Abstract
We have developed a simple quantitative computational approach for objective analysis of cis-regulatory sequences in promoters of coregulated genes. The program, designated MotifFinder, identifies oligo sequences that are overrepresented in promoters of coregulated genes. We used this approach to analyze promoter sequences of Viviparous1 (VP1)/abscisic acid (ABA)-regulated genes and cold-regulated genes, respectively, of Arabidopsis (Arabidopsis thaliana). We detected significantly enriched sequences in up-regulated genes but not in down-regulated genes. This result suggests that gene activation but not repression is mediated by specific and common sequence elements in promoters. The enriched motifs include several known cis-regulatory sequences as well as previously unidentified motifs. With respect to known cis-elements, we dissected the flanking nucleotides of the core sequences of Sph element, ABA response elements (ABREs), and the C repeat/dehydration-responsive element. This analysis identified the motif variants that may correlate with qualitative and quantitative differences in gene expression. While both VP1 and cold responses are mediated in part by ABA signaling via ABREs, these responses correlate with unique ABRE variants distinguished by nucleotides flanking the ACGT core. ABRE and Sph motifs are tightly associated uniquely in the coregulated set of genes showing a strict dependence on VP1 and ABA signaling. Finally, analysis of distribution of the enriched sequences revealed a striking concentration of enriched motifs in a proximal 200-base region of VP1/ABA and cold-regulated promoters. Overall, each class of coregulated genes possesses a discrete set of the enriched motifs with unique distributions in their promoters that may account for the specificity of gene regulation.
Abscisic acid (ABA) is one of the classic plant hormones that regulates a variety of development and responses, such as seed maturation and drought tolerance (Zeevaart and Creelman, 1988; Giraudat, 1995; McCarty, 1995). During these processes, endogenous ABA level is known to increase, thereby leading to physiological changes in plant cells (Zeevaart and Creelman, 1988; Thomashow, 1999; Xiong et al., 2002). Expression of a large number of genes is modulated mainly by transcriptional control to the elevated ABA level (Shinozaki et al., 2003). Transcription factors, which directly or indirectly bind to promoter elements of the target genes, are the key determinants of transcriptional control. In order to understand the mechanisms of gene expression regulated by ABA signaling during plant development, identification and characterization of transcription factors as well as their target cis-sequences are essential.
A number of transcription factors have been identified and characterized as key mediators of ABA signaling (Finkelstein et al., 2002). The well-characterized transcription factors include ABA-Insensitive3 (ABI3), ABI4, and ABI5 proteins of Arabidopsis (Arabidopsis thaliana; Giraudat et al., 1992; Finkelstein et al., 1998; Finkelstein and Lynch, 2000). These transcription factors were first identified through genetic screening for ABA-insensitive seeds during germination (Koornneef et al., 1984; Finkelstein, 1994). The ABI3 gene encodes a B3 domain transcription factor that is orthologous to maize (Zea mays) Viviparous1 (VP1; McCarty et al., 1991). These transcription factors directly bind to the Sph (RY) element in promoters through its C-terminal B3 DNA binding domain (Suzuki et al., 1997; Monke et al., 2004). Moreover, ABI3 interacts physically with ABI5 bZIP DNA binding protein, thereby recruiting the B3 transcription factor to the promoters of the target genes via a protein-protein interaction (Nakamura et al., 2001). Consequently, ABI3 and ABI5 regulate gene expression synergistically in a highly ABA-dependent manner (Gampala et al., 2002; Casaretto and Ho, 2003). ABI5, which is both transcriptionally and posttranscriptionally regulated by ABA (Lopez-Molina et al., 2001, 2003; Brocard et al., 2002), belongs to a gene family consisting of 13 related bZIP genes in Arabidopsis (Bensmihen et al., 2002). These proteins bind sequence elements known as ABA response elements (ABREs) that contain an ACGT-core motif (Choi et al., 2000; Uno et al., 2000; Bensmihen et al., 2002; Carles et al., 2002; Kim et al., 2002). ABI4 encodes an AP2 DNA binding domain transcription factor (Finkelstein et al., 1998) that binds to the Coupling Element 1 (CE1) motif of ABA-regulated genes (Niu et al., 2002). Whereas the ABI3 and ABI5 functions are implicated mostly with seed development, ABI4 has been shown to regulate ABA and sugar signaling in vegetative tissues as well (Soderman et al., 2000).
In addition to the ABI factors, a growing number of other transcription factors have been implicated in ABA signaling (Finkelstein et al., 2002). These include a variety of classes of transcription factors, including bZIP, Zn-finger, AP2/EREBP, MYB, bHLH, NAC, and WRKY. Opaque2-like bZIP factors and a WRKY transcription factor were recently shown to interact with ABI3 and activate expression of ABA-regulated genes (Lara et al., 2003; Zou et al., 2004), thereby suggesting that additional transcription factors are yet to be identified in ABA signaling during seed development. Moreover, several transcription factors in these classes are known to be activated by exogenous application of ABA as well as abiotic stress conditions, such as cold and drought (Urao et al., 1993; Soderman et al., 1996, 1999; Lee and Chun, 1998; Choi et al., 2000; Uno et al., 2000; Kim et al., 2001; Denekamp and Smeekens, 2003; Shen et al., 2003; Sakamoto et al., 2004; Tran et al., 2004; Villalobos et al., 2004; Xue and Loveridge, 2004). Ectopic expression of these transcription factors is often sufficient to modulate ABA signaling (Himmelbach et al., 2002; Kang et al., 2002; Abe et al., 2003; Fujita et al., 2004; Villalobos et al., 2004). Among the abiotic stresses, drought and cold responses of plants are extensively studied and implicated in ABA signaling. Both stresses are able to promote biosynthesis of ABA (Zeevaart and Creelman, 1988; Thomashow, 1999; Xiong et al., 2002) and change the global expression of genes (Shinozaki et al., 2003), suggesting that ABA signaling plays a significant role for these responses. In drought stress responses, some transcription factors, such as bZIPs, AtMYC2, AtMYB2, and RD26, have been described as key mediators of the ABA-dependent signaling pathway (Uno et al., 2000; Abe et al., 2003; Fujita et al., 2004). In contrast, roles of ABA-mediated signaling are thought to be minor in cold responses (Thomashow, 1999; Shinozaki et al., 2003). Whereas some cold-regulated genes, including CBF/DREB1 transcription factors (Haake et al., 2002; Kizis and Pages, 2002; Knight et al., 2004), are regulated by ABA, the biological significance of the ABA signaling in the cold response is still unclear.
Combinatorial arrangements of cis-elements have been described in several ABA-regulated promoters (Baker et al., 1994; Yamaguchi-Shinozaki and Shinozaki, 1994; Kao et al., 1996; Shen et al., 1996; Hobo et al., 1999; Delseny et al., 2001; Chandrasekharan et al., 2003) that provide some insight into interactions of transcription factors that mediate ABA signaling. Global expression studies based on microarrays allow identification and classification of large numbers of genes into coregulated sets. Not surprisingly, perhaps, given the diversity of transcription factors involved in ABA signaling, a global expression analysis of ABA- and VP1-regulated genes reveals a complex hierarchy of response classes (Suzuki et al., 2003). This large collection of genes provides an opportunity for discovery of patterns that determine ABA response classes.
