Abstract
Background
The gut microbiome has been identified as a pivotal factor in ulcerative colitis (UC), given its role as the main reservoir of microbes in the body. This community of microorganisms, present in variable concentrations in the digestive tract, makes a wide range of beneficial roles for the host. However, the role of the gut microbiome in patients with refractory UC is still significant, so this study aimed to further investigate the role of these bacteria in patients with refractory UC.
Methods
This case-control study was conducted on stool samples from four distinct groups: the first group comprised new patients diagnosed with ulcerative colitis (all of them had responded to treatment after follow-up) (N = 24); the second group consisted of patients with treatment-resistant ulcerative colitis (N = 23); the third group included first-degree relatives of group 1 patients (N = 24); and the fourth group consisted of first-degree relatives of group 2 patients (N = 23). The research tools employed in this study included a questionnaire, quantitative real-time PCR (qPCR) test, and culture on stool samples.
Result
The mean age of patients in groups 1 and 2 was 45.88 ± 18.51 and 41.30 ± 13.01 years, while the mean age of controls in groups 3 and 4 was 37.29 ± 9.62 and 40.96 ± 13.01 years, respectively. Stool culture results for pathogenic bacteria were negative in all four groups. The of history of consuming dairy products containing probiotics was highest in Group 1, with 22 (91.67%) subjects, while the lowest was observed in Group 3, with 16 (66.67%). The highest history of self-administered antibiotic use was observed in Group 2, with 13 cases (56.52%), while the lowest was noted in Group 3, with 4 cases (16.67%). The findings indicated a statistically significant relationship (P < 0.05) between Groups 2 and 4 with respect to the E. coli and Bifidobacterium ssp. microbial population. Additionally, a significant relationship was identified between the Lactobacillus ssp., Bifidobacterium ssp., and Bacteroides ssp. microbial community between groups 1 and 2 (P < 0.05).
Conclusion
The findings of this study demonstrated that several intestinal microbiomes have a substantial impact on the management of ulcerative colitis. The results of this study suggest that by comparing the gut microbiome of treatment-resistant and individuals newly diagnosed with ulcerative colitis, we can gain a better understanding of microbiome differences that may influence treatment outcomes. The results of this study may also lead to the identification of new therapeutic strategies that are based on regulating the gut microbiome. These strategies could include the use of fecal microbiome transplantation (FMT), probiotics, prebiotics, or specific bacteria-based therapies.
Keywords: Ulcerative colitis, Gut microbiome, qPCR
Introduction
Ulcerative colitis (UC) is a type of inflammatory bowel disease (IBD) that causes inflammation and ulcers in the colon [1–2]. This condition can lead to symptoms such as abdominal pain, bloody diarrhea, weight loss, and fatigue [3]. The exact cause of UC is unknown, but it appears that genetic, environmental, and immune factors play a role in its onset [1, 4]. The gut microbiome has garnered significant attention as an important factor in this disease [5–6]. The gut microbiota coexists with its host in varying concentrations in the gastrointestinal tract, with their amounts in the colon reaching 1011 to 1012 cells per gram of gastrointestinal contents [7]. This community makes a wide range of beneficial roles for the host, containing digesting substrates inaccessible to host enzymes, enhancing the immune system, and preventing colonization by harmful microorganisms [8–9]. Several studies have shown that these gut bacteria may have protective effects against inflammatory bowel diseases, including ulcerative colitis [10–12]. In patients with UC, significant changes in the diversity and composition of the gut microbiome, with reduced microbial diversity compared to healthy individuals [10–12]. Certain bacterial species, such as Bacteroides vulgatus, are strongly associated with the severity of UC, to the extent have been observed that the proteolytic activity of this bacterium can exacerbate disease symptoms [13]. The gut microbiome also plays an important role in the production of short-chain fatty acids (SCFAs), especially butyrate, which helps maintain the health and integrity of the intestinal epithelium. A reduction in these SCFAs can lead to increased intestinal permeability and inflammation [14–17]. Probiotics, as a promising therapeutic approach, can be very helpful in treating ulcerative colitis by changing the intestinal microbiota, so that in one study, the consumption of Bifidobacterium infantis reduced inflammation in this disease, and therefore the consumption of such probiotics can reduce the risk of developing this type of disease [18]. However, the effectiveness of probiotics depends on the type of bacterial strain and the results in different studies vary due to heterogeneity in bacterial formulation and host factors. Significant advancements in molecular technologies technology over the past decade have facilitated research on the microbiome in various diseases, including ulcerative colitis. A substantial corpus of research has been dedicated to the examination of the role of the gut microbiome in ulcerative colitis (UC). Therefore, this study aimed to further investigate the role of these bacteria in two important groups of patients with treatment-resistant UC and new patients who have ultimately responded to treatment. The reason we chose these two specific groups is because comparing them could help identify microbiome differences that may contribute to treatment resistance. These differences could include changes in microbial diversity, microbial composition, and microbial function. By identifying these differences, we can gain a better understanding of the mechanisms of treatment resistance and find new ways to improve treatment for this disease.
