCCS gene in the MIPS database is predicted to contain an additional 66 amino acids carrying a putative chloroplastic targeting signal (designated hereinafter as AtCCSchloroplastic [AtCCScp]). However, the upstream ATG was not found in four cDNA clones encoding AtCCS (AF179371, AF061517, AY050357, and GSLTSIL10ZC12) identified in the MIPS database. In our results, the upstream ATG is present in a 979-bp reverse transcription (RT)-PCR product amplified from rosette leaves and flowers using a specific primer set, ACCS-15 and ACCS-12 (Fig. 1, A and B). This confirmed the existence of the upstream ATG in the AtCCS transcript. Sequence comparison between the genomic PCR and RT-PCR products amplified by the same primer pair positioned the exons and introns as shown in Figure 1A. Thirteen cDNA clones obtained by 5′-RACE were randomly selected for sequencing. The results indicated that the first ATG existed in the 5′-end of all 13 AtCCS transcripts. Moreover, 10 clones were mapped to one nucleotide upstream, while the other clones were mapped to four, 14, and 26 nucleotides upstream of the first ATG, respectively (Fig. 1C, arrows).
Figure 1.
Sequence and expression of the AtCCS gene. A, The AtCCS gene structure. Black boxes represent the predicted exons and the gray box flanked by two ATGs represents part of the first exon not predicted in TAIR database. Lines between the boxes represent introns. ACCS-15 and ACCS-12 are primers designed for cloning the AtCCScp cDNA (accession no. DQ003058). ACCS-8 is the primer used for the 5′-RACE experiment. ACCS-1 is the primer used for RT-PCR analysis. B, Expression of the AtCCS gene in rosette leaves (RL) and flowers (F) of wild-type plants analyzed by RT-PCR. The same primers (ACCS-15 and ACCS-12) were also used for genomic PCR (G-PCR). Actin2 (ACT2), shown in the inset section, was used as an internal control. The size of DNA markers (M) in kilobases (kb) was shown at the left. C, The 5′-end sequence of AtCCS. The positions of the two in-frame ATGs are boxed and the sequences corresponding to the primer ACCS-8 are underlined. Arrows indicate the 5′-end of the different clones obtained by 5′-RACE and the accession numbers for the four clones are DQ003054, DQ003055, DQ003056, and DQ003057, respectively. D, The deduced amino acid sequence and domain structure of AtCCS gene. Amino acid numbers are indicated at the right. Residues conserved in all CCS homologs are shown in bold. The putative chloroplast-targeting peptide and peroxisomal-targeting signal are boxed and underlined, respectively. The two conserved copper-binding motifs are shown with gray boxes.
In Figure 1D, bioinformatic searches (on Web sites of http://www.cbs.dtu.dk/services/ChloroP and http://www.cbs.dtu.dk/services/TargetP) revealed that the additional 66 amino acids encoded by the sequence between the first and second ATG comprise a potential chloroplast-targeting transit peptide. The following 67th to 161st amino acids contain a highly conserved region, MXCXXC, which is a homologous domain of the ATX1 copper chaperone. The central domain (162nd to 290th amino acids) shares 19.1% sequence similarity to CSD1, and 22.7% similarity to both CSD2 and CSD3 of Arabidopsis. The last 30 residues of the C-terminal domain are unique to CCS and contain another highly conserved CXC motif.
CuZnSOD Activity and AtCCS Expression Pattern
The pattern for SOD activity in Arabidopsis is shown in Figure 2A. In addition to the two CuZnSOD activities as reported in Kliebenstein et al. (1998), an extra SOD activity migrating ahead the two CuZnSODs was also identified in reproductive tissues (CSD3, Fig. 2A). This activity band was characterized as a CuZnSOD due to the inhibition of the activity by KCN (data not shown). Based on the analysis of SOD activity in the csd1 knockout mutant (SALK_109389) and transgenic Arabidopsis plants containing an RNA interference CSD2 construct (C.-C. Chu, S.-M. Pan, and T.-L. Jinn, unpublished data), the three CuZnSODs activity bands were identified. The band with slowest mobility is CSD2, the second band is CSD1, and the fastest band is CSD3. CSD1 and CSD2 activities were detected in all tissues tested with the highest CSD1 activity detected in reproductive tissues. CSD3 activity was observed only in flowers and siliques, not in vegetative tissues (Fig. 2A).
Figure 2.
Expression patterns of the AtCCS gene and CuZnSOD activity in wild-type plants. A, SOD activity assay in different tissues and immunoblot detection of AtCCS. The positions of the MnSOD1 (MSD1), FeSOD (FSD), and CuZnSODs (CSD1, CSD2, and CSD3) are indicated at the left. In the right section (western) is an immunoblot of a similar nondenaturing gel probed with anti-AtCCS antibody to reveal the relative position of AtCCS to SODs. The arrowheads indicate the signals of the AtCCS protein and the asterisk indicates a nonspecific signal which was also detected in the Atccs knockout mutant (data not shown). B, AtCCS expression pattern in different tissues analyzed by northern blots. The labeled AtCCS and ACT2 cDNA probes were used for cohybridization. ACT2 was used as a loading control of RNA. C, AtCCS expression pattern in different tissues analyzed by RT-PCR. AtCCS was amplified with primers ACCS-1 and ACCS-8 (Fig. 1A) and ubiquitin 10 (UBQ-10) was amplified at the same time as a control. D, Immunoblot detection of AtCCS in different tissues on a nondenaturing gel. RL, rosette leaves; CL, cauline leaves, St, stem; F, flowers. E, Immunoblot detection of AtCCS in rosette leaves of the wild type (WT) and a transgenic line overexpressing AtCCScyt (AtCCScyt/WT).
We then analyzed the expression of the AtCCS gene. In northern-blot analyses, the AtCCS cDNA probe detected only one hybridization signal in flowers but not in rosette leaves (Fig. 2B). However, low levels of expression could be detected in rosette and cauline leaves by RT-PCR (Fig. 2C). In immunoblot analyses for protein expression, for some unknown reasons, our anti-AtCCS serum could detect AtCCS only when used on nondenaturing, but not on denaturing, PAGE (data not shown). As shown in Figure 2D, the anti-AtCCS antiserum detected several signals in stems and flowers when used in nondenaturing PAGE. Very weak signals were sometimes also observed in leaves (Fig. 2, D and E). The AtCCS signals did not result from antiserum cross-hybridization with SOD since they migrated at different positions in the native gels (Fig. 2A, western). Furthermore, overexpression of a cDNA encoding AtCCScyt (AtCCScyt/wild type) increased the AtCCS signals detected (Fig. 2E), supporting that the bands detected on the native gels were indeed AtCCS.