Various computational approaches have been developed to identify putative regulatory sequences that correlate with expression data (Wolfsberg et al., 1999; Hughes et al., 2000; Bussemaker et al., 2001; Fujibuchi et al., 2001; Ohler and Niemann, 2001; Thijs et al., 2002; Aerts et al., 2003). Here, we describe a comprehensive statistical analysis of cis-elements in genes identified by two independent microarray experiments. As a starting point, we implement an algorithm (in a program designated MotifFinder) that performs a complete search of a dictionary of oligonucleotides of defined length and degeneracy for motifs that are statistically enriched in a set of coregulated promoters. We have applied this program to identify overrepresented sequences in a set of VP1- and ABA-regulated genes (Suzuki et al., 2003) and in a set of cold-regulated genes (Fowler and Thomashow, 2002), respectively. We show that MotifFinder successfully identifies known cis-elements implicated in ABA and cold signaling as well as novel elements that may discriminate subclasses of coregulated genes. We extend the analysis to identify patterns of flanking nucleotide variation and combinatorial arrangements of cis-elements that correlate with qualitative and quantitative differences in gene regulation.
RESULTS AND DISCUSSION
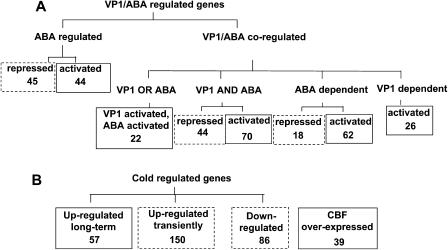
We analyzed two independent sets of microarray experiments based on the Affymetrix 8 K GeneChip (Fig. 1): VP1- and ABA-regulated genes (Suzuki et al., 2003) and cold-regulated genes (Fowler and Thomashow, 2002), respectively. The affected genes were classified by response patterns as summarized in Figure 1. Critical cis-elements are known for both sets of genes, including Sph, the binding site for B3 DNA binding domain of VP1 protein (Suzuki et al., 1997), and the CRT/DRE binding site for the CBF transcription factors (Stockinger et al., 1997; Liu et al., 1998). Moreover, a subset of VP1/ABA-regulated genes and cold-regulated genes possesses ABREs broadly implicated in ABA signaling (Yamaguchi-Shinozaki and Shinozaki, 1994; McCarty, 1995). These known cis-elements served as positive controls to evaluate the efficacy of the MotifFinder program.
Figure 1.
Classification of VP1/ABA- and cold-regulated genes selected for MotifFinder analysis. A, VP1/ABA-regulated genes (Suzuki et al., 2003). B, Cold-regulated genes (Fowler and Thomashow, 2002). Based on bootstrap analysis (Table I), the groups outlined by solid lines were selected for further analysis.
Analysis of VP1/ABA-Regulated Genes
From VP1/ABA-regulated genes, we selected eight response classes that contained at least 18 genes for statistical analysis (Fig. 1A). Motif frequencies were evaluated in triplicate experiments using independent random control sets. Based on the bootstrap results, we selected motifs having P < 10−5 as being significant for the ABA-regulated activated, VP1-dependent activated, ABA-dependent activated, and VP1- and ABA-dependent activated classes (Table I). Because the number of motifs having P < 10−5 for the VP1- or ABA-activated class was within the standard deviation of the average bootstrap value, those enriched motifs are viewed with caution. By contrast, the number of enriched motifs from the three repressed classes, ABA-dependent repressed, ABA-regulated repressed, and VP1- and ABA-dependent repressed, were comparable to the bootstrap values, indicating a likelihood that these motifs are due to chance. For this reason, we excluded the repressed classes from further analysis. These results suggest that gene activation but not repression is mediated by specific combinations of cis-elements. As a consequence of the bootstrap analysis, we decided to further evaluate the motifs from the five coregulated classes. For convenience, we designated each class with the simplified annotation shown in Table II.
Table I.
Bootstrap analysis for statistically significant 8-mer motifs from VP1/ABA-regulated genes
| Coregulated Class | Motifs (P < 10−5) Extracted by MotifFinder | Random Motifs (Average of 10 Replications) | Size of the Class |
|---|---|---|---|
| Activated | |||
| VP1 and ABA dependent | 141 | 3 ± 2.8 | 70 |
| ABA dependent | 25 | 6.1 ± 3.8 | 62 |
| VP1 dependent | 49 | 11.6 ± 7.8 | 26 |
| VP1 or ABA dependent | 17 | 10.8 ± 6.4 | 22 |
| ABA regulated | 45 | 6.1 ± 3.0 | 44 |
| Repressed | |||
| VP1 and ABA dependent | 2 | 6.1 ± 3.0 | 44 |
| ABA dependent | 7 | 9.9 ± 6.4 | 18 |
| ABA regulated | 4 | 5.7 ± 4.2 | 45 |
Table II.
Annotation and coregulation of the five classes of VP1/ABA-activated genes
| Coregulated Class | Annotation | ABA | VP1 | VP1 + ABA |
|---|---|---|---|---|
| ABA regulated | ABR | +a | =b | + |
| ABA dependent | ABD | + | = | ++c |
| VP1 dependent | VPD | = | + | ++ |
| VP1 and ABA dependent | VAA | = | = | ++ |
| VP1 or ABA dependent | VOA | + | + | + |
Activated.
Neutral.
Activated.
All the motifs satisfying a P < 10−5 cutoff for each response class were mapped on the promoters sequences (Supplemental Data 1–5 online). In many cases, the significant 8-mer motifs formed overlapping clusters that highlight presumptive enhancer elements (summarized in Table III). Strong clustering over highly conserved consensus elements is expected because the 8-mers are fairly degenerate (i.e. 28-fold for a perfectly conserved 8-mer motif). In all five activated classes of VP1/ABA-regulated genes, eight base motifs containing an ACGT sequence were the most highly enriched. This result is consistent with the fact that regulation of gene expression in VP1 and ABA response is mediated by ABREs that possess the ACGT-core sequences (Hattori et al., 1995; Vasil et al., 1995; Busk and Pages, 1997). In addition to the ACGT-core motifs, MotifFinder extracted other significant motifs in these activated classes. A motif matching the CE3 ABRE (Shen et al., 1996; Hobo et al., 1999), GTGTC, was enriched in three classes (VAA, VPD, and ABR). A similar sequence, designated as proximal B-box motif, has also been shown to be required for seed-specific expression of the napA gene (Ezcurra et al., 1999). A related motif, CGTGT, was enriched in the ABD class. Recently, Chung et al. (2005) detected nuclear protein complexes with the distal element in the carrot (Daucus carota) Dc3 promoter. This element was designated as TCGTGT and was shown to mediate ABA-inducible gene expression.
Table III.