Materials and methods
Study community
With a power of 80% and a type I error rate of 0.05, and assuming an effect size of 0.80, the required sample size was calculated to be 21 individuals per group. However, our study included more participants than this minimum requirement. Additionally, by incorporating three control groups, we anticipate that the effect size estimates will be more precise and the risk of type II errors will be reduced. This study was conducted as a case-control study on stool samples from four distinct groups: Group 1: individuals newly diagnosed with ulcerative colitis (all of them had responded to treatment after follow-up) (24 individuals). Group 2: individuals with treatment-resistant ulcerative colitis (23 individuals). Group 3: first-degree relatives of individuals newly diagnosed with ulcerative colitis (these individuals were healthy and numbered 24). Group 4: first-degree relatives of individuals with treatment-resistant ulcerative colitis (23 individuals), who were also healthy. It should be noted that in this study, the definition of first-degree relatives includes siblings, children, and biological parents who live in the same household and are genetically identical because they share the same diet and lifestyle, reducing potential confounding factors that may affect the composition of the gut microbiome.
The inclusion criteria encompassed patients who had been recently diagnosed with ulcerative colitis, patients with treatment-resistant ulcerative colitis, and healthy individuals who did not have ulcerative colitis. Conversely, individuals with concurrent conditions such as diabetes, rheumatism, colon cancer, or immunodeficiency, and history of antibiotic use six months before sampling were excluded from the study.
Diagnosis of ulcerative colitis
The diagnosis of ulcerative colitis was made on the basis of clinical symptoms, biochemical tests, pathology, and colonoscopy, and was subsequently confirmed by a gastroenterology specialist.
Diagnosis of treatment-resistant ulcerative colitis
In this study, treatment-resistant ulcerative colitis (UC) patients were defined as those who did not respond to three stages of treatment: the first stage involving 5-aminosalicylic acid, the second stage involving glucocorticoids and azathioprine, and the third stage involving anti-TNF-α therapy. Conversely, patients who responded to any of these three stages of treatment were categorized as treated individuals. This classification allows for a comparative analysis between the microbiomes of treatment-resistant and treatment-responsive UC patients.
Collecting samples
Following the identification of subjects for the study from each group, two stool samples were collected and 5 cc of blood was taken to separate the serum for test C-reactive protein (CRP). These samples were then transferred to the laboratory in a refrigerated. Diagnostic culture tests were performed on one of the stool samples for pathogen bacterial identification, while the second sample was stored at -70 °C for subsequent molecular testing. To measure CRP the commercial kit of Pishtaz Teb Zaman Diagnostics (Tehran, Iran) was used.
Stool culture
In order to identify pathogenic bacteria, a culture test was performed, for which Hektoen Enteric Agar, Xylose Lysine Deoxycholate agar, and Salmonella Shigella Agar selective media, as well as differential media such as Triple Sugar Iron Agar, Lysine Iron Agar, Sulfide Indole Motility, Methyl Red, and Voges-Proskauer reactions, and Simmons Citrate Agar, were used [18].
DNA extraction
Stool DNA extraction was performed using the FavorPrep™ kit (FAVORGEN Biotech Corporation, Taiwan) according to the kit’s protocol. The extracted DNA samples were subsequently stored at -20 °C until the molecular tests were conducted.
Standard curve preparation to determine the copy number of the studied bacteria
The standard curve was obtained by utilizing standard bacteria. These bacteria were obtained from the Pasteur Institute of Iran and the Iranian Biological Resource Center. DNA extraction was performed on bacteria using the Sinaclon extraction kit (Tehran, Iran). Subsequently, a polymerase chain reaction (PCR) was conducted employing the primers shown in Table 1 to amplify the 16 S rRNA gene of the bacteria. Then gene purification was carried out using the Sinaclon gene purification kit. The concentration of purified DNA was measured with a Nanodrop (BioTek Synergy HTX Reader, USA), and serial dilutions ranging from 10− 1 to 10− 6 were prepared for utilization as standards in quantitative real-time PCR (qPCR). The following formula was used to estimate the copy number of bacteria [19]:
Table 1.
The of primers for detection of bacteria used in qPCR
| Target taxon | Primer/sequences (5 − 3) | PCR product (bp) |
Reference |
|---|---|---|---|
| Firmicutes |
F: GGAGATGTGGTTTAATTCGAAGCA R: AGCTGACGACAACCATGCAC |
126 | [20] |
| Bacteroides spp. |
F: GAAGGTCCCCCACATTG R: CAATCGGAGTTCTTCGTG |
410 | [21] |
| E. coli |
F: TCCAGGTGTAGCGGTGAAAT R: TGAGTTTTAACCTTGCGGCC |
236 | [22] |
| Lactobacillus spp. |
F: GAGGCAGCAGTAGGGAATCTTC R: GGCCAGTTACTACCTCTATCCTTCTTC |
132 | [21] |
| Clostridium coccoides group |
F: AAATGACGGTACCTGACTAA R: CTTTGAGTTTCATTCTTGCGAA |
440 | [23] |
| Bifidobacterium spp. |
F: GCGTGCTTAACACATGCAAGTC R: CACCCGTTTCCAGGAGCTATT |
442 | [24] |
| Akkermansia muciniphila |
F: CAGCACGTGAAGGTGGGGAC R: CCTTGCGGTTGGCTTCAGAT |
327 | [25] |
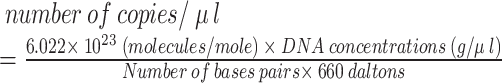 |
Quantitative real-time PCR
For qPCR, the quantitative method utilized 16 S rRNA primers (Table 1) and a 2X-PCR master mix from Syber (Yekta Tajhiz Azma, Iran). Each reaction contained 5 µL of 2X Syber green Master Mix, 2 µL of DNA, 1µL of forward primer, 1µL of reverse primer, and 1µL of RNase-free water. The amplification program was conducted on a Rotor Gen 6000 machine (Germany) with the following steps: initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s, extension at 72 °C for 45 s, and a final extension at 72 °C for 5 min.