Characterization of the Atccs Knockout Mutant
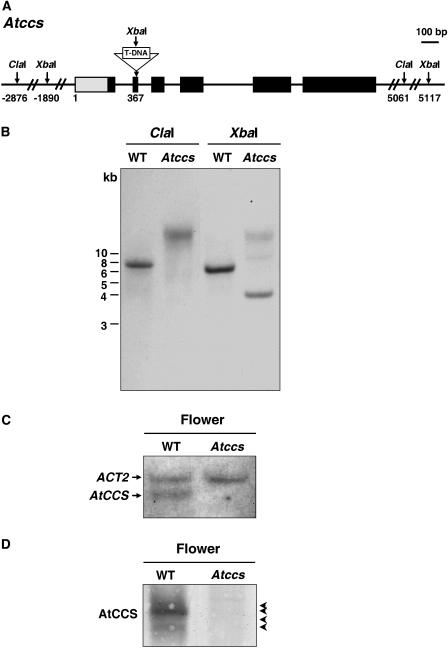
An Atccs mutant (SALK_025986), caused by a T-DNA insertion in the second exon (Fig. 3A), was obtained. Lines homozygous for the T-DNA insertion were identified by PCR and confirmed by Southern-blot analyses (Fig. 3B). Incomplete digestion might be the cause of higher Mr bands seen in the Atccs sample digested with XbaI. In this Atccs mutant, neither the AtCCS transcript nor the AtCCS protein was detected (Fig. 3, C and D).
Figure 3.
The Atccs knockout mutant. A, Schematic representation of the positions of T-DNA insertion and related restriction enzyme sites in the AtCCS gene. B, Southern-blot analyses of Atccs homozygous T-DNA-insertion mutant. The genomic DNA of the wild type (WT) and Atccs mutant was digested with ClaI and XbaI and the AtCCS genomic clone was used as the probe. Size markers of DNA in kilobases (kb) are shown at the left. C, Northern-blot analyses of AtCCS gene expression in flowers from the wild type (WT) and Atccs mutant. The labeled AtCCS and ACT2 cDNA probes were used for cohybridization. ACT2 was used as a loading control of RNA. D, Immunoblot analyses of AtCCS in flowers of the wild type (WT) and Atccs mutant by nondenaturing PAGE. Arrowheads indicate the AtCCS signals.
Almost no CuZnSODs activity was detected in the Atccs mutant (Fig. 4A) even in the presence of CuZnSOD transcripts (Fig. 4B) and proteins (Fig. 4C), which indicated that AtCCS was necessary for the activation of all types of CuZnSOD activity. The protein levels of CSD1 and CSD2 in the Atccs mutant were decreased in all tissues tested compared to the levels in the wild type (Fig. 4C). However, the steady-state level of CSD1 and CSD2 transcripts were not reduced in the Atccs mutant (Fig. 4B).
Figure 4.
CuZnSOD RNA, protein, and enzyme activity in the Atccs mutant. A, CuZnSOD activity analyses in the Atccs mutant and wild type (WT). The positions of the CSD1, CSD2, and CSD3 activities are indicated at the left. B, Northern-blot analyses of CSD1 and CSD2 expression in flowers with CSD1 and CSD2 cDNA as probes. ACT2 and ethidium bromide-stained rRNAs were used as loading controls. C, Immunoblot analyses of CSD1 and CSD2 by SDS-PAGE with antibodies against CSD1 and CSD2.
Different Recovery of CuZnSOD Activities in the Atccs Mutant Complemented with AtCCScyt, AtCCScp, and AtCCScp-mutant
Because CSD1, CSD2, and CSD3 activities could be simultaneously detected in flowers, in subsequent studies only results from flower tissue were presented. The Atccs mutant was transformed with AtCCScyt, the TAIR-predicted 254-amino acid open reading frame of the AtCCS gene (the transformants were designated as AtCCScyt/Atccs), or with a 1.64-kb genomic fragment (Fig. 1, A and B) that was sufficient to encode the MIPS-predicted 320-amino acid open reading frame (designated as AtCCScp/Atccs), to test for the recovery of the CuZnSOD activities. Both constructs were driven by the cauliflower mosaic virus 35S promoter. Immunoblot analyses revealed that AtCCS proteins were indeed expressed in AtCCScyt/Atccs and AtCCScp/Atcc transgenic plants (Fig. 5B). The CuZnSOD activity profile in different AtCCScyt/Atccs transgenic lines was the same as that of the wild type except that the chloroplastic CSD2 activity was never recovered (Fig. 5A, AtCCScyt/Atccs 1 to 3). However, T1 transgenic individuals transformed with AtCCScp were able to recover all three types of CuZnSOD activity (Fig. 5A, AtCCScp/Atccs 1 to 3). Recovery of all three types of CuZnSOD activity was also observed when the Atccs mutant was transformed with the AtCCScp cDNA driven by the 35S promoter (data not shown). These results indicated that AtCCScp, but not AtCCScyt, was sufficient to recover all three CuZnSOD activities in the Atccs mutant. To further test the importance of the 66 amino acids at the N terminus of AtCCScp, a mutant construct (AtCCScp-mut, Fig. 6A) was generated by introducing a guanine nucleotide before the second ATG through site-directed mutagenesis. CuZnSOD activity profile in the AtCCScp-mut/Atccs T1 transgenic plants was again missing the CDS2 band (Fig. 6B). This result suggested that the additional 66 amino acids in AtCCScp comprised a plastidic transit peptide necessary to restore the chloroplastic CSD2 activity.
Figure 5.
CuZnSOD activity patterns and the AtCCS protein profile in the AtCCScyt/Atccs and AtCCScp/Atccs transgenic plants. A and B, CuZnSOD activity (A) and immunoblot detection of AtCCS proteins (B) in flowers from wild type (WT), different T1 lines (1 to 3) of AtCCScyt/Atccs and AtCCScp/Atccs transgenic plants, and the Atccs mutant. Positions of CSD1, CSD2, and CSD3 activities are indicated at the left. The arrowheads indicate AtCCS signals.
Figure 6.