Statistically enriched motifs from VP1/ABA-activated genes
| Coregulated Class | Motifs (P < 10−5) | Genes with the Motif |
|---|---|---|
| ABA regulated (ABR) | ACGT-core ABRE | 84% |
| GTGTC | 45% | |
| ABA dependent (ABD) | ACGT-core ABRE | 71% |
| CGTGT | 69% | |
| TGTCG | 34% | |
| VP1 dependent (VPD) | ACGT-core ABRE | 77% |
| GTGTC | 62% | |
| VP1 and ABA dependent (VAA) | ACGT-core ABRE | 77% |
| CATGCA (Sph) | 40% | |
| GTGTC | 67% | |
| VP1 or ABA dependent (VOA) | ACGT-core ABRE | 68% |
| TCGGC | 41% |
In addition to shared motifs, the MotifFinder identified enriched sequences that are unique to specific activated subclasses. The Sph motif, CATGCA, was enriched specifically in the VAA class as we reported previously (Suzuki et al., 2003). Sph is known to be a key cis-element for expression of many seed-specific genes (Baumlein et al., 1992; Chamberland et al., 1992; Hattori et al., 1992; Fujiwara and Beachy, 1994; Bobb et al., 1997; Ezcurra et al., 1999, 2000) and is specifically recognized by the B3 domain of VP1 and ABI3 (Suzuki et al., 1997; Monke et al., 2004). The lack of enrichment of B3 binding sites in other VP1-regulated classes (VPD and VOA) suggests that B3 binding is a key determinant of the interaction of VP1 with ABA signaling. Whereas the C-terminal B3 domain is required for activation of a subset of VP1-regulated genes (Carson et al., 1997), the N-terminal coactivation/repression (COAR) domain is necessary and sufficient for activation of ABA-regulated genes as well as repression of germination-specific genes (Hoecker et al., 1995; Nambara et al., 1995, 2002; Carson et al., 1997) of VP1/ABI3. The COAR domain physically interacts with bZIP factors, such as ABI5 (Nakamura et al., 2001), and confers VP1-mediated gene expression. Therefore, interactions of VP1 with ABRE binding proteins mediating the COAR domain are likely responsible for recruitment to genes that lack Sph elements. Other novel sequences, TGTCG and TCGGC, were uniquely enriched in the ABD and VOA classes, respectively. Although the significance of these novel elements remains to be determined, they may discriminate the respective response classes. Interestingly, our analysis of genes ectopically activated by VP1 in vegetative tissues detected no evidence for enrichment of the CE1-like motif (CCACC or CACCG) implicated as the binding site of ABI4 (Niu et al., 2002; Shen et al., 2004). Although the overlap of ABI4 and ABI3/ABI5 functions has been shown in regulation of some seed-specific genes (Finkelstein et al., 1998; Finkelstein and Lynch, 2000; Soderman et al., 2000; Brocard et al., 2002), this result suggests that ABI4 may function independently of ABI3 and ABI5 in this context.
Analysis of Cold-Regulated Genes
As an independent test of the MotifFinder approach, we analyzed a large set of cold-regulated genes that were classified according to microarray expression data by Fowler and Thomashow (2002). We analyzed promoters of 293 genes representing three response classes: up-regulated long term by cold, up-regulated transiently by cold, and down-regulated by cold. We also analyzed the promoters of 39 genes up-regulated by CBF/DREB1 overexpression (Fig. 1B). Based on bootstrap analysis (data not shown), we chose two positively coregulated sets for detailed study: up-regulated long term (ColdUp) and up-regulated by CBF/DREB1 overexpression (CBFox). As was the case for repressed classes of VP1/ABA-regulated genes, we failed to identify significant shared motifs for cold down-regulated genes.
Among 57 genes of the ColdUp class and 39 genes of the CBFox class, the CBF/DREB1 binding motif, CCGAC, and ACGT-type ABREs were highly significant with P < 10−5 (Table IV; see Supplemental Data 6 and 7 online for the mapped motifs). The CCGAC motifs were detected as the most enriched, supporting that the CBF/DREB1 transcription factors play central roles in cold response (Thomashow, 2001; Xiong et al., 2002; Shinozaki et al., 2003). By comparison, P values of ABRE elements in cold-regulated genes were generally higher (less significant) than in the VP1- and ABA-regulated genes (see supplemental data online), consistent with ABA signaling playing a minor role in the cold response (Thomashow, 1999; Xiong et al., 2002; Shinozaki et al., 2003). In addition to the known ABREs and CBF/DREB1 binding sequences, we identified three novel consensus motifs in the ColdUp genes. One of these motifs, CTCCGCGT, resembles the ICEr2 motif (ACTCCG) found in CBF2 promoter (Zarka et al., 2003). Our analysis detected three additional significant bases (CTCCGCGT) at the 3′ end of the motif but did not include the A in the ICEr2. A second novel motif, AGTNGGTCC, was found in 47% of the ColdUp promoters, and a third motif, GATATNNT, occurs in at least two copies in 19 genes. Interestingly, however, the three motifs (CTCCGCGT, AGTNGGTCC, and GATATNNT) were not detected in the CBFox genes, suggesting that these elements may be independent of CBF/DREB1 function. Conversely, the TGGCCNA/G motif was enriched only in the CBFox genes. Thus, this element may be an accessory to CBF/DREB1 function.
Table IV.
Statistically enriched motifs from cold/CBF up-regulated genes
| Coregulated Class | Motifs (P < 10−5) | Genes with the Motif |
|---|---|---|
| Cold long-term up-regulated (ColdUp) | ACGT-core ABRE | 82% |
| CCGAC (CRT/DRE) | 40% | |
| CTCCGCG | 21% | |
| AGTNGGTCC | 47% | |
| GATATNNT | 74% | |
| CBF overexpression (CBFox) | ACGT-core ABRE | 67% |
| CCGAC (CRT/DRE) | 64% | |
| TGGCCNA/G | 33% |
Comparison of MotifFinder with Other cis-Element Detection Programs
The MotifFinder program uses a simple enumerative (i.e. word counting) algorithm, whereas most other programs for motif analysis are based on Gibbs sampling algorithms (Hughes et al., 2000; Liu et al., 2001, 2002, 2004). To compare effectiveness of MotifFinder to other similar programs, we analyzed the VAA and ColdUp coregulated promoters using AlignACE (Hughes et al., 2000), BioProspector, and MDScan (Liu et al., 2001, 2002, 2004). While AlignACE, a widely used first generation program based on Gibbs sampling (Hughes et al., 2000), detected a subset of ACGT core ABREs in VAA-regulated genes, the majority of extracted motifs consisted of low complexity sequences (e.g. A/T rich repeats; data not shown). The web-based BioProspector and MDScan programs (Liu et al., 2001, 2002, 2004) detected consensus motifs that were similar to those identified by the MotifFinder, though each program extracted somewhat different sets of enriched motifs (Supplemental Data 8 online). The Sph motif was detected in VAA promoters by all three programs, whereas the ACGT core ABRE was detected by MotifFinder and MDScan but surprisingly not by BioProspector. Conversely, the CBF/DREB1 binding motif, CCGAC, was identified in ColdUp genes by MotifFinder and BioProspector but not by MDScan. In addition, each program detected distinct oligosequences that were not extracted by the other programs. In the ColdUp promoter set, the ACGT core ABRE and ICEr2-like elements were detected exclusively by MotifFinder. BioProspector extracted several unique sequences from both VAA and ColdUp genes. These results indicate that MotifFinder is at least comparable if not superior to the other methods in detecting consensus elements. Used in combination, the enumerative and Gibbs sampling analyses can provide complementary information.