Statistical analysis
The statistical analyses were conducted using SPSS 20. A t-test was utilized to compare the control group with the case group. In instances where the normality assumption was not fulfilled, the Mann-Whitney test was employed. Fisher’s exact and chi-squared tests evaluated the relationship between categorical variables based on frequency distribution, with p-values less than 0.05 indicating significant differences.
Results
The mean age of patients in groups 1 and 2 was 45.88 ± 18.51 and 41.30 ± 13.01 years, respectively, while the mean age of controls in groups 3 and 4 was 37.29 ± 9.62 and 40.96 ± 13.01 years. The patient groups included 16 (34.05%) women and 31 (65.95%) men, while the control groups comprised 31 (65.95%) women and 16 (34.05%) men.
The stool culture results for pathogenic bacteria were negative for all groups. The highest history of consuming dairy products containing probiotics use was observed in Group 1 with 22 (91.67%) subjects while the lowest use was reported in Group 3 with 16 (66.67%) subjects. The highest history of self-administered antibiotic use was observed in Group 2 with 13(56.52%) cases while the lowest was noted in Group 3 with 4 (16.67%) cases. The findings of this study demonstrated a significant relationship between Groups 2 and 4 with respect to the E. coli and Bifidobacterium spp. microbial population (P < 0.05). Additionally, a significant relationship was identified between the Lactobacillus spp., Bifidobacterium spp., and Bacteroides spp. microbial community in Groups 1 and 2 (P < 0.05).
Furthermore, a significant correlation was observed between Clostridium coccoides in Groups 1 and 3 (P < 0.05). The average copy numbers (mean ± SD) of bacterial in stool samples of the four studied groups is shown in Table 2.
Table 2.
The relationship between the copy numbers of bacteria with variables in all groups
| Variable | Group1; N = 24; frequency (%) | Group3; N = 24; frequency (%) |
P-Value a | Group2; N = 23; frequency (%) |
Group4; N = 23; frequency (%) |
P-Value b | P-Value c |
|---|---|---|---|---|---|---|---|
| History of consuming dairy products containing probiotics | |||||||
| No | 2 (8.33) | 8 (33.33) | 0.036* | 4 (17.39) | 5 (21.74) | 0.500* | 0.312* |
| Yes | 22 (91.67) | 16 (66.67) | 19 (82.61) | 18 (78.26) | |||
| Consumption of local dairy products | |||||||
| No | 1 (4.17) | 2 (8.33) | 1.00* | 1 (4.35) | 3 (13.04) | 0.608* | 1.00* |
| Yes | 23 (95.83) | 22 (91.67) | 22 (95.65) | 20 (86.96) | |||
| History of self-medication with antibiotics | |||||||
| No | 15 (62.50) | 20 (83.33) | 0.193* | 10 (43.48) | 16 (69.57) | 0.074** | 0.191** |
| Yes | 9 (37.50) | 4 (16.67) | 13 (56.52) | 7 (30.43) | |||
|
E.coli; Median [IQR(Q1,Q3)] |
4.26 × 109 [1.95 × 1010 ( 1.51 × 109, 2.10 × 1010)] |
2.90 × 109 [1.19 × 1010 ( 1.37 × 109, 1.33 × 1010 ] |
0.354‡ |
3.02 × 109 [4.99 × 109 ( 2.40 × 109, 7.40 × 109)] |
3 × 1010 [4.88 × 1010( 2.08 × 1010, 6.97 × 1010)] |
< 0.001‡ | 0.395‡ |
|
Akkermansia muciniphila; Median [IQR(Q1,Q3)] |
6.54 × 108 [3.27 × 109 ( 2.92 × 108, 3.560 × 109)] |
3.39 × 109 [2.84 × 1011( 5.03 × 108, 2.85 × 1011)] |
0.070‡ |
3.15 × 109 [6.66 × 109 ( 5.59 × 108, 7.02 × 109)] |
9.87 × 108 [1.99 × 1010( 6.47 × 108, 2.05 × 1010)] |
0.489‡ | 0.360‡ |
| Lactobacillus spp, Median [IQR(Q1,Q3)] |
2.79 × 1010 [1.11 × 1011 ( 5.62 × 108, 1.11 × 1011)] |
2.23 × 1010 [2.86 × 1011( 3.79 × 108, 2.87 × 1011)] |
0.483‡ |
1.88 × 1011 [5 × 1011 ( 5.21 × 1010, 5.53 × 1011)] |
1.66 × 1011 ]4.06 × 1011( 4.35 × 1010, 4.49 × 1011)] |
0.701‡ | < 0.001‡ |
| Bacteroides spp; Median [IQR(Q1,Q3)] |
1.69 × 109 [2.27 × 1010 ( 1.93 × 108, 2.29 × 1010)] |
3.69 × 109 [4.79 × 1010( 6.81 × 108, 4.86 × 1010)] |
0.409‡ |
3.80 × 1011 [1.75 × 1012 ( 9.18 × 109, 1.76 × 1012)] |
7.08 × 109 [2.86 × 1012( 3.74 × 108, 2.86 × 1012)] |
0.531‡ | 0.007‡ |
| Bifidobacterium spp; Median [IQR(Q1,Q3)] |
3.79 × 1010 [6.89 × 1011( 5.72 × 109, 6.89 × 10110)] |
1.82 × 1011 [1.97 × 1011( 5.13 × 1010, 2.48 × 1011)] |
0.224‡ |
3.21 × 1012 [3.76 × 1012 ( 5.33 × 1010, 3.81 × 1012)] |
5.08 × 109 [4.73 × 1010( 2.35 × 109, 4.97 × 1010)] |
< 0.001‡ | 0.008‡ |
|
Clostridium coccoides; Median [IQR(Q1,Q3)] |
15.97 × 1010 [2.45 × 1011 ( 8.10 × 109, 2.53 × 1011)] |
5.92 × 1011 [1.75 × 1012( 1.77 × 1011, 1.93 × 1012)] |
0.003‡ |
2.89 × 1011 [2.43 × 1012 ( 9.75 × 109, 2.44 × 1012)] |