The CuZnSOD activity pattern in AtCCScp-mut/Atccs transgenic plants. A, The partial 5′-end sequence of the AtCCScp-mut is presented. The two ATGs were boxed. The extra nucleotide, G, inserted is indicated by the arrow, and the stop codon created before the second ATG by the insertion was underlined. B, CuZnSOD activity in flowers from the wild type (WT), different T1 lines (1 to 6) of AtCCScp-mut/Atccs transgenic plants and Atccs mutant. Positions of CSD1, CSD2, and CSD3 activities are indicated at the left.
Chloroplastic Localization of AtCCScp
To investigate if the first 66 amino acids of AtCCScp function as a chloroplast-targeting transit peptide, 35S-labeled AtCCScyt and AtCCScp were synthesized by in vitro transcription/translation and incubated with isolated pea (Pisum sativum) chloroplasts under import conditions. The AtCCScp cDNA directed the synthesis of two proteins, one 34 kD and one 29 kD (Fig. 7, lane 5). The AtCCScyt cDNA directed the synthesis of only the 29-kD protein (Fig. 7, lane 1), suggesting that the 29-kD product from AtCCScp was a result of internal initiation of translation from the second ATG. The 29-kD protein synthesized from AtCCScyt could not be imported into chloroplasts (Fig. 7, lane 3). In contrast, when the protein products from AtCCScp were incubated with chloroplasts, a 29-kD protein was produced (Fig. 7, lane 7), and this 29-kD protein was fully resistant to thermolysin (Fig. 7, lane 8), indicating its localization within the chloroplasts. Since the 29-kD protein synthesized from AtCCScyt could not be imported into chloroplasts (Fig. 7, lane 3), the 29-kD chloroplast-localized protein produced after import of AtCCScp (Fig. 7, lane 7) most likely resulted from import of the 34-kD protein directed by AtCCScp. These results indicated that the first 66 amino acids of AtCCScp were necessary for the chloroplast import of AtCCScp.
Figure 7.
Import of AtCCS into isolated chloroplasts. In vitro-translated and [35S]Met-labeled AtCCScyt (lane 1) and AtCCScp (lane 5) proteins were treated with thermolysin directly (lanes 2 and 6) or incubated with isolated pea chloroplasts under import conditions. The reisolated chloroplasts were further incubated in import buffer (lanes 3 and 7) or in import buffer containing thermolysin (lanes 4 and 8). Chloroplasts were reisolated, solubilized in sample buffer, and analyzed by SDS-PAGE and fluorography. Arrow indicates the precursor form of AtCCScp and arrowhead indicates the imported mature form of AtCCScp, which is the same size as AtCCScyt. Asterisk indicates the thermolysin-resistant fragment from both AtCCScp and AtCCScyt. The protein molecular mass markers in kD are shown at the left.
Interaction between AtCCScyt and CSD1
We further investigated the possibility of direct interaction between AtCCScyt and CSD1 using the yeast two-hybrid assay. The AtCCScyt cDNA and CSD1 cDNA were fused with GAL4 DNA binding domain (BD-AtCCScyt) and activation domain (AD-CSD1), respectively. Cells containing both the BD and AD constructs were produced by mating of haploid cells containing individual plasmids. Diploid cells were then streaked on selection media to test for protein interaction. AD-CSD1 and BD-AtCCScyt together resulted in growth of the yeast transformants while cells transformed with individual binding or activation construct or with control plasmids did not grow (Fig. 8). This result suggested that AtCCScyt physically interacted with CSD1.
Figure 8.
Analyses of AtCCScyt and CSD1 interaction by yeast two-hybrid assays. Chimeric proteins were constructed in which the open reading frame of AtCCScyt was fused to the GAL4 DNA-binding domain (BD-AtCCScyt) and that of CSD1 fused to the GAL4 activation domain (AD-CSD1). Haploid strains transformed with individual plasmids were mated and selected on the medium without Leu, Trp, His, and Ade. + for positive control and − for negative control as described in “Materials and Methods.”
Importance of the ATX1-Like Domain in Conferring CuZnSOD Activity
A truncated AtCCS gene containing only the central and C-terminal domains (hereinafter referred as AtCCSD2D3, encoding the 162th–320th amino acids, Fig. 1D) was transformed into the Atccs mutant (designated as AtCCSD2D3/Atccs lines) to test for the importance of the ATX1-like region. The expression of the AtCCSD2D3 transcript in AtCCSD2D3/Atccs T1 transgenic lines was confirmed by northern-blot analyses (data not shown). Among 70 AtCCSD2D3/Atccs T1 transgenic lines, no CuZnSOD activity was observed in any of the plants even when protein extracts were prepared from detached leaf stalks that had been first incubated with a 1 mm CuSO4 solution at 25°C for 4 h (Fig. 9A). However, a weak CuZnSOD activity did appear if the leaf protein extracts were prepared first and the extracts were incubated directly in a 1 mm CuSO4 solution at 25°C for 4 h (Fig. 9B). This partial restoration of CuZnSOD activity was due to AtCCSD2D3 since no activity was observed in the Atccs mutant without the transgene (Fig. 9B, Atccs). Furthermore, the CSD3 activity, never observed in vegetative tissues, was detected in the leaf extracts of wild type plants under such copper-supplemented conditions (Fig. 9, A and B, wild type, compare control and +CuSO4).
Figure 9.
The importance of the AtCCS ATX1-like domain for CuZnSOD activation. The activity of CuZnSODs was analyzed in the wild type (WT), different T1 transgenic lines of AtCCSD2D3/Atccs (1 to 3) and Atccs mutant. A and B, Detached rosette leaves (A) and leaf protein extracts (B) were treated with a 1 mm CuSO4 solution at 25°C for 4 h (water treatment for controls). All samples were then analyzed for CuZnSOD activity. Positions of the CSD1, CSD2, and CSD3 activities are indicated at the left.
DISCUSSION
One AtCCS gene was demonstrated to activate three forms of CuZnSODs localized in different subcellular locations in this study. The results obtained from the AtCCScyt/Atccs, AtCCScp/Atccs, and AtCCScp-mut/Atccs transgenic plants and the in vitro protein import analysis indicated that the open reading frame from the first ATG encoded a plastidic AtCCS containing a chloroplastic transit peptide and was responsible for CSD2 activation (Figs. 5–7). Import of cytosolically synthesized precursor proteins into chloroplasts is usually very efficient and no precursor proteins are normally detected in the cytosol (Sun et al., 2001). Furthermore, the AtCCS precursor with the chloroplast-targeting transit peptide may not possess the chaperone function before the removal of the transit peptide. Therefore, a cytosolic form of AtCCS without the transit peptide is most likely also produced from AtCCScp from the second ATG for the activation of CSD1 and CSD3. Results from AtCCScyt/Atccs and AtCCScp-mut/Atccs transgenic plants also support that the second ATG has the potential to be used as a translation initiation site. However, detection of the two proteins synthesized from AtCCScp will be difficult since the cytosolic form and the processed mature form in plastids would be almost the same size (Fig. 7).