The enumerative approach has several advantages for in-depth analysis of motif patterns that correlate with gene regulation. A key feature is that MotifFinder outputs the significance of every motif in the degenerate dictionary in a format that can be used to create a statistical profile of each promoter sequence in the coregulated set facilitating visualization of patterns (Suzuki et al., 2003; see supplemental data online). Also, in contrast to Gibbs sampling methods, an enumerative analysis of a set of promoters with a particular user-defined dictionary is guaranteed to be complete and exactly reproducible. The resulting transparency gives the user exact control of the sequence space searched by the program and simplifies the statistical treatment. This feature is particularly useful in evaluating frequencies of sequence variants.
Analysis of Flanking Sequences of the Sph and CRT/DRE Motifs
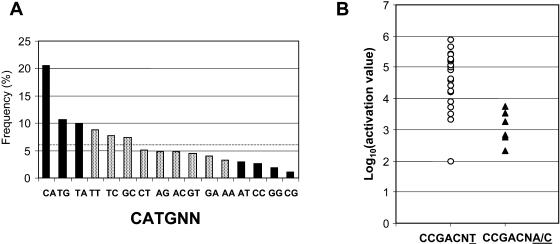
Statistically enriched 8-mers tend to form clusters that overlap strong consensus elements (see supplemental data online) and therefore include information about less-conserved flanking nucleotides that may alter specificity of transcription factor binding. Taking an advantage of this feature, we first analyzed the two nucleotides flanking the CATG core of Sph-related sequences (Fig. 2A). Among the 268 pairs of two base flanking nucleotides from 134 significant CATG motifs, the CATGCA motif occurred in 21% (P < 9× 10−22), whereas, CATGCA, CATGTG, and CATGTA variants together accounted for 42% of the total (P < 3 × 10−18, including both orientations). Moreover, 60% of all the CATG motifs include one of these three flanking dinucleotides in at least one orientation. These results are consistent with the RY repeat consensus determined from seed-expressed genes (Baumlein et al., 1992; Chamberland et al., 1992; Hattori et al., 1992; Fujiwara and Beachy, 1994; Bobb et al., 1997; Ezcurra et al., 1999). An interesting exception to the RY rule is the strong depletion of CATGCG variants relative to other (P < 8 × 10−4) RY variants. Second, we analyzed flanking nucleotides of CCGAC motif in the ColdUp genes (Fig. 2B). Among 23 genes containing the motif, 18 genes possessed CCGACNT, whereas the remaining genes have either CCGACNA or CCGACNG. We examined a correlation between these variations and level of gene activation (induction×absolute difference) using values in the study by Fowler and Thomashow (2002). As shown in Figure 2B, genes with CCGACNT variant are activated more strongly than those with CCGACNA/G. Inspection of the outlier CCGACNT-containing gene with exceptionally low activation (At1g14170) revealed that the motif actually occurs in the first intron based on annotation of a full-length cDNA sequence (Seki et al., 2002; www.gsc.riken.go.jp). No other CCGAC motifs occur within 600 bp of the transcription start site of this gene. This result correlates nicely with recent results of Maruyama et al. (2004) showing that CBF3/DREB1A binds the CCGACNT variant with higher affinity than CCGACNA/C/G.
Figure 2.
Analysis of nucleotides flanking consensus CATG motifs and CCGAC motifs in VAA- and cold-regulated promoters. A, All 8-mer oligos enriched the VAA promoter set that included CATG were evaluated for the two flanking nucleotides in both orientations. The dotted line shows expected frequency by random distribution. The black bars represent the sequences with a significantly skewed frequency (P < 0.05), whereas the shaded bars represent those with P > 0.05. B, All the CCGAC sequences within significant 8-mers from cold long-term up-regulated genes were examined for the flanking nucleotides. If more than two distinct CCGAC motifs were present in the promoters, then CCGACNT was taken for the evaluation. Activation level was calculated by peak average difference × induction at 24 h, according to the values reported by Fowler and Thomashow (2002).
Analysis of Flanking Sequences of the Widely Distributed ACGT-Core Motifs
As shown in Table III, ACGT-type elements were common among all the activated classes of VP1/ABA-regulated genes. The bZIP transcription factors implicated in ABA signaling are known to bind the ACGT-core sequences (Choi et al., 2000; Uno et al., 2000; Bensmihen et al., 2002; Carles et al., 2002; Kim et al., 2002). However, it is not known how the specificity of gene expression is conferred by the widely distributed sequences and their binding proteins. Several independent studies have shown that flanking nucleotides of the ACGT affect binding affinities of bZIP proteins (Schindler et al., 1992a, 1992b; Izawa et al., 1993; Foster et al., 1994; Hong et al., 1995; Carles et al., 2002) as well as regulation of gene expression qualitatively and quantitatively (Hattori et al., 2002; Shen et al., 2004).
In order to analyze whether the flanking sequences of the core ACGT discriminate different activated classes, we analyzed the frequency of all possible ACGT-core sequences consisting of seven bases with three flanking bases (NNNACGT, both orientations) in each response class (Table V). We first determined the frequencies of all ACGT motif sequences in the entire promoter database. We then evaluated the frequency of the motifs in each of the five activated classes of VP1/ABA-regulated genes using χ2 tests.
Table V.
Flanking nucleotides analysis of the ACGT-cored motifs in VP1/ABA- and cold-activated genes
P values are shown in the parentheses.
