Abstract
In breast cancer related to the BRCA1 mutation, luminal progenitor cells are believed to be the cells of origin, yet how these cells transform into invasive cancer cells remain poorly understood. Here, we combine single-cell epigenomic and transcriptomic data to reconstitute sequences of events in luminal cells that lead to tumorigenesis. Upon deletion of Trp53 and Brca1, we find that luminal progenitors display an extensive epigenomic disorder associated with a loss of cell identity. These cells then progress to tumor formation through a partial epithelial-to-mesenchymal transition, orchestrated by Snail and the timely activation of immunosuppressive and FGF signaling with their microenvironment. In human samples, pre-tumoral changes can be detected in early stage, basal-like tumors, which rarely recur, as well as in normal-like mammary glands of BRCA1 mutation carriers who have had cancer. Our study fills critical gaps in our understanding of BRCA1-driven tumorigenesis, opening perspectives for the early monitoring of individuals with high cancer risk.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12943-025-02331-9.
Introduction
Mutations in oncogenes or tumor suppressors can accumulate over time in healthy tissues [1–3]. The potential for tumor initiation must therefore be determined not only by accumulated genetic aberrations but also by the capacity of each cell and tissue to deal with them. Some tissues and specific cell lineages are more susceptible to a rupture of tissue homeostasis. For instance, women with BRCA1 deficiency mainly develop ovarian and breast cancers [4], and in breast cancers, BRCA1 mutations specifically lead to the transformation of luminal progenitor cells [5, 6]. A large fraction of breast tumors with BRCA1 mutation (BRCA1 m) carrier cells harbor basal-like characteristics, suggesting that luminal identity is disrupted during tumor formation [4]. However, reconstituting the cell states that are acquired by a luminal progenitor cell up to tumor initiation still needs in-depth mapping.
Our ability to study tumor initiation and identify pre-tumoral cell states based on human observations is limited. Studying tumorigenesis solely based on samples from individuals who already have cancer is complex, as tumor samples are entangled stacks of molecular alterations that occur over time. An alternative is to examine normal tissues from individuals predisposed to cancer. For instance, mammary gland tissue removed during prophylactic surgeries in individuals carrying BRCA1 or BRCA2 germline mutations show early anomalies in"normal-like"tissue, based on cytometry or single-cell transcriptomics [7–10]. Dysregulation of the microenvironment and epithelial tissue homeostasis disruption appear to be steppingstones toward tumorigenesis. However, BRCA1/2-deficient cells within prophylactic surgeries might be far from initiating tumors. The timing of tumor emergence in these mammary glands remains uncertain and may span from months to decades, limiting our ability to study tumor initiation.
Genetically engineered mouse models offer a valuable alternative for studying breast tumorigenesis [5, 11]. Mouse models allow tumor suppressor genes to be deleted, or oncogenes to be activated, in multiple cells, increasing the likelihood of detecting cells in transitioning from a normal-like state to a breast cancer phenotype. These models have been instrumental in identifying the cell-of-origin of basal-like breast tumors [12]. Mice with Brca1 and Trp53 deficiencies in luminal progenitors recapitulate the formation of human basal-like breast cancers [5]. These mice display aberrant alveolar differentiation of luminal progenitors [13], suggesting that mis-control of cell identity could be an element of tumor initiation.
Here, we study the non-genetic determinants of breast tumor initiation by combining mice and human studies. We first leverage single-cell epigenomics and transcriptomics to map the non-genetic evolution of epithelial cells towards transformation and to identify critical cell states and markers of tumorigenesis. We identify a recurrent pre-tumoral cell state, characterized by partial EMT and cell cycle defects, and specific activation of FGF and immuno-suppressive signaling. We further show that we can detect features of the pre-tumoral state in human tissues of BRCA1 mutation carriers.