1.85 × 1011 [2.91 × 1011( 1.52 × 1010, 3.06 × 1011)] |
0.362‡ | 0.115‡ |
|
Firmicutes; Median [IQR(Q1,Q3)] |
1.22 × 1011 [2.43 × 1011 ( 3.89 × 1010, 2.82 × 1011)] |
2.54 × 1011 [4.81 × 1011( 5.28 × 1010, 5.34 × 1011)] |
0.433‡ |
1.56 × 1011 [5.73 × 1011( 3.72 × 1010, 6.10 × 1011)] |
1.87 × 1011 [1 × 1012( 4.85 × 1010,1 × 1012)] |
0.328‡ | 0.349‡ |
†: From t-test
*: From Fisher’s exact test
‡: From Mann–Whitney test
**: From Pearson’s chi-squared
a: P-value from the comparison of Group 1 and Group 3
b: P-value from the comparison of Group 2 and of Group 4
c: P-value from the comparison of Group 1 and Group 2
IQR: Interquartile range
The investigation into the correlation between the classification and extent of ulcerative colitis (which encompasses four distinct types: distal colitis, extensive colitis, left colitis, and pancolitis) and the composition of the gut microbiome revealed that the Firmicutes microbial community exhibits a substantial association with distal colitis. Conversely, the Lactobacillus spp. microbial community demonstrated a significant association with two types (extensive colitis and pancolitis), of involvement while the E. coli microbial community exhibited a significant association with all four types of involvement. Figure 1 illustrates the relationship between the gut microbial populations and the four ulcerative colitis involvement types.
Fig. 1.
Graph of gut microbial populations by types of ulcerative colitis involvement. D: distal colitis; P: pancolitis; E: extensive colitis; L: left colitis; *: P-value < 0.05;**: P-value < 0.001;***: P-value < 0.0001; ns: no significant
Furthermore, the findings of this study demonstrated a substantial correlation between self-administered antibiotic utilization and Clostridium coccoides bacteria (P.value: 0.04). Conversely, no substantial correlation was observed between BMI and probiotic consumption with the gut microbiome. Table 3 presents the findings related to the association between BMI, history of self-administered antibiotic use, and history of consuming dairy products containing probiotics with the gut microbiome.
Table 3.
The relationship between the copy number of bacterial with BMI, history of self-administered antibiotic use and history of consuming dairy products containing probiotics
| Variable | BMI | History of consuming dairy products containing probiotics | History of self-medication with antibiotics | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pairwise corrolatons | P. value* |
No | Yes | P. value‡ |
No | Yes | P. value‡ |
|||
| copy number; Median [IQR(Q1,Q3)] | copy number; Median [IQR(Q1,Q3)] | |||||||||
| E.coli | 0.096 | 0.355 |
3 × 1010 [6.66 × 1010 ( 4.86 × 109, 7.15 × 1010) ] |
4.80 × 109 [3 × 1010 ( 1.99 × 109, 3.32 × 1010) ] |
0.241 |
9.94 × 109 [3.69 × 1010 ( 1.87 × 109, 3.88 × 1010) ] |
4.24 × 109 [3.18 × 1010 ( 2.46 × 109, 3.46 × 1010) ] |
0.683 | ||
| Akkermansia muciniphila | 0.102 | 0.327 |
3.63 × 109 [1.59 × 1010 ( 2.63 × 109, 1.85 × 1010) ] |
9.62 × 108 [2.62 × 1010 ( 3.84 × 108, 2.65 × 1010) ] |
0.449 |
1.06 × 109 [1.78 × 1010 ( 4.10 × 108, 1.82 × 1010) ] |
9.87 × 108 [5 × 1010 ( 3.95 × 108, 5 × 1010) ] |
0.545 | ||
| Lactobacillus spp. | 0.062 | 0.556 |
1.14 × 1011 [2.59 × 1012 ( 6.37 × 109, 2.66 × 1011) ] |
4.88 × 1010 [2.72 × 1011 ( 5.71 × 109, 2.77 × 1011) ] |
0.344 |
4.75 × 1010 [2.64 × 1011 ( 2.74 × 109, 2.67 × 1011) ] |
7.27 × 1010 [3.43 × 1011 ( 1.52 × 1010, 3.59 × 1011) ] |
0.392 | ||
| Bacteroides spp. | 0.006 | 0.953 |
1 × 1011 [1.72 × 1012 ( 3.12 × 109, 1.73 × 1012) ] |
6.30 × 109 [1.86 × 1011 ( 2.01 × 108, 1.86 × 1011) ] |
0.249 |
4.52 × 109 [1.63 × 1011 ( 2.45 × 108, 1.63 × 1011) ] |
1.67 × 1010 [3.57 × 1011 ( 1.67 × 108, 3.57 × 1011) ] |
0.877 | ||
| Bifidobacterium spp. | 0.009 | 0.935 |
1.28 × 1011 [2.85 × 1012 ( 3.64 × 109, 2.85 × 1012) ] |
5.67 × 1010 [6.64 × 1011 ( 6.14 × 109, 6.70 × 1011) ] |
0.133 |
9.07 × 1010 [5.38 × 1011 ( 5.61 × 109, 5.43 × 1011) ] |
5 × 1010 [8.62 × 1011 ( 7.73 × 109, 8.70 × 1011) ] |
0.940 | ||
| Clostridium coccoides group | 0.066 | 0.525 |
3.12 × 1011 [8.70 × 1011 ( 2.31 × 1011, 1.10 × 1012)] |
1.85 × 1011 [1.06 × 1012 ( 1.66 × 1010, 1.08 × 1012) ] |
0.212 |
2.94 × 1011 [1.11 × 1012 ( 3.28 × 1010, 1.41 × 1012) ] |
6.61 × 1010 [3.48 × 1011 ( 9.75 × 109, 3.58 × 1011) ] |
0.047 | ||
| Firmicutes | 0.041 | 0.692 |
5.34 × 1011 [8.33 × 1011 ( 1.39 × 1011, 9.73 × 1011)] |