| Class | ABA Regulated (ABR) | ABA Dependent (ABD) | VP1 Dependent (VPD) | VP1 and ABA Dependent (VAA) | VP1 or ABA Dependent (VOA) | Cold/VP1/ABA | Cold (ColdUp) | Cold Only | |
|---|---|---|---|---|---|---|---|---|---|
| Enriched | CCC (0.00002) | AAC (0.00007) | AAA (0.003) | GAC (0.0001) | GAC (0.0000003) | GAC (0.002) | CCG (0.00007) | CCC (0.03) | CCC (0.007) |
| GAC (0.0002) | GAC (0.002) | TTA (0.005) | GCT (0.001) | GCC (0.00001) | TAC (0.03) | CAT (0.02) | GCC (0.02) | ||
| CCG (0.02) | TGC (0.002) | AAT (0.009) | GCC (0.004) | TAC (0.003) | ACC (0.03) | ||||
| TGG (0.002) | ATA (0.01) | GCG (0.03) | AAC (0.005) | AAC (0.04) | |||||
| GTT (0.002) | GAA (0.03) | CCC (0.04) | AGC (0.008) | GAC (0.05) | |||||
| CGC (0.03) | |||||||||
| Depleted | ACA (0.05) | CTG (0.01) | TAA (0.02) | TAA (0.02) | AAA (0.03) | ||||
| ATG (0.05) | GCT (0.02) | GCG (0.04) | GTC (0.05) | ||||||
| GCA (0.02) | |||||||||
Of the 64 possible variants, 23 distinct motifs were identified as either enriched or depleted in the activated genes using a cutoff of P < 0.05 supported by bootstrap analysis. One variant, GACACGT, was enriched in all classes and particularly frequent in the VAA genes. GCCACGT was enriched in two VP1-dependent classes, VAA and VPD, respectively. These two variants are identical to the ABREs that are most effective in ABA induction of OsVP1-regulated Osem gene expression (Hattori et al., 2002). A third variant, AACACGT, was prominent in ABA-dependent response classes, including the VAA group. The CCCACGT motif was more frequent in ABA-regulated genes compared to the VP1-regulated classes. Thirteen variants were uniquely enriched in a particular response class. Interestingly, two variants (GCTACGT and GCGACGT) that were enriched only in the VPD class were significantly underrepresented in the ABD class compared to all promoters in the database, suggesting that the biased distribution of these variants may be a determinant of the differential regulation of these classes. AGCACGT and CGCACGT were enriched solely in the VAA class, whereas CCGACGT was enriched exclusively in the ABR class. Eight of the 12 unique variants were identified in the ABD class. Hence, these unique distributions of the ABRE variants may contribute specificity to ABA signaling and VP1 function. Soybean (Glycine max) GBF-1 as well as cold-induced and ABA-inducible bZIP proteins have been shown to bind to the ACGT-core variants with different affinities (Kim et al., 2001). This GBF1 has highest affinity for GCCACGT and GACACGT variants. Other bZIPs implicated in ABA signaling, including Arabidopsis ABI5 and related factors and OPAQUE2-related factors (Choi et al., 2000; Uno et al., 2000; Bensmihen et al., 2002; Carles et al., 2002; Kim et al., 2002; Lara et al., 2003), may also bind variants of ABRE with different affinities.
A small subset of cold-regulated genes has been shown to be regulated by ABA through ABREs (Pla et al., 1993; Baker et al., 1994; Yamaguchi-Shinozaki and Shinozaki, 1994). As shown in Table IV, we detected ACGT-core sequences among the enriched motifs in the ColdUp class. We detected a single variant, CCCACGT, as enriched in these genes overall (Table V). Interestingly, this particular variant is more enriched in a subset of genes that are up-regulated by cold but not by VP1 or ABA (cold only: 24 of 57 genes). By contrast, the CCGACGT variant was more enriched in the ColdUp genes that are also regulated by VP1 or ABA (Cold/VP1/ABA: 23 of 57 genes). These results indicate that ABA signaling is mediated by distinct ABREs in cold/ABA-responsive and VP1/ABA-regulated genes.
Combinatorial Relationships of ACGT and Non-ACGT Motifs
Promoter structure is often complex with multiple cis-regulatory elements acting combinatorially. Modular organization of cis-elements has been shown in many promoters, including those of ABA-regulated genes (Kao et al., 1996; Shen et al., 1996; Hobo et al., 1999; Delseny et al., 2001; Chandrasekharan et al., 2003). In addition to variation of flanking nucleotides from the ACGT sequence of ABREs (Table IV), physical associations with other cis-elements in promoters may determine specificity of gene expression.
In order to search for nearby conserved elements, we analyzed sequences that are enriched in the vicinity of the ACGT-core motifs in the five activated classes of VP1/ABA-regulated genes and in the ColdUp genes, respectively. We extracted 20 nucleotides located immediately upstream and downstream of the two most significant ACGT variants in the promoters of each response class. MotifFinder was used to identify enriched motifs within the short flanking regions relative to randomly selected 100-nucleotide segments in the promoters.
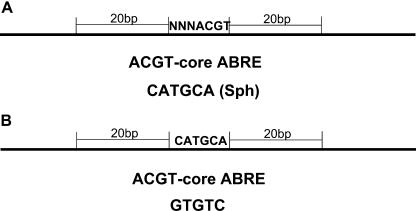
Among the five activated groups of VP1/ABA-regulated genes, we detected significant enrichment of motifs in the vicinity of ACGT elements (P < 10−4) in promoters of the VAA class but no other groups (Fig. 3A). Not surprisingly, ABREs were associated with other ABREs, indicating that these elements tend to occur in clusters. In addition, we identified significant enrichment of Sph-related (RY) elements in close proximity to ABREs. This is consistent with the known synergistic interactions of VP1 and bZIPs in regulation of this response class (Gampala et al., 2002; Casaretto and Ho, 2003). In order to extend this result, we also analyzed enriched motifs that are associated with the Sph element in the same manner (Fig. 3B). As expected, we identified the ACGT-core ABREs as enriched associated motifs. In addition, we recovered CE3-like motifs containing GTGTC, suggesting that this sequence may be correlated with a distinct Sph function. In the ColdUp class, we detected numerous AT-rich motifs with P < 10−5. These motifs, however, did not show a strong consensus due to the low complexity of the sequences. Moreover, the analysis did not detect significant association of CCGAC and ABREs within this group.
Figure 3.
Motifs closely associated with the ABRE and Sph elements in the VAA class of promoters. A, Motifs associated with ABREs. B, Motifs associated with Sph elements.
While combinatorial interactions between cis-elements are frequently invoked to explain specificity of gene regulation, the only short range combinatorial relationship between different consensus cis-elements detected in our analysis is an association between Sph and ABRE elements exclusively in the VAA class of VP1/ABA-regulated genes. However, detection of this particular combination is strongly favored by the design of the original microarray experiment (Suzuki et al., 2003) because the VAA class is comprised of those genes that show the strongest interdependence on the two principle treatments, ectopic VP1 and ABA. An implication is that treatment interactions can be very effective in discerning combinatorial patterns in promoters in other contexts. While we failed to detect a correlation between CBF consensus binding sites and ABRE elements, for example, the cold data set did not include explicit treatments combining ABA, cold, and/or CBF overexpression. Hence, the resulting classification of genes may not be optimal for detecting that pattern, if it exists.
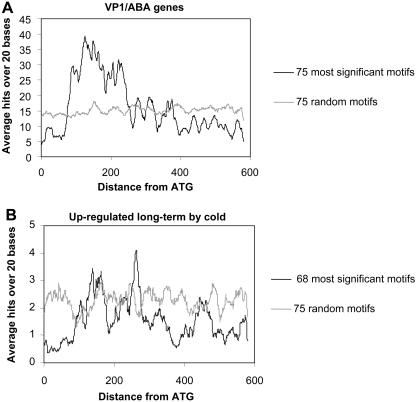
In order to analyze distribution of the cis-elements broadly in the 5′ upstream regions of the VP1/ABA-regulated genes and cold-regulated genes, we selected and mapped 75 8-mer motifs and 68 motifs that are most enriched in the promoters of these genes, respectively. Overall, the enriched motifs showed a striking enrichment in the proximal region within 300 bp upstream of the annotated ATG codon (Fig. 4). By comparison, 75 randomly selected motifs from the motif dictionary are evenly distributed. We tested 15 distinct sets of the 75 random motifs and obtained similar distributions (data not shown).