Several mechanisms have been reported for translation of two proteins from a single gene (Danpure, 1995; Small et al., 1998; Silva-Filho, 2003). With respect to AtCCS, it is not clear whether the two forms of AtCCS proteins resulted from two different transcripts or from alternative translation initiation from the same transcript. Sequences obtained from the 5′-RACE suggested that there might be at least two different types of AtCCS transcripts (Fig. 1C). Due to the possibility of mRNA degradation and abortion of the reverse transcriptase before reaching the 5′-end of mRNA, one would expect that the clones obtained from 5′-RACE should contain different lengths of 5′-untranslated region. However, 10 out of 13 of the clones from 5′-RACE extended only one nucleotide beyond the first ATG, and this high ratio suggested that this type of transcript existed in vivo. The size of the 5′-untranslated region in this transcript is too short, and thus the 40S subunit of ribosome may escape the first ATG (Kozak, 1991) and use the downstream ATG as a translation initiation site, resulting in the production of the cytosolic AtCCS. Meanwhile, the three clones with four, 14, and 26 nucleotides upstream of the first ATG suggested that there should be a longer AtCCS transcript that would encode a chloroplastic form of AtCCS. Our proposal is consistent with the hypothesis of Wintz and Vulpe (2002). The authors suggested that the expression of the cytosolic and plastidic forms of AtCCS involves two transcription initiation sites in the AtCCS gene.
Rizhsky et al. (2003) reported that CSD2 knockdown plants had reduced CSD2 expression at the RNA and activity levels. Growth of the CSD2 knockdown plants was suppressed and the flowering time was delayed under a constant light of 55 to 100 μmol m−2 s−1. However, under a 16-h light/8-h dark photoperiod cycle at 100 μmol m−2 s−1, we did not observe any obvious difference in growth among the wild type, the Atccs mutant, the AtCCScyt/Atccs, and the AtCCScp-mut/Atccs transgenic plants. In the Atccs mutant, approximately 6% of CuZnSOD activity compared with that of the wild type was observed in flowers when 30 μg of proteins were applied to the SOD activity assay (Fig. 4A). A very small amount of CuZnSOD activity was also observed in rosette leaves, cauline leaves, and stems when the protein loading was increased to 120 μg (data not shown). These results revealed the existence of a CCS-independent pathway in addition to the CCS-dependent pathway for CuZnSOD activity. The CCS-independent pathway might activate a small amount of CSD2 that is sufficient to overcome the oxidative stress under normal growth condition. In yeast, ySOD1 activation is completely dependent on CCS (Culotta et al., 1997) due to the presence of the specific dual Pros at the 142nd and 144th amino acid in the ySOD1 protein sequence (Carroll et al., 2004). Whereas, the mammalian SOD1 can obtain some copper independent of CCS (Wong et al., 2000) and this CCS-independent pathway involves the reduced form of glutathione (Carroll et al., 2004). The specific dual Pros are largely restricted to Ascomycota fungi, (Carroll et al., 2004) and Arabidopsis CuZnSOD sequences do not contain these two specific Pro residues.
In the copper-deficient mice and the CCS knockout mice, the level of SOD1 protein was significantly reduced to different levels in different tissues, even though the level of SOD1 transcript was not altered (Prohaska et al., 2003). The authors suggested a posttranscriptional mechanism for the reduction of SOD1 protein when copper is limited. In Arabidopsis, the Atccs mutant showed a reduced protein level of the two forms of CuZnSOD, but the amount of CuZnSODs transcripts was not reduced (Fig. 4, B and C). This result is consistent with that from mice. However, a transcriptional regulation of CuZnSODs expression cannot be ruled out in Arabidopsis.
Protein translocation into chloroplasts and peroxisomes occur posttranslationally (Schnell and Hebert, 2003; Soll and Schleiff, 2004). Therefore, there is the possibility that activation of different CuZnSODs could take place in the cytosol before CuZnSOD proteins are delivered to different subcellular locations. However, the CSD2 activity was detected only in AtCCScp/Atccs but not in AtCCScyt/Atccs and AtCCScp-mut/Atccs transgenic plants (Figs. 5 and 6). These results indicated that CSD2 was activated in chloroplasts by chloroplast-localized AtCCS. On the other hand, although AtCCS contains a Ser-Lys-Val tripeptide at its C terminus, this tripeptide has been reported to function as a peroxisomal-targeting signal only in yeast, but not yet in higher plants (Elgersma et al., 1996; Hayashi et al., 1996; Mullen et al., 1997a, 1997b; Reumann, 2004). In addition, proteins can be fully folded in the cytosol before they are transported into the peroxisome (Walton et al., 1995), and therefore CSD3 could be fully assembled with copper and zinc before it is transported into peroxisomes. Therefore, whether the activation of CSD3 occurred in the cytosol or peroxisomes still remains to be demonstrated.
The importance of the ATX1-like domain in copper binding has been studied in yeast (Schmidt et al., 1999). In yeast, the free copper-ion concentration in an unstressed yeast cell is less than 10−18 m (Rae et al., 1999) and the ATX1-like domain is necessary only under such strict copper-limited conditions (Schmidt et al., 1999). If the growth media is supplemented with CuSO4, yCCS without the ATX1-like domain could fully restore SOD1 activity (Schmidt et al., 1999). The authors have proposed that a copper-binding CXC motif in the C-terminal domain is sufficient for copper binding. However, cooperation with the ATX1-like domain results in maximal yCCS activity. In this study, the CuZnSOD activity was not restored when the rosette leaves from AtCCSD2D3/Atccs transgenic plants were immersed in 1 mm CuSO4 solution (Fig. 9A). However, when protein extracts from the AtCCSD2D3/Atccs transgenic plants was directly mixed with 1 mm CuSO4, a weak CuZnSOD activity was restored (Fig. 9B). Because direct incubation of the extracts with CuSO4 would provide a higher concentration of copper to the AtCCSD2D3 mutant, our results agree with the results from yeast that AtCCSD2D3 can still function if copper is readily available and the ATX1-like domain is required when the availability of copper is limited. Copper content in Arabidopsis leaves has been reported to be 15.7 mg/kg dry weight (Abdel-Ghany et al., 2005), but the free copper-ion concentration is still unclear. The result that the AtCCSD2D3 mutant failed to restore SOD activities suggests that the concentration of available copper is very low under normal growth conditions.