1.26 × 1011 [5.43 × 1011 ( 4.10 × 1010, 5.84 × 1011) ] |
0.294 |
1.46 × 1011 [6.20 × 1011 ( 4.79 × 1010, 6.68 × 1011) ] |
1 × 1011 [4.22 × 1011 ( 3.94 × 1010, 4.62 × 1011) ] |
0.524 | ||
*: From Pearson correlation coefficient
‡: From Mann–Whitney test
IQR: Interquartile range
The analysis of gut microbiome data in conjunction with C-reactive protein (CRP) and the geographical location of subjects revealed an absence of a statistically significant correlation between the two variables (Table 4).
Table 4.
The relationship between CRP, residence location of individuals and the copy number of bacterial
| Variable | CRP | Residence location | ||||||
|---|---|---|---|---|---|---|---|---|
| Positive | Negative | P-value‡ | Urban | Rural | P-value‡ | |||
| copy number; Median [IQR(Q1,Q3)] | copy number; Median [IQR(Q1,Q3)] | |||||||
| E.coli |
3.50 × 109 [3.74 × 1010 ( 1.34 × 109, 3.88 × 1010) ] |
9.21 × 109 [3.17 × 1010 ( 2.61 × 109, 3.43 × 1010) ] |
0.204 |
4.82 × 109 [3.57 × 1010 ( 1.77 × 109, 3.75 × 1010) ] |
1.18 × 1010 [3.32 × 1010 ( 2.73 × 109, 3.60 × 1010) ] |
0.483 | ||
| Akkermansia muciniphila |
1.24 × 109 [4.81 × 1010 ( 4.08 × 108, 4.85 × 1010) ] |
9.87 × 108 [1.78 × 1010 ( 4.10 × 108, 1.82 × 1010) ] |
0.834 |
1.25 × 109 [4.76 × 1010 ( 3.99 × 108, 4.80 × 1010) ] |
1.02 × 109 [5.39 × 109 ( 4.46 × 108, 5.84 × 109) ] |
0.822 | ||
| Lactobacillus spp. |
4.69 × 1010 [1.98 × 1011 ( 2.19 × 109, 2 × 1011) ] |
1 × 1011 [3.18 × 1011 ( 8.34 × 109, 3.27 × 1011) ] |
0.381 |
5.75 × 1010 [3.42 × 1011 ( 3.57 × 109, 3.46 × 1011) ] |
5.34 × 1010 [1.86 × 1011 ( 1.42 × 1010, 2 × 1011) ] |
0.502 | ||
| Bacteroides spp. |
3.12 × 109 [8.56 × 1010 ( 2.22 × 108, 8.58 × 1010) ] |
1.67 × 1010 [3.66 × 1011 ( 2.21 × 108, 3.66 × 1011) ] |
0.445 |
6.86 × 109 [3.34 × 1011 ( 2.16 × 108, 3.34 × 1011) ] |
7.05 × 109 [8.06 × 1010 ( 2.66 × 108, 8.09 × 1010) ] |
0.372 | ||
| Bifidobacterium spp. |
5.02 × 1010 [2.01 × 1011 ( 3.34 × 109, 2.04 × 1011) ] |
1.09 × 1011 [9.11 × 1011 ( 9.14 × 109, 9.20 × 1011) ] |
0.118 |
9.43 × 1010 [3.13 × 1011 ( 5.63 × 109, 3.19 × 1011) ] |
5.76 × 1010 [9.42 × 1011 ( 9.05 × 109, 9.51 × 1011) ] |
0.652 | ||
| Clostridium coccoides group |
2.31 × 1011 [1.11 × 1012 ( 1.92 × 1010, 1.13 × 1012) ] |
2.07 × 1011 [8.87 × 1011 ( 1.78 × 1010, 9.05 × 1011) ]] |
0.701 |
2.10 × 1011 [1.15 × 1012 ( 1.63 × 1010, 1.16 × 1012) ] |
1.71 × 1011 [6.78 × 1011 ( 2.30 × 1010, 7.01 × 1011) ] |
0.856 | ||
| Firmicutes |
1.39 × 1011 [6.18 × 1011 ( 3.72 × 1010, 6.56 × 1011) ] |
1.41 × 1011 [5.68 × 1011 ( 4.28 × 1010, 6.10 × 1011) ] |
0.946 |
1.40 × 1011 [6.26 × 1011 ( 3.50 × 1010, 6.61 × 1011) ] |
1.38 × 1011 [4.66 × 1011 ( 4.96 × 1010, 5.16 × 1011) ] |
0.941 | ||
‡: From Mann–Whitney test
IQR: Interquartile range
Discussion
In this study, the role of the gut microbiome was examined and analyzed in four groups, two of which were considered patients (groups 1 and 2) and the other two as healthy individuals (groups 3 and 4). Unlike many other studies, the healthy individuals were selected from first-degree relatives of the patients. The reason for selecting these individuals was that they lived in the same place and community and had used the same dietary system throughout their lives. Also, the reason for choosing these microbiomes in our study is that in previous studies, they have been investigated as key bacteria in inflammatory diseases of the digestive tract due to their role and close relationship with intestinal microbiota dysbiosis and disease progression. The difference is that in our study, they were simultaneously investigated in 4 different groups so that targeted modulation of these bacteria could improve the therapeutic potential in this disease, because the reduction of these bacteria disrupts immune homeostasis, the barrier function of intestinal epithelial cells, and the production of metabolites, and causes chronic inflammation. Our study results showed that Lactobacillus ssp., Bifidobacterium ssp., and Bacteroides ssp.. could play a very important role in the treatment process of ulcerative colitis. On the other hand, comparing the microbial community between groups 1 and 3 (individuals who had just been diagnosed with ulcerative colitis and their first-degree relatives) showed that the Clostridium coccoides microbial community was different. This is while in Ostadmohammadi’s 2021 study, the role of the Firmicutes and