Figure 4.
Biased distribution of enriched motifs in the promoters of VP1/ABA and cold long-term up-regulated genes. A, The distribution of the 75 most significant motifs in promoters (600 bp upstream of ATG) of 353 VP1/ABA-regulated genes (black line) compared to 75 random motifs (gray line). B, The distribution of the 68 most significant motifs in promoters of 58 ColdUp genes (black line) compared to 75 random motifs (gray line). The plots show the average number of significant bases in a 20-bp sliding window.
This analysis reveals a higher order organization of conserved cis-elements in VP1/ABA-regulated genes (Fig. 4A). The enriched sequence motifs show a striking concentration in the −100- to −300-nucleotide interval upstream of the translation initiation site, indicating that the key activators bind immediately upstream of the transcription initiation site. Interestingly, the 200-nucleotide interval spanned by the enriched motifs is roughly the span of a nucleosome. This observation is consistent with evidence that VP1/ABI3 factors specifically direct remodeling of a nucleosome positioned over TATA (Li et al., 1999, 2001; Grace et al., 2004). A proximal bias of enriched motifs is much less pronounced in ColdUp genes (Fig. 4B) and CBFox genes (data not shown), implying that the activators may be less constrained in cold-regulated genes than in VP1/ABA-regulated genes. However, we cannot rule out the possibility that the more diffuse distribution in cold-regulated promoters is related to the fact that topmost cold-enriched motifs overall had a lower statistical significance compared to top 75 VP1/ABA-enriched motifs.
CONCLUSION
In this report, we show that the MotifFinder algorithm based on a complete search of a motif dictionary is effective for identifying putative cis-regulatory elements in sets of coregulated genes. We applied this program to the two independent sets of global expression data from microarray experiments. In addition to detecting known regulatory elements, the analysis delineated distinct variants and combinations of conserved elements that distinguish subclasses of coregulated genes. Our results reveal at least three sources of specificity for transcriptional regulation: (1) presence or absence of consensus cis-elements recognized by distinct transcription factors, (2) sequence variants of consensus binding sites that may differentially affect TF binding affinity, and (3) combinatorial interactions among cis-elements.
MATERIALS AND METHODS
MotifFinder Program
Promoter sequences analyzed in this study were selected on the basis of two independent microarray experiments using the Affymetrix 8 K Gene Chip (Affymetrix). A database was constructed by extracting a 600-nucleotide segment of 5′ flanking genomic DNA sequence for each of the 7,402 genes represented on the array (www.tigr.org). We then constructed a complete, nonredundant dictionary consisting of the 43,168 possible eight-nucleotide motifs that contain two degenerate bases where reverse complement motifs are considered equivalent. The frequency of promoters containing at least one copy of each motif in either orientation was determined for each set of coregulated promoters and compared to frequency of the motif in the entire database. We used a simple χ2 test to identify motifs that were enriched in the test set. This approach assures that all elements matching a particular definition are analyzed. A prototype of this program was used to analyze Sph motif and specific types of ABREs in a subset of VP1/ABA coregulated genes (Suzuki et al., 2003). The source code of the MotifFinder program is available for download by e-mailing a request to drm@ufl.edu.
Bootstrap Analysis
To determine the appropriate level of statistical significance for the MotifFinder analysis, a bootstrap procedure based on 10 replicates of independently selected random sets of test promoters was used to empirically estimate that likelihood of false-positive motifs for each size of coregulated set (Table I). The frequencies of three nucleotide motifs flanking the ACGT and Sph core elements in coregulated promoters were compared to total frequencies of these variants in the promoter database (7,402 promoters). Bootstrap estimates for flanking nucleotide distributions were based on 100 replicates of random promoter sets. A larger number of bootstrap replicates were used to establish conservative P value confidence levels for trinucleotide frequencies because empirical observations showed greater variation in random frequencies than observed in the 8-mer bootstrap replicates.
Supplementary Material
Acknowledgments
We thank Dr. Michael Popp and Joint Shands Cancer Center-Interdisciplinary Center for Biotechnology Research at University of Florida for assistance in analysis of microarray data.
This work was supported by the National Science Foundation (award nos. 0080175 and 0322005 to M.S. and D.R.M.) and the Florida Agricultural Experiment Station (journal series no. R-11025).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.058412.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aerts S, Thijs G, Coessens B, Staes M, Moreau Y, De Moor B (2003) Toucan: deciphering the cis-regulatory logic of coregulated genes. Nucleic Acids Res 31: 1753–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SS, Wilhelm KS, Thomashow MF (1994) The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol 24: 701–713 [DOI] [PubMed] [Google Scholar]
- Baumlein H, Nagy I, Villarroel R, Inze D, Wobus U (1992) Cis-analysis of a seed protein gene promoter: The conservative RY repeat CATGCATG within the legumin box is essential for tissue-specific expression of a legumin gene. Plant J 2: 233–239 [PubMed] [Google Scholar]
- Bensmihen S, Rippa S, Lambert G, Jublot D, Pautot V, Granier F, Giraudat J, Parcy F (2002) The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14: 1391–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobb AJ, Chern MS, Bustos MM (1997) Conserved RY-repeats mediate transactivation of seed-specific promoters by the developmental regulator PvALF. Nucleic Acids Res 25: 641–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard IM, Lynch TJ, Finkelstein RR (2002) Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol 129: 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busk PK, Pages M (1997) Protein binding to the abscisic acid-responsive element is independent of VIVIPAROUS1 in vivo. Plant Cell 9: 2261–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussemaker HJ, Li H, Siggia ED (2001) Regulatory element detection using correlation with expression. Nat Genet 27: 167–171 [DOI] [PubMed] [Google Scholar]
- Carles C, Bies-Etheve N, Aspart L, Leon-Kloosterziel KM, Koornneef M, Echeverria M, Delseny M (2002) Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J 30: 373–383 [DOI] [PubMed] [Google Scholar]
- Carson CB, Hattori T, Rosenkrans L, Vasil V, Vasil IK, Peterson PA, McCarty DR (1997) The quiescent/colorless alleles of viviparous1 show that the conserved B3 domain of VP1 is not essential for ABA-regulated gene expression in the seed. Plant J 12: 1231–1240 [DOI] [PubMed] [Google Scholar]
- Casaretto J, Ho TD (2003) The transcription factors HvABI5 and HvVP1 are required for the abscisic acid induction of gene expression in barley aleurone cells. Plant Cell 15: 271–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberland S, Daigle N, Bernier F (1992) The legumin boxes and the 3′ part of a soybean beta-conglycinin promoter are involved in seed gene expression in transgenic tobacco plants. Plant Mol Biol 19: 937–949 [DOI] [PubMed] [Google Scholar]