Results
Epigenomic integrity and cell identity is disrupted in Brca1/Trp53-deficient mammary glands
To map cell state transitions in the mammary gland prior to tumor formation, we performed a time-series analysis on virgin Blg-Cre+/– Trp53fl/fl Brca1fl/fl (CreP) female mice, combining transcriptomics via single-cell RNA sequencing (scRNA-seq) with single-cell epigenomic profiling (snCUT&Tag)(Fig. 1a). CreP mice developed mammary tumors at a median age of 5.4 months (Extended Data Supplementary Fig. 1a). Early abnormalities were observed starting at 3 months, characterized by abnormal gland structures and carcinoma in situ (CIS), with irregular nuclei and disrupted duct organization (Fig. 1a). By 5 months, CreP mice displayed multiple CIS, which were not detectable before dissection. To enhance our chances of detecting tumor initiation events, we especially focused on tumor-free CreP mice from a litter in which at least one other mouse already had a tumor (n = 3 animals), as we reasoned that these mice would be on the verge of developing tumors. We included mammary gland tissue from control mice that did not express the Cre-recombinase (Blg-Cre–/– Trp53fl/fl Brca1fl/fl; CreN), as well as tumors from CreP mice (Fig. 1a and Supplementary Table 1). To increase the likelihood of identifying rare phenotypic states, we enriched part of the collected samples for the epithelial fraction (Supplementary Table 1; see Methods).
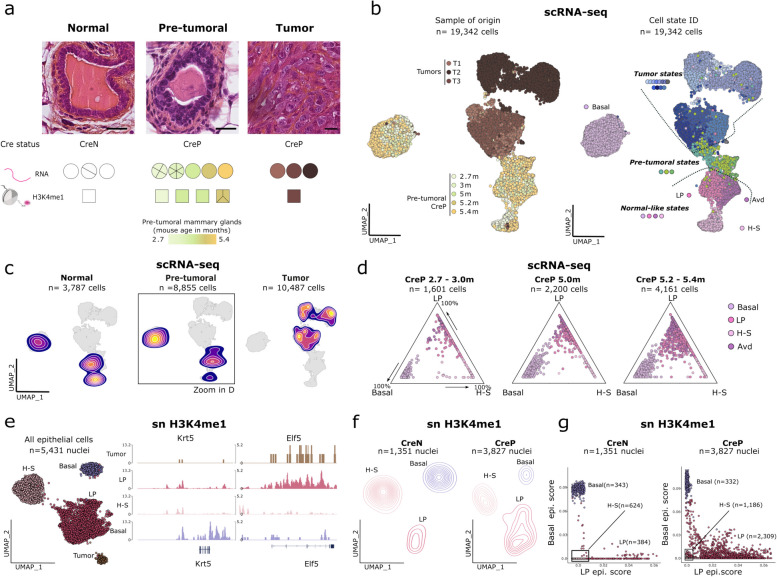
Fig. 1.
Transcriptomic and epigenomic profiling reveals non-genetic loss of cell identity in pre-tumoral Brca1/Trp53 deficient mammary glands. a Top: representative immuno-histochemistry for normal, pre-tumoral and tumor tissues, scale bars correspond to 20 µm. Bottom: Cre-recombinase–positive (CreP) or –negative (CreN) state in normal, pre-tumoral and tumor bearing mice, showing the number of samples used for scRNA-seq and snH3K4me1 profiling. For each sample, the number of slices within the circle/square corresponds to the number of mice used. For tumor-free mammary glands from CreP mice, color codes represent the age of the mouse. b Left: UMAP representation of scRNA-seq datasets for CreP epithelial cells. Cells are colored according to the sample of origin. Right: UMAP representation of scRNA-seq datasets for CreP epithelial cells; cells are colored according to the cluster of origin. Clusters are classified into states according to the sample of origin of cells, whereby tumor states correspond to clusters originating from tumor samples only; pre-tumoral states, from pre-tumoral CreP mice; and normal-like states, from both CreN and CreP mice. c Density plots representing the distribution of single cells according to their sample of origin grouped by genotype and presence or not of a tumor in the mouse, as in (a). d Focus on epithelial cells in normal-like states in CreP mice. Cells in normal-like state were grouped into 3 categories depending on the age of their mouse-of-origin. Ternary plots represent cell populations along three axes representing basal, luminal progenitor (LP) and luminal hormone-sensing (H–S) cells. Signatures (extreme poles) were established based on the top marker genes of each cell population in CreN animals. e UMAP representation of single nuclei H3K4me1 profiles of epithelial cells from CreN and CreP mice with and without tumors. Nuclei are colored according to their cell type. Right: representative snapshots of pseudo-bulk snH3K4me1 profiles for each cell type for the Krt5 and Elf5 genes. f Density UMAP plot representing the distribution of CreN and pre-tumoral CreP cells, similar as in (e). g Scatterplot representing the epigenomic basal and luminal progenitor scores for CreN and pre-tumoral CreP cells. Basal and LP epigenomic signatures were defined based on CreN basal and LP epigenomic profiles