Enterobacteriaceae microbial community was more prominent [26], and this difference in the microbial community could be due to the type of healthy individuals selected, as the healthy individuals in our study were first-degree relatives of the patients. In another study conducted by Kabeerdoss J in 2015, the frequency of Bacteroides ssp. and Lactobacillus ssp.. was higher in UC patients compared to the control group, which was healthy individuals [27]. However, in our study, the frequency of Bacteroides ssp. and Lactobacillus ssp. was different between people with refractory ulcerative colitis and people who had just been diagnosed with ulcerative colitis, not between people with ulcerative colitis and healthy individuals. This was probably due to the difference in the participants in the study and also the type of sample tested. In our study, stool samples were examined, but in their study, biopsy samples were analyzed. Luma Al-Bayati’s study in 2023 showed that there is a significant relationship between the microbial community of Faecalibacterium prausnitzii, Provetella and Peptostreptococcus with ulcerative colitis [28]. The results of their study were contrary to our study, and this may be due to the selection of the type of gut microbiome community for analysis and the type of groups participating in our study. The results of our study showed that the gut microbial community in the two groups of patients (groups 1 and 2) also differs based on the type of ulcerative colitis involvement, so the type of patient involvement can also affect the decrease or increase in the gut microbiome population. A notable point in our study was that there was no significant association between place of residence and history probiotic dairy products with the gut microbiome community, while various studies have shown that probiotic consumption can affect the gut microbial population [29–31]. This difference in results could possibly be due to the fact that we analyzed history of dairy products containing probiotics in the study, not pharmaceutical probiotics consumption during the study. Studies have shown that multi-strain probiotics can have very variable effects on diseases, because these compounds are composed of several types of beneficial bacteria, each of which can have different effects on the immune system and gut microbiome, and these variable effects are due to reasons such as differences in the mechanism of action, interaction with the gut microbiome, effect on the immune system and differences in individual resistance. Therefore, the effectiveness of multi-strain probiotics can be highly variable, with very favorable results in some cases and less effective in others [32–33]. Analysis of the history of spontaneous antibiotic use among the participants in our study showed that the spontaneous use of antibiotics only affected the Clostridium coccoide population and did not affect other microbiomes. Although many studies have shown that the use of antibiotics has a significant impact on the intestinal microbiome [34–35], it should be noted that this impact can be based on the type of antibiotic and its dose. On the other hand, sampling of the people in our study was not done immediately after taking antibiotics and their samples were taken 6 months before taking antibiotics. In general, what the results of this study show is that a number of intestinal microbiomes can play an important role in the treatment process of ulcerative colitis and they can be used as a line of treatment in clinical trials for this disease. The limitations of this study are manifold, including the lack of investigation of the role of bacteriophages, the utilization of a specific probiotic treatment protocol for patients, the utilization of non-family groups, the investigation of other microbiomes, and the investigation of metabolic factors. Addressing these limitations in subsequent studies such as the concurrent examination of the gut microbiome and phages utilizing metagenomics technique given the established role of phages in regulating the gut microbial community and clinical trial studies such as the administration of probiotics and using fecal microbiota transplant (FMT) in patients, also studying patients’ metabolites alongside the gut microbiome will contribute to the advancement of treatment methodologies for this condition.