MATERIALS AND METHODS
Plants, Growth Condition, and CuSO4 Treatment
The Atccs mutant (SALK_025986) of Arabidopsis (Arabidopsis thaliana) Columbia ecotype was obtained from the Arabidopsis Biological Resource Center (The Ohio State University). Seeds were sown in BIO-MIXTTING SUBSTRATUM (Agricultural Materials Company), incubated in the dark for 2 to 4 d at 4°C, and transferred to a growth chamber for germination. The seedlings were grown under 16-h light/8-h dark at 23°C/21°C at a light intensity of 60 to 100 μmol m−2 s−1.
For experiments shown in Figure 9, the detached rosette leaf stalks were immersed in a 1 mm CuSO4 solution, or crude protein extracts prepared from rosette leaves were incubated with 1 mm CuSO4. Both treatments were performed at 25°C for 4 h. Protein extracts were then prepared from the detached leaves and samples from both treatments were assayed for CuZnSOD activity as described by Pan et al. (2001).
Analysis of the CCS Gene Sequence
Sequence information of genes, proteins, and cDNAs were retrieved by searching public databases with the BLAST algorithm (Altschul et al., 1997) at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), TAIR (http://www.arabidopsis.org), and MIPS Arabidopsis database (http://mips.gsf.de/proj/thal/db/index.html). The chloroplast-targeting signal was predicted with CHLOROP version 1.1 (Emanuelsson et al., 1999; http://www.cbs.dtu.dk/services/ChloroP) and TargetP version 1.01 (Emanuelsson et al., 2000; http://www.cbs.dtu.dk/services/TargetP).
Southern-Blot Analysis
Genomic DNA was isolated from rosette leaves according to Dellaporta (1993). DNA was digested with restriction enzymes, separated on a 1% (w/v) agarose gel, transferred to a nylon membrane (Roche), and hybridized with the AtCCS-genomic fragment labeled with Digoxigenin-11-dUTP by PCR with the primers ACCS-15, 5′-GCTCTAGACTGAAGAAGCAAGATAATG-3′ (the XbaI site is underlined and the first ATG was boxed) and ACCS-12, 5′-GCGAGCTCTTAAACCTTACTGGCCACG-3′ (the SacI site is underlined). The membrane was washed twice at 65°C for 10 min in 2× sodium chloride/sodium phosphate/EDTA buffer supplemented with 0.1% (w/v) SDS. Hybridization signals were detected by the chemiluminescent reaction with disodium 3-(4-methoxyspiro{1,2,-dioxetane-3,2′-(5′-chloro) tricyclo[3.3.1.1]decan}-4-yl)phenyl phosphate; Roche) according to the manufacturer's instructions.
Northern-Blot Analysis
Total RNA was isolated with TRIZOL reagent (Invitrogen) and 10 μg total RNA was separated on a 1.2%-formaldehyde agarose gel and hybridized with AtCCS, CSD1, CSD2, and ACT2 cDNA fragments labeled with Digoxigenin-11-dUTP by PCR. Hybridization signals were detected as described for Southern-blot analyses. The cDNA probes were amplified by PCR with the following gene-specific primers: AtCCS, ACCS-15 and ACCS-12 (as described in Southern-blot analyses); CSD1, 5′-CGCCATGGCGAAAGGAGTTGCAGTTT-3′ (the NcoI site is underlined) and 5′-ATCCCGGGGCCCTGGAGACCAATGAT-3′ (the SmaI site is underlined); CSD2, 5′-ACGGATCCATCCTCGCATTCTCATCTCCTT-3′ (the BamHI site is underlined) and 5′-ACTCTAGAGACGGCACTCATCTTCTGG-3′ (the XbaI site is underlined); ACT2, 5′-AGCTCCCGGGCTAAGCTCTCAAGATCAAAGGCTTA-3′ (ACT2-376s) and 5′-AGCTCCCGGGTTAACATTGCAAAGAGTTTCAAGGT-3′ (ACT2-3′N3).
The ACT2 and AtCCS probes detected only one signal each on northern-blot analyses with sizes of approximately 1.7 and 1.1 kb, respectively. Thus, the two probes were mixed together to hybridize with the same membrane.
RT-PCR and 5′-RACE of the AtCCS Gene
For RT-PCR, Superscript II (Invitrogen) kit was used according to the manufacturer's instructions. Total RNA (1 μg) was used and primed with the oligo (dT) primer for cDNA synthesis. The RT reaction mixture (1 μL) was used for subsequent PCR with the following gene-specific primers: AtCCS, ACCS-15 and ACCS-12, or ACCS-1, 5′-ATTCTAGAGCCTCTGCGATTCCCATC-3′ (the XbaI site is underlined) and ACCS-8, 5′-CTGTGCTGGCTGCTCCGTTTGT-3′ (Fig. 1A), and internal control ACT2, ACT2-376 s and ACT2-3′N3, or UBQ10, 5′-GATCTTTGCCGGAAAACAATTGGAGGATGGT-3′ (UBQ1) and 5′-CGACTTGTCATTAGAAAGAAAGAGATAACAGG-3′ (UBQ2).
The 5′-terminal sequence of the AtCCS transcript was obtained using the BD SMART RACE cDNA Amplification kit (CLONTECH) according to the manufacturer's instructions (5′-RACE). Total RNA (1 μg) from the flowers in the wild type was used for cDNA synthesis. The subsequent PCR was amplified with the universal primers mix provided in the kit and the gene-specific reverse primer, ACCS-8. The PCR products of the 5′-RACE were cloned into the yT&A vector (Yeastern Biotech) for DNA sequencing.
Protein Extraction and Quantification
Tissues were ground (tissue:medium ratio 1:3, w/v) with 150 mm Tris-HCl (pH 7.2), and the homogenate was centrifuged at 13,000g at 4°C for 10 min. The protein concentration was determined by the method of Bradford (1976) with the Bio-Rad protein assay reagent (Bio-Rad) and bovine serum albumin was used as the protein standard.