- Chandrasekharan MB, Bishop KJ, Hall TC (2003) Module-specific regulation of the beta-phaseolin promoter during embryogenesis. Plant J 33: 853–866 [DOI] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275: 1723–1730 [DOI] [PubMed] [Google Scholar]
- Chung HJ, Fu HY, Thomas TL (2005) Abscisic acid-inducible nuclear proteins bind to bipartite promoter elements required for ABA response and embryo-regulated expression of the carrot Dc3 gene. Planta 220: 424–433 [DOI] [PubMed] [Google Scholar]
- Delseny M, Bies-Ethève N, Carles C, Hull G, Vicient C, Raynal M, Grellet F, Aspart L (2001) Late abundant (LEA) protein gene regulation during Arabidopsis seed maturation. J Plant Physiol 158: 419–427 [Google Scholar]
- Denekamp M, Smeekens SC (2003) Integration of wounding and osmotic stress signals determines the expression of the AtMYB102 transcription factor gene. Plant Physiol 132: 1415–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezcurra I, Ellerström M, Wycliffe P, Stålberg K, Rask L (1999) Interaction between composite elements in the napA promoter: Both the B-box ABA-responsive complex and the RY/G complex are necessary for seed-specific expression. Plant Mol Biol 40: 699–709 [DOI] [PubMed] [Google Scholar]
- Ezcurra I, Wycliffe P, Nehlin L, Ellerstrom M, Rask L (2000) Transactivation of the Brassica napus napin promoter by ABI3 requires interaction of the conserved B2 and B3 domains of ABI3 with different cis-elements: B2 mediates activation through an ABRE, whereas B3 interacts with an RY/G-box. Plant J 24: 57–66 [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Lynch T (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR (1994) Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J 5: 765–771 [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell (Suppl) 14: S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster R, Izawa T, Chua NH (1994) Plant bZIP proteins gather at ACGT elements. FASEB J 8: 192–200 [DOI] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujibuchi W, Anderson JSJ, Landsman D (2001) PROSPECT improves cis-acting regulatory element prediction by integrating expression profile data with consensus pattern searches. Nucleic Acids Res 29: 3988–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LS, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39: 863–876 [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Beachy RN (1994) Tissue-specific and temporal regulation of a beta-conglycinin gene: roles of the RY repeat and other cis-acting elements. Plant Mol Biol 24: 261–272 [DOI] [PubMed] [Google Scholar]
- Gampala SS, Finkelstein RR, Sun SS, Rock CD (2002) ABI5 interacts with abscisic acid signaling effectors in rice protoplasts. J Biol Chem 277: 1689–1694 [DOI] [PubMed] [Google Scholar]
- Giraudat J (1995) Abscisic acid signaling. Curr Opin Cell Biol 7: 232–238 [DOI] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis AB13 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace ML, Chandrasekharan MB, Hall TC, Crowe AJ (2004) Sequence and spacing of TATA box elements are critical for accurate initiation from the beta-phaseolin promoter. J Biol Chem 279: 8102–8110 [DOI] [PubMed] [Google Scholar]
- Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ (2002) Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 130: 39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori T, Terada T, Hamasuna S (1995) Regulation of the Osem gene by abscisic acid and the transcriptional activator VP1: analysis of cis-acting promoter elements required for regulation by abscisic acid and VP1. Plant J 7: 913–925 [DOI] [PubMed] [Google Scholar]
- Hattori T, Totsuka M, Hobo T, Kagaya Y, Yamamoto-Toyoda A (2002) Experimentally determined sequence requirement of ACGT-containing abscisic acid response element. Plant Cell Physiol 43: 136–140 [DOI] [PubMed] [Google Scholar]
- Hattori T, Vasil V, Rosenkrans L, Hannah LC, McCarty DR, Vasil IK (1992) The Viviparous-1 gene and abscisic acid activate the C1 regulatory gene for anthocyanin biosynthesis during seed maturation in maize. Genes Dev 6: 609–618 [DOI] [PubMed] [Google Scholar]
- Himmelbach A, Hoffmann T, Leube M, Hohener B, Grill E (2002) Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J 21: 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobo T, Asada M, Kowyama Y, Hattori T (1999) ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J 19: 679–689 [DOI] [PubMed] [Google Scholar]
- Hoecker U, Vasil IK, McCarty DR (1995) Integrated control of seed maturation and germination programs by activator and repressor functions of Viviparous-1 of maize. Genes Dev 9: 2459–2469 [DOI] [PubMed] [Google Scholar]
- Hong JC, Cheong YH, Nagao RT, Bahk JD, Key JL, Cho MJ (1995) Isolation of two soybean G-box binding factors which interact with a G-box sequence of an auxin-responsive gene. Plant J 8: 199–211 [DOI] [PubMed] [Google Scholar]
- Hughes JD, Estep PW, Tavazoie S, Church GM (2000) Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J Mol Biol 296: 1205–1214 [DOI] [PubMed] [Google Scholar]
- Izawa T, Foster R, Chua NH (1993) Plant bZIP protein DNA binding specificity. J Mol Biol 230: 1131–1144 [DOI] [PubMed] [Google Scholar]
- Kang J, Choi H, Im MY, Kim SY (2002) Arabidopsis basic leucine zipper proteins mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CY, Cocciolone SM, Vasil IK, McCarty DR (1996) Localization and interaction of the cis-acting elements for abscisic acid, VIVIPAROUS1, and light activation of the C1 gene of maize. Plant Cell 8: 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JC, Lee SH, Cheong YH, Yoo CM, Lee SI, Chun HJ, Yun DJ, Hong JC, Lee SY, Lim CO, Cho MJ (2001) A novel cold-inducible zinc finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plants. Plant J 25: 247–259 [DOI] [PubMed] [Google Scholar]
- Kim SY, Ma J, Perret P, Li Z, Thomas TL (2002) Arabidopsis ABI5 subfamily members have fistinct DNA-binding and transcriptional activities. Plant Physiol 130: 688–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizis D, Pages M (2002) Maize DRE-binding proteins DBF1 and DBF2 are involved in rab17 regulation through the drought-responsive element in an ABA-dependent pathway. Plant J 30: 679–689 [DOI] [PubMed] [Google Scholar]
- Knight H, Zarka DG, Okamoto H, Thomashow MF, Knight MR (2004) Abscisic acid induces CBF gene transcription and subsequent induction of cold-regulated genes via the CRT promoter element. Plant Physiol 135: 1710–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM (1984) The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis. Physiol Plant 6: 377–383 [Google Scholar]
- Lara P, Onate-Sanchez L, Abraham Z, Ferrandiz C, Diaz I, Carbonero P, Vicente-Carbajosa J (2003) Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. J Biol Chem 278: 21003–21011 [DOI] [PubMed] [Google Scholar]
- Lee YH, Chun JY (1998) A new homeodomain-leucine zipper gene from Arabidopsis thaliana induced by water stress and abscisic acid treatment. Plant Mol Biol 37: 377–384 [DOI] [PubMed] [Google Scholar]
- Li G, Bishop KJ, Chandrasekharan MB, Hall TC (1999) β-Phaseolin gene activation is a two-step process: PvALF-facilitated chromatin modification followed by abscisic acid-mediated gene activation. Proc Natl Acad Sci USA 96: 7104–7109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Chandrasekharan MB, Wolffe AP, Hall TC (2001) Chromatin structure and phaseolin gene regulation. Plant Mol Biol 46: 121–129 [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Brutlag DL, Liu JS (2001) BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac Symp Biocomput 127–138 [PubMed]