Conclusion
The findings of this study demonstrated that several intestinal microbiomes have a substantial impact on the management of ulcerative colitis. The results of this study suggest that by comparing the gut microbiome of treatment-resistant and individuals newly diagnosed with ulcerative coliti, we can gain a better understanding of microbiome differences that may influence treatment outcomes. The results of this study may also lead to the identification of new therapeutic strategies that are based on regulating the gut microbiome. These strategies could include the use of fecal microbiota transplantation (FMT), probiotics, prebiotics, or specific bacteria-based therapies.
Acknowledgements
The authors express their gratitude to the Vice-Chancellor of Research and Technology of Hamadan University of Medical Sciences, Hamadan, Iran, and the National Institute for Medical Research Development (NIMAD), Tehran, Iran for advocating this research.
Abbreviations
- qPCR
Quantitative Real-Time PCR
- PCR
Polymerase chain reaction
- UC
Ulcerative colitis
- IBD
Inflammatory bowel disease
- SCFAs
Short-chain fatty acids
- FMT
Fecal microbiota transplantation
- CRP
C-reactive protein
Author contributions
MYA and AA designed and supervised the study. HK, FS and LS were responsible for data collection and doing experiments. MYA, AA and MKM performed data interpretation. BN and AM performed clinical examination. MYA and RY analyzing the statistical results of the study. MYA and RY writing and editing the original draft. All authors approved the final version of the manuscript.
Funding
Funding provided by the Vice-Chancellor of Research and Technology of Hamadan University of Medical Sciences, Hamadan, Iran (grant numbers: 140110279103 and 140110279102) and the National Institute for Medical Research Development (NIMAD), Tehran, Iran (Grant number: 4003032).
Data availability
All data generated or analyzed during this study were included in this article but the raw data are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Hamadan University of Medical Sciences, Hamadan, Iran (Ethic approval codes: IR.UMSHA.REC.1401. 787 and IR.UMSHA.REC.1401. 595). All research was performed following relevant guidelines according to the Declaration of Helsinki (https://www.wma.net/policies-post/wma-declaration-of-helsinki/). Written informed consent was obtained from all participants.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mohammad Yousef Alikhani, Email: alikhani43@yahoo.com, Email: alikhani@umsha.ac.ir.
Rasoul Yousefimashouf, Email: yousefimash@yahoo.com.
References
- 1.Kobayashi T, Siegmund B, Le Berre C, Wei SC, Ferrante M, Shen B, Bernstein CN, Danese S, Peyrin-Biroulet L, Hibi T. Ulcerative colitis (primer). Nat Reviews Disease Primers. 2020;6(1):74. [DOI] [PubMed] [Google Scholar]
- 2.Gros B, Kaplan GG. Ulcerative colitis in adults: A review. JAMA. 2023;330(10):951–65. [DOI] [PubMed] [Google Scholar]
- 3.Du L, Ha C. Epidemiology and pathogenesis of ulcerative colitis. Gastroenterol Clin. 2020;49(4):643–54. [DOI] [PubMed] [Google Scholar]
- 4.Segal JP, LeBlanc JF, Hart AL. Ulcerative colitis: an update. Clin Med. 2021;21(2):135–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo XY, Liu XJ, Hao JY. Gut microbiota in ulcerative colitis: insights on pathogenesis and treatment. J Dig Dis. 2020;21(3):147–59. [DOI] [PubMed] [Google Scholar]
- 6.Qiu P, Ishimoto T, Fu L, Zhang J, Zhang Z, Liu Y. The gut microbiota in inflammatory bowel disease. Front Cell Infect Microbiol. 2022;12:733992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dave M, Higgins PD, Middha S, Rioux KP. The human gut microbiome: current knowledge, challenges, and future directions. Translational Res. 2012;160(4):246–57. [DOI] [PubMed] [Google Scholar]
- 8.Seo D-O, Holtzman DM. Gut microbiota: from the forgotten organ to a potential key player in the pathology of Alzheimer’s disease. Journals Gerontology: Ser A. 2020;75(7):1232–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quaglio AE, Grillo TG, De Oliveira EC, Di Stasi LC, Sassaki LY. Gut microbiota, inflammatory bowel disease and colorectal cancer. World J Gastroenterol. 2022;28(30):4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen Z-H, Zhu C-X, Quan Y-S, Yang Z-Y, Wu S, Luo W-W, et al. Relationship between intestinal microbiota and ulcerative colitis: mechanisms and clinical application of probiotics and fecal microbiota transplantation. World J Gastroenterol. 2018;24(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. 2021;19(1):55–71. [DOI] [PubMed] [Google Scholar]
- 12.Haneishi Y, Furuya Y, Hasegawa M, Picarelli A, Rossi M, Miyamoto J. Inflammatory bowel diseases and gut microbiota. Int J Mol Sci. 2023;24(4):3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mills RH, Dulai PS, Vázquez-Baeza Y, Sauceda C, Daniel N, Gerner RR, Batachari LE, Malfavon M, Zhu Q, Weldon K, Humphrey G. Multi-omics analyses of the ulcerative colitis gut Microbiome link Bacteroides vulgatus proteases with disease severity. Nat Microbiol. 2022;7(2):262–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63(8):1275–83. [DOI] [PubMed] [Google Scholar]