Electrophoresis and SOD Activity Analysis
Total protein (30 μg) was separated on a 10% nondenaturing polyacrylamide gel in Tris-Gly buffer (pH 8.3). A photochemical method modified from Beauchamp and Fridovich (1971) was used to visualize SOD activity. The gel was soaked in 0.1% (w/v) nitroblue tetrazolium solution for 15 min, rinsed with distilled water, and transferred to 100 mm potassium phosphate buffer (pH 7.0) containing 0.028 mm riboflavin and 28 mm TEMED (N,N,N′,N′-tetramethyl-ethylenediamine) for another 15 min. After being washed with distilled water, gel was illuminated on a light box, with a light intensity of 30 μmol m−2 s−1 for 15 min to initiate the photochemical reaction. The SOD activity was verified by KCN and H2O2 as described by Pan et al. (1999). KCN is an inhibitor of CuZnSOD, whereas H2O2 inhibits both CuZnSOD and FeSOD. The MnSOD activity is not inhibited by either chemical.
Anti-AtCCS Serum Preparation
The coding region of AtCCS gene predicted in TAIR database was amplified by PCR with the gene-specific primers 5′-TCTGGATCCATGGCGACTGCTCTCACT-3′ (the BamHI site is underlined) and 5′-TCTCTCGAGGTTATTAAACCTTACTGG-3′ (the XhoI site is underlined) that introduced the two restriction sites on the two ends. The PCR fragment was cloned into the pGEX6P-1 (Amersham Biosciences) for recombinant protein expression. The induction and purification of the fusion protein were performed as described by Pan et al. (1999). The purified glutathione S-transferase-AtCCS fusion protein was used for induction of the AtCCS antiserum in rabbits.
Immunoblot Analyses
For the detection of AtCCS, 60 μg of total protein was separated on a 10% nondenaturing polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Amersham Biosciences), blocked with 5% nonfat-milk in 1× PBST (phosphate-buffered saline containing 0.05% Tween 20) for 1 h, and then incubated with anti-AtCCS antiserum (1:1,000 dilution) for another hour. The washed membrane was incubated with 1:3,000 diluted goat anti-rabbit IgG conjugated with alkaline phosphatase (Perkin-Elmer); the signals in the membrane were detected by colorimetric reaction with 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium (PerkinElmer). For CuZnSOD immunoblot assay, 30 μg of total protein was separated on a 10% SDS-PAGE and transferred to a polyvinylidene difluoride membrane with the anti-CSD1 (1:1,000 dilution) and -CSD2 (1:2,000 dilution) anti-sera as described in Kliebenstein et al. (1998).
In Vitro Transcription/Translation of AtCCScyt and AtCCScp Proteins, and Protein Import into Isolated Chloroplasts
The AtCCScyt and AtCCScp cDNA fragments were amplified by RT-PCR with the following specific primers: AtCCScyt, ACCS-1 and ACCS-12; and AtCCScp, ACCS-15 and ACCS-12 (Fig. 1A). The RT-PCR products were cloned into the yT&A vector for DNA sequencing. Then the AtCCScyt and AtCCScp cDNA fragments were subcloned into the KpnI and BamHI sites of pBluescript SK+ (Stratagene). These constructs were used as templates for in vitro transcription/translation using the TNT T7 coupled wheat germ extract system (Promega) in the presence of [35S]Met according to the manufacturer's instructions. Isolation of chloroplasts from pea seedlings (Pisum sativum cv Little Marvel) and import of the labeled proteins into chloroplasts were performed according to Perry et al. (1991). Reisolated chloroplasts were treated with thermolysin as described (Smeekens et al., 1986) and analyzed by SDS-PAGE as described (Tu and Li, 2000).
Yeast Two-Hybrid Analysis
The AtCCScyt cDNA and the Arabidopsis CSD1 cDNA were amplified by PCR and subcloned into the pGBKT7 and pACT2 yeast (Saccharomyces cerevisiae) expression vectors (Matchmaker 3, CLONTECH), in-framed with the GAL4 DNA BD and the GAL4 AD, respectively. The resulting BD-AtCCScyt and AD-CSD1 vectors were then transformed into the yeast strain AH109 (MAT a) and Y187 (MAT α) using a modified lithium acetate method. For positive and negative controls, yeast cells were transformed with the plasmids pACT2-T antigen (AD-T antigen) and pGBKT7-53 (BD-p53), or with the plasmids pACT2-T antigen (AD-T antigen) and pGBKT7-Lam (DB-Lamin C), respectively. Yeast cells were inoculated into yeast peptone dextrose medium and cultured overnight. Cells were than streaked on synthetic dextrose medium lacking Leu, Trp, His, and Ade.
Gene Construction and Plant Transformation
The pPZP200GB with β-glucuronidase and BAR (BASTA resistance gene) cassettes was derived from pBI221 (CLONTECH) and pSK-35S-BAR (John Walker, University of Missouri Columbia). This binary vector was used to clone AtCCScyt and AtCCSD2D3. The cDNA fragment corresponding to AtCCScyt and AtCCSD2D3 was amplified by PCR with the following gene-specific primers: AtCCScyt, 5′-ATTCTAGAATGGCGACTGCTCTCACT-3′ (the XbaI site is underlined) and 5′-GCGAGCTCTTAAACCTTACTGGCCACG-3′ (the SacI site is underlined); and AtCCSD2D3, 5′-TCTGGATCCATGTCAGCTGCAGTAGCAGAATTC-3′ (the BamHI site is underlined) and 5′-GCGAGCTCTTAAACCTTACTGGCCACG-3′ (the SacI site is underlined). The PCR products were cloned into the XbaI/SacI sites of pPZP200GB by replacing the β-glucuronidase fragment. The obtained plasmids were named pPZP200GB-AtCCScyt and pPZP200GB-AtCCSD2D3, respectively.
The AtCCScp and AtCCScp-mut genomic fragments were amplified by PCR with the following specific primers: AtCCScp, ACCS-15 and ACCS-12; AtCCScp-mut, ACCS-17, 5′-ATAATGGCATCAAGTTCTCAGGTCAGT-3′ (the first ATG is boxed and the introduced extra G nucleotide is underlined), and ACCS-12. The PCR products were cloned into the yT&A vector for DNA sequencing. Then the KpnI/SacI fragments corresponding to AtCCScp and AtCCScp-mut were cloned into pBIB-HYG (Becker et al., 1992) for plant transformation. The obtained plasmids were named pBIB-HYG-AtCCScp and pBIB-HYG-AtCCScp-mut, respectively.