- Liu XS, Brutlag DL, Liu JS (2002) An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat Biotechnol 8: 835–839 [DOI] [PubMed] [Google Scholar]
- Liu Y, Wei L, Batzoglou S, Brutlag DL, Liu JS, Liu XS (2004) A suite of web-based programs to search for transcriptional regulatory motifs. Nucleic Acids Res 32: W204–W207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Kinoshita N, Chua NH (2003) AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev 17: 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, Shimada Y, Yoshida S, Shinozaki K, Yamagichi-Shinozaki K (2004) Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J 38: 982–993 [DOI] [PubMed] [Google Scholar]
- McCarty DR (1995) Genetic control and intergration of maturation and germination pathways in seed development. Annu Rev Plant Physiol Plant Mol Biol 46: 71–93 [Google Scholar]
- McCarty DR, Hattori T, Carson CB, Vasil V, Lazar M, Vasil IK (1991) The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66: 895–905 [DOI] [PubMed] [Google Scholar]
- Monke G, Altschmied L, Tewes A, Reidt W, Mock HP, Conrad U (2004) Seed-specific transcription factors ABI3 and FUS3: molecular interaction with DNA. Planta 219: 158–166 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Lynch TJ, Finkelstein RR (2001) Physical interactions between ABA response loci of Arabidopsis. Plant J 26: 627–635 [DOI] [PubMed] [Google Scholar]
- Nambara E, Keith K, McCourt P, Naito S (1995) A regulatory role for the ABI3 gene in the establishment of embryo maturation in Arabidopsis thaliana. Development 121: 629–636 [Google Scholar]
- Nambara E, Suzuki M, Abrams S, McCarty DR, Kamiya Y, McCourt P (2002) A screen for genes that function in abscisic acid signaling in Arabidopsis thaliana. Genetics 161: 1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X, Helentjaris T, Bate NJ (2002) Maize ABI4 binds Coupling Element1 in abscisic acid and sugar response genes. Plant Cell 14: 2565–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U, Niemann H (2001) Identification and analysis of eukaryotic promoters: recent computational approaches. Trends Genet 17: 56–60 [DOI] [PubMed] [Google Scholar]
- Pla M, Vilardell J, Guiltinan MJ, Marcotte WR, Niogret MF, Quatrano RS, Pages M (1993) The cis-regulatory element CCACGTGG is involved in ABA and water-stress responses of the maize gene rab28. Plant Mol Biol 21: 259–266 [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol 136: 2734–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler U, Menkens AE, Beckmann H, Ecker JR, Cashmore AR (1992. b) Heterodimerization between light-regulated and ubiquitously expressed Arabidopsis GBF bZIP proteins. EMBO J 11: 1261–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler U, Terzaghi W, Beckmann H, Kadesch T, Cashmore AR (1992. a) DNA binding site preferences and transcriptional activation properties of the Arabidopsis transcription factor GBF1. EMBO J 11: 1275–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y, et al (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145 [DOI] [PubMed] [Google Scholar]
- Shen Q, Zhang P, Ho T-HD (1996) Modular nature of abscisic acid (ABA) response complexes: composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell 8: 1107–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QJ, Casaretto JA, Zhang P, Ho T-HD (2004) Functional definition of ABA-response complexes: the promoter units necessary and sufficient for ABA induction of gene expression in barley (Hordeum vulgare L.). Plant Mol Biol 54: 111–124 [DOI] [PubMed] [Google Scholar]
- Shen YG, Zhang WK, He SJ, Zhang JS, Liu Q, Chen SY (2003) An EREBP/AP2-type protein in Triticum aestivum was a DRE-binding transcription factor induced by cold, dehydration and ABA stress. Theor Appl Genet 106: 923–930 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 6: 410–417 [DOI] [PubMed] [Google Scholar]
- Soderman E, Hjellstrom M, Fahleson J, Engstrom P (1999) The HD-Zip gene ATHB6 in Arabidopsis is expressed in developing leaves, roots and carpels and up-regulated by water deficit conditions. Plant Mol Biol 40: 1073–1083 [DOI] [PubMed] [Google Scholar]
- Soderman E, Mattsson J, Engstrom P (1996) The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and by abscisic acid. Plant J 10: 375–381 [DOI] [PubMed] [Google Scholar]
- Soderman EM, Brocard IM, Lynch TJ, Finkelstein RR (2000) Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol 124: 1752–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Kao CY, McCarty DR (1997) The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 9: 799–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Ketterling MG, Li Q-B, McCarty DR (2003) VP1 alters global gene expression patterns through regulation of ABA signaling. Plant Physiol 132: 1664–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thijs G, Marchal K, Lescot M, Rombauts S, De Moor B, Rouzé P, Moreau Y (2002) A Gibbs sampling method to detect overrepresented motifs in the upstream regions of coexpressed genes. J Comput Biol 9: 447–464 [DOI] [PubMed] [Google Scholar]
- Thomashow MF (1999) PLANT COLD ACCLIMATION: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Thomashow MF (2001) So what's new in the field of plant cold acclimation? Lots! Plant Physiol 125: 89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Furuhata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K (1993) An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5: 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasil V, Marcotte WR Jr, Rosenkrans L, Cocciolone SM, Vasil IK, Quatrano RS, McCarty DR (1995) Overlap of Viviparous 1 (VP1) and abscisic acid response elements in the Em promoter: G-box elements are sufficient but not necessary for VP1 transactivation. Plant Cell 7: 1511–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalobos MA, Bartels D, Iturriaga G (2004) Stress tolerance and glucose insensitive phenotypes in Arabidopsis overexpressing the CpMYB10 transcription factor gene. Plant Physiol 135: 309–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfsberg TG, Gabrielian AE, Campbell MJ, Cho RJ, Spouge JL, Landsman D (1999) Candidate regulatory sequence elements for cell cycle-dependent transcription in Saccharomyces cerevisiae. Genome Res 9: 775–792 [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK (2002) Cell signaling during cold, drought, and salt stress. Plant Cell (Suppl) 14: S165–S183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue GP, Loveridge CW (2004) HvDRF1 is involved in abscisic acid-mediated gene regulation in barley and produces two forms of AP2 transcriptional activators, interacting preferably with a CT-rich element. Plant J 37: 326–339 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarka DG, Vogel JT, Cook D, Thomashow MF (2003) Cold induction of Arabidopsis CBF genes involves multiple ICE (Inducer of CBF Expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiol 133: 910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD, Creelman RA (1988) Metabolism and physiology of abscisic acid. Annu Rev Plant Physiol Plant Mol Biol 39: 439–473 [Google Scholar]
- Zou X, Seemann JR, Neuman D, Shen QJ (2004) A WRKY gene from creosote bush encodes an activator of the ABA signaling pathway. J Biol Chem 279: 55770–55779 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.