- 15.Duncan SH, Hold GL, Barcenilla A, Stewart CS, Flint HJ. Roseburia intestinalis Sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int J Syst Evol MicroBiol. 2002;52(5):1615–20. [DOI] [PubMed] [Google Scholar]
- 16.Silva YP, Bernardi A, Frozza RL. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front Endocrinol. 2020;11:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu L, Xu L-Z, Zhao S, Shen Z-F, Shen H, Zhan L-B. Protective effect of Baicalin on the regulation of Treg/Th17 balance, gut microbiota and short-chain fatty acids in rats with ulcerative colitis. Appl Microbiol Biotechnol. 2020;104(12):5449–60. [DOI] [PubMed] [Google Scholar]
- 18.Han T, Hu X, Li K, Zhang D, Zhang Y, Li J. Bifidobacterium infantis maintains genome stability in ulcerative colitis via regulating anaphase-promoting complex subunit 7. Front Microbiol. 2021;12:761113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morello JA. Bailey and Scott’s diagnostic microbiology. JAMA. 1987;257(8):1112–3. [Google Scholar]
- 20.Dhanasekaran S, Doherty TM, Kenneth J, TB Trials Study Group. Comparison of different standards for real-time PCR-based absolute quantification. J Immunol Methods. 2010;354(1–2):34–9. [DOI] [PubMed] [Google Scholar]
- 21.García-López M, Meier-Kolthoff JP, Tindall BJ, Gronow S, Woyke T, Kyrpides NC, Hahnke RL, Göker M. Analysis of 1,000 type-strain genomes improves taxonomic classification of Bacteroidetes. Front Microbiol. 2019;10:2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marathe N, Shetty S, Lanjekar V, Ranade D, Shouche Y. Changes in human gut flora with age: an Indian Familial study. BMC Microbiol. 2012;12:1–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuki T, et al. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl Environ Microbiol. 2002;68:5445–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yadav H, Jain S, Nagpal R, Marotta F. Increased fecal viral content associated with obesity in mice. World J Diabetes. 2016;7(15):316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl Environ Microbiol. 2007;73:7767–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostadmohammadi S, Azimirad M, Houri H, Naseri K, Javanmard E, Mirjalali H, et al. Characterization of the gut microbiota in patients with primary sclerosing cholangitis compared to inflammatory bowel disease and healthy controls. Mol Biol Rep. 2021;48(7):5519–29. [DOI] [PubMed] [Google Scholar]
- 27.Kabeerdoss J, Jayakanthan P, Pugazhendhi S, Ramakrishna BS. Alterations of mucosal microbiota in the colon of patients with inflammatory bowel disease revealed by real time polymerase chain reaction amplification of 16S ribosomal ribonucleic acid. Indian J Med Res. 2015;142(1):23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al-Bayati L, Fasaei BN, Merat S, Bahonar A, Ghoddusi A. Quantitative analysis of the three gut microbiota in UC and non-UC patients using real-time PCR. Microb Pathog. 2023;181:106198. [DOI] [PubMed] [Google Scholar]
- 29.Hajela N, Ramakrishna BS, Nair GB, Abraham P, Gopalan S, Ganguly NK. Gut microbiome, gut function, and probiotics: implications for health. Indian J Gastroenterol. 2015;34(2):93–107. [DOI] [PubMed] [Google Scholar]
- 30.Bodke H, Jogdand S. Role of probiotics in human health. Cureus. 2022;14(11). [DOI] [PMC free article] [PubMed]
- 31.Wierzbicka A, Mańkowska-Wierzbicka D, Mardas M, Stelmach-Mardas M. Role of probiotics in modulating human gut microbiota populations and activities in patients with colorectal cancer—a systematic review of clinical trials. Nutrients. 2021;13(4):1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McFarland LV. Efficacy of single-strain probiotics versus multi-strain mixtures: systematic review of strain and disease specificity. Dig Dis Sci. 2021;66:694–704. [DOI] [PubMed] [Google Scholar]
- 33.Dias JA, Alves LL, Barros FA, Cordeiro CA, Meneses JO, Santos TB, Santos CC, Paixão PE, Nogueira Filho RM, Martins ML, Pereira SA. Comparative effects of using a single strain probiotic and multi-strain probiotic on the productive performance and disease resistance in Oreochromis niloticus. Aquaculture. 2022;550:737855. [Google Scholar]
- 34.Konstantinidis T, Tsigalou C, Karvelas A, Stavropoulou E, Voidarou C, Bezirtzoglou E. Effects of antibiotics upon the gut microbiome: a review of the literature. Biomedicines. 2020;8(11):502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hao WZ, Li XJ, Zhang PW, Chen JX. A review of antibiotics, depression, and the gut Microbiome. Psychiatry Res. 2020;284:112691. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study were included in this article but the raw data are available from the corresponding author on reasonable request.