Plasmids for plant transformation were transformed into Agrobacterium tumefaciens C58 by electroporation. Agrobacterium cells containing each plasmid were transformed into the Atccs mutant by the floral-dipping method (Clough and Bent, 1998). Transgenic plants of AtCCScyt/Atccs and AtCCSD2D3/Atccs were selected by spraying seedlings at 7, 9, and 11 d after germination with a solution of 0.4% BASTA herbicide (McDowell et al., 1998). The AtCCScp/Atccs and AtCCScp-mut/Atccs transgenic plants were selected by half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) containing 25 μg mL−1 hygromycin.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ003054 to DQ003058.
Acknowledgments
We thank Dr. Hans Bohnert (University of Illinois, Urbana-Champaign) and Dr. John Walker (University of Missouri, Columbia) for providing the AtCCS cDNA clone (BE038022) and the pSK-35S-BAR plasmid, respectively. We also thank Dr. Robert Last (Cornell University, Ithaca, NY) for kindly providing the Arabidopsis SOD anti-sera, and the Arabidopsis Biological Resource Center for the Atccs mutant (SALK_025986). We are grateful to Dr. Chu-Yung Lin and Dr. Wen-Ju Yang (National Taiwan University, Taipei) and Dr. John Walker and Dr. Kevin Lease (University of Missouri, Columbia) for critically reading and for revising the manuscript.
This paper is dedicated to memory of the late Dr. Shu-Mei Pan.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.065284.
References
- Abdel-Ghany SE, Müller-Moulé P, Niyogi KK, Pilon M, Shikanai T (2005) Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 7: 1233–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp C, Fridovich I (1971) Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem 44: 276–287 [DOI] [PubMed] [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20: 1195–1197 [DOI] [PubMed] [Google Scholar]
- Beem KM, Rich WE, Rajagopalan KV (1974) Total reconstitution of copper-zinc superoxide dismutase. J Biol Chem 249: 7298–7305 [PubMed] [Google Scholar]
- Beyer W, Imlay J, Fridovich I (1991) Superoxide dismutases. Prog Nucleic Acid Res Mol Biol 40: 221–253 [DOI] [PubMed] [Google Scholar]
- Bowler C, Van Montagu M, Inzé D (1992) Superoxide dismutase and stress tolerance. Annu Rev Plant Physiol Plant Mol Biol 43: 83–116 [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bueno P, Varela J, Gimeenez-Gallego G, del Rio LA (1995) Peroxisomal copper, zinc superoxide dismutase: characterization of the isoenzyme from watermelon cotyledons. Plant Physiol 108: 1151–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll MC, Girouard JB, Ulloa JL, Subramaniam JR, Wong PC, Valentine JS, Culotta VC (2004) Mechanisms for activating Cu- and Zn-containing superoxide dismutase in the absence of the CCS Cu chaperone. Proc Natl Acad Sci USA 101: 5964–5969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casareno RL, Waggoner D, Gitlin JD (1998) The copper chaperone CCS directly interacts with copper/zinc superoxide dismutase. J Biol Chem 273: 23625–23628 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Culotta VC, Klomp LW, Strain J, Casareno RL, Krems B, Gitlin JD (1997) The copper chaperone for superoxide dismutase. J Biol Chem 272: 23469–23472 [DOI] [PubMed] [Google Scholar]
- Danpure CJ (1995) How can the products of a single gene be localized to more than one intracellular compartment? Trends Cell Biol 5: 230–238 [DOI] [PubMed] [Google Scholar]
- Dellaporta SL (1993) Plant DNA miniprep and microprep: versions 2.1-2.3. In M Freeling, V Walbot, eds, The Maize Handbook. Springer-Verlag, New York, pp 522–525
- Elgersma Y, Vos A, van den Berg M, van Roermund CW, van der Sluijs P, Distel B, Tabak HF (1996) Analysis of the carboxyl-terminal peroxisomal targeting signal 1 in a homologous context in Saccharomyces cerevisiae. J Biol Chem 271: 26375–26382 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman HJ, Fridovich I (1973) On the stability of bovine superoxide dismutase: the effects of metals. J Biol Chem 248: 2645–2649 [PubMed] [Google Scholar]
- Fridovich I (1978) The biology of oxygen radicals. Science 201: 875–880 [DOI] [PubMed] [Google Scholar]
- Glerum DM, Shtanko A, Tzagoloff A (1996) Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J Biol Chem 271: 14504–14509 [DOI] [PubMed] [Google Scholar]
- Harrison MD, Jones CE, Dameron CT (1999) Copper chaperones: function, structure and copper-binding properties. J Biol Inorg Chem 4: 145–153 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Aoki M, Kato A, Kondo M, Nishimura M (1996) Transport of chimeric proteins that contain a carboxy-terminal targeting signal into plant microbodies. Plant J 10: 225–234 [DOI] [PubMed] [Google Scholar]
- Horecka J, Kinsey PT, Sprague GF Jr (1995) Cloning and characterization of the Saccharomyces cerevisiae LYS7 gene: evidence for function outside of lysine biosynthesis. Gene 162: 87–92 [DOI] [PubMed] [Google Scholar]
- Imlay JA, Linn S (1988) DNA damage and oxygen radical toxicity. Science 240: 1302–1309 [DOI] [PubMed] [Google Scholar]
- Jackson C, Dench J, Moore AL, Halliwell B, Foyer CH, Hall DO (1978) Subcellular localisation and identification of superoxide dismutase in the leaves of higher plants. Eur J Biochem 91: 339–344 [DOI] [PubMed] [Google Scholar]
- Kanematsu S, Asada K (1989) CuZn-superoxide dismutase in rice: occurrence of an active, monomeric enzyme and two types of isozyme in leaf and non-photosynthetic tissues. Plant Cell Physiol 30: 381–391 [Google Scholar]
- Kliebenstein DJ, Monde RA, Last RL (1998) Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol 118: 637–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (1991) Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem 266: 19867–19870 [PubMed] [Google Scholar]
- Lamb AL, Torres AS, O'Halloran TV, Rosenzweig AC (2000) Heterodimer formation between superoxide dismutase and its copper chaperone. Biochemistry 39: 14720–14727 [DOI] [PubMed] [Google Scholar]
- Lamb AL, Torres AS, O'Halloran TV, Rosenzweig AC (2001) Heterodimeric structure of superoxide dismutase in complex with its metallochaperone. Nat Struct Biol 8: 751–755 [DOI] [PubMed] [Google Scholar]
- Lin SJ, Culotta VC (1995) The ATX1 gene of Saccharomyces cerevisiae encodes a small metal homeostasis factor that protects cells against reactive oxygen toxicity. Proc Natl Acad Sci USA 92: 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Pufahl RA, Dancis A, O'Halloran TV, Culotta VC (1997) A role for the Saccharomyces cerevisiae ATX1 gene in copper trafficking and iron transport. J Biol Chem 272: 9215–9220 [PubMed] [Google Scholar]
- McCord JM, Fridovich I (1969) Superoxide dismutase: an enzymic function for erythrocuprein (hemocuprein). J Biol Chem 244: 6049–6055 [PubMed] [Google Scholar]
- McDowell JM, Dhandaydham M, Long TA, Aarts MG, Goff S, Holub EB, Dangl JL (1998) Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell 10: 1861–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehdy MC (1994) Active oxygen species in plant defense against pathogens. Plant Physiol 105: 467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen RT, Lee MS, Flynn CR, Trelease RN (1997. a) Diverse amino acid residues function within the type 1 peroxisomal targeting signal: implications for the role of accessory residues upstream of the type 1 peroxisomal targeting signal. Plant Physiol 115: 881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen RT, Lee MS, Trelease RN (1997. b) Identification of the peroxisomal targeting signal for cottonseed catalase. Plant J 12: 313–322 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 437–497 [Google Scholar]
- Pan SM, Chen MK, Chung MH, Lee KW, Chen IC (2001) Expression and characterization of monocot rice cytosolic CuZnSOD protein in dicot Arabidopsis. Transgenic Res 10: 343–351 [DOI] [PubMed] [Google Scholar]
- Pan SM, Hwang GB, Liu HC (1999) Over-expression and characterization of copper/zinc-superoxide dismutase from rice in Escherichia coli. Bot Bull Acad Sin (Taipei) 40: 275–281 [Google Scholar]
- Perry SE, Li HM, Keegstra K (1991) In vitro reconstitution of protein transport into chloroplasts. Methods Cell Biol 34: 327–344 [DOI] [PubMed] [Google Scholar]
- Prohaska JR, Geissler J, Brokate B, Broderius M (2003) Copper, zinc-superoxide dismutase protein but not mRNA is lower in copper-deficient mice and mice lacking the copper chaperone for superoxide dismutase. Exp Biol Med 228: 959–966 [DOI] [PubMed] [Google Scholar]
- Rae TD, Schmidt PJ, Pufahl RA, Culotta VC, O'Halloran TV (1999) Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science 284: 805–808 [DOI] [PubMed] [Google Scholar]
- Reumann S (2004) Specification of the peroxisome targeting signals type 1 and type 2 of plant peroxisomes by bioinformatics analyses. Plant Physiol 135: 783–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Mittler R (2003) The water-water cycle is essential for chloroplast protection in the absence of stress. J Biol Chem 278: 38921–38925 [DOI] [PubMed] [Google Scholar]
- Schmidt PJ, Kunst C, Culotta VC (2000) Copper activation of superoxide dismutase 1 (SOD1) in vivo: role for protein-protein interactions with the copper chaperone for SOD1. J Biol Chem 275: 33771–33776 [DOI] [PubMed] [Google Scholar]
- Schmidt PJ, Rae TD, Pufahl RA, Hamma T, Strain J, O'Halloran TV, Culotta VC (1999) Multiple protein domains contribute to the action of the copper chaperone for superoxide dismutase. J Biol Chem 274: 23719–23725 [DOI] [PubMed] [Google Scholar]
- Schnell DJ, Hebert DN (2003) Protein translocons: multifunctional mediators of protein translocation across membranes. Cell 112: 491–505 [DOI] [PubMed] [Google Scholar]
- Silva-Filho MC (2003) One ticket for multiple destinations: dual targeting of proteins to distinct subcellular locations. Curr Opin Plant Biol 6: 589–595 [DOI] [PubMed] [Google Scholar]
- Small I, Wintz H, Akashi K, Mireau H (1998) Two birds with one stone: genes that encode products targeted to two or more compartments. Plant Mol Biol 38: 265–277 [PubMed] [Google Scholar]
- Smeekens S, Bauerle C, Hageman J, Keegstra K, Weisbeek P (1986) The role of the transit peptide in the routing of precursors toward different chloroplast compartments. Cell 46: 365–375 [DOI] [PubMed] [Google Scholar]
- Soll J, Schleiff E (2004) Protein import into chloroplasts. Nat Rev Mol Cell Biol 5: 198–208 [DOI] [PubMed] [Google Scholar]
- Sun CW, Chen LJ, Lin LC, Li HM (2001) Leaf-specific upregulation of chloroplast translocon genes by a CCT motif-containing protein, CIA2. Plant Cell 13: 2053–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trindade LM, Horvath BM, Bergervoet MJ, Visser RG (2003) Isolation of a gene encoding a copper chaperone for copper/zinc superoxide dismutase and characterization of its promoter in potato. Plant Physiol 133: 618–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu SL, Li HM (2000) Insertion of OEP14 into the outer envelope membrane is mediated by proteinaceous components of chloroplasts. Plant Cell 12: 1951–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine JS, Gralla EB (1997) Delivering copper inside yeast and human cells. Science 278: 817–818 [DOI] [PubMed] [Google Scholar]
- Walton PA, Hill PE, Subramani S (1995) Import of stably folded proteins into peroxisomes. Mol Biol Cell 6: 675–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintz H, Vulpe C (2002) Plant copper chaperones. Biochem Soc Trans 30: 732–735 [DOI] [PubMed] [Google Scholar]
- Wong PC, Waggoner D, Subramaniam JR, Tessarollo L, Bartnikas TB, Culotta VC, Price DL, Rothstein J, Gitlin JD (2000) Copper chaperone for superoxide dismutase is essential to activate mammalian Cu/Zn superoxide dismutase. Proc Natl Acad Sci USA 97: 2886–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Shipp E, Sanchez RJ, Liba A, Stine JE, Hart PJ, Gralla EB, Nersissian AM, Valentine JS (2000) Cobalt (2+) binding to human and tomato copper chaperone for superoxide dismutase: implications for the metal ion transfer mechanism. Biochemistry 39: 5413–5421 [DOI] [PubMed] [Google Scholar]