For the scRNA-seq dataset, we collected 43,084 cells from 20 mice, including 23,129 epithelial cells (19,342 from CreP mice, and 3,787 cells from CreN mice) (Fig. 1b and Extended Data Supplementary Fig. 1b). To identify distinct cell states, we first performed unsupervised graph-based clustering (Fig. 1b) and then annotated cell clusters based on the level of expression of physiological markers and their samples of origin (Extended Data Supplementary Fig. 1c,d and Supplementary Table 2), whereby cluster was named if > 50% of cells in it belonged to a single sample (e.g. the T1a cluster has > 50% cells from tumor T1; Extended Data Supplementary Fig. 1e). We then grouped clusters into three states according to their distribution within CreN, CreP and tumor-bearing mice (Fig. 1b,c): (i)"normal-like"states corresponding to clusters in tumor-free CreN and CreP mice, (ii) tumor states, found only in tumors, and (iii) pre-tumoral states, corresponding to clusters found in tumor-free CreP mice and tumors but not in control CreN mice. We identified a series of normal-like states corresponding to well-known mammary gland cell populations: basal cells (Krt5) and clusters of luminal cells (Krt8), including luminal hormone-sensing (H–S) cells (Prlr), luminal progenitor (LP) cells (Aldh1a3) and secretory alveolar-differentiated (Avd) cells (Csn2) (Extended Data Supplementary Fig. 1c,d). Of particular interest, secretory Avd cells were abnormally enriched in CreP virgin mice at all time points, consistent with previous reports of deregulated luminal progenitor differentiation in Brca1/Trp53-mediated tumorigenesis [13].
To distinguish the earliest non-genetic defects occurring in Brca1/Trp53 deficient mammary glands, we first studied normal-like states using both transcriptomics and epigenomic information. Building on previous work identifying LP differentiation defects [13], we quantified and monitored the cell lineage integrity of epithelial cells over time, based on our detailed mapping of tumorigenesis in CreP mice, from 2.7 to 5.4 months. We measured lineage integrity based on coordinates of cells within a ternary plot, whereby each pole represented a reference epithelial cell type—basal, LP or luminal H–S—based on marker genes from CreN mice. At three months, basal cells from CreP mice were segregated near each pole, while LP and H–S cells displayed a continuum between LP and H–S poles, reflecting physiological differentiation from LP to H–S lineage (Fig. 1d). In contrast, starting from 5 months, basal, LP and Avd CreP cells accumulated in the center of the plot, with a continuum of expression profiles from basal to LP reference cell states (Fig. 1d).
To elucidate the non-genetic determinants of this loss of cell lineage integrity, we analysed single-cell epigenomic features of CreP mammary glands using H3K4me1 profiling (Fig. 1a). H3K4me1 accumulates at primed and activated enhancers and promoters [14, 15], offering insights on cell state encoding beyond gene transcription. We adapted snCUT&Tag [16] to mammary glands, achieving a median coverage of 890 unique fragments per nucleus (n = 7,045 nuclei; Extended Data Supplementary Fig. 2a,b). We then in silico sorted cells based on their H3K4me1 enrichment at marker genes (Extended Data Supplementary Fig. 2c–d and Fig. 1e). Hierarchical clustering showed that tumor cells have an epigenome closely related to that of LP cells (Extended Data Supplementary Fig. 1 d), in line with LP cells being the cell of origin of these tumors. It also demonstrated that tumor cells "keep track" of their cell of origin through epigenomic features, even in a highly genomically-rearranged setting. Next, we compared epigenomic profiles of epithelial cells in CreP and CreN mice (Fig. 1f,g). CreP LP cells exhibited a broader range of epigenomic states than CreN LP cells, illustrated by a broader spread of cells over a 2D space (Fig. 1f). To determine whether this variability was associated with the loss of cell lineage integrity that we observed between LP and basal cells, we projected single-cell epigenomic profiles into a 2D plot, with poles representing the reference epigenomes of LP and basal cells obtained from CreN cells (Fig. 1g). Similar to gene expression patterns, we observed a continuum of epigenomic profiles from LP to H–S cells, testifying to a physiological differentiation route in CreN cells. Further, we observed highly disordered epigenomic landscapes for CreP LP cells, with 27% (75 th quantile) of cells occupying intermediary states between basal and LP epigenomes (Fig. 1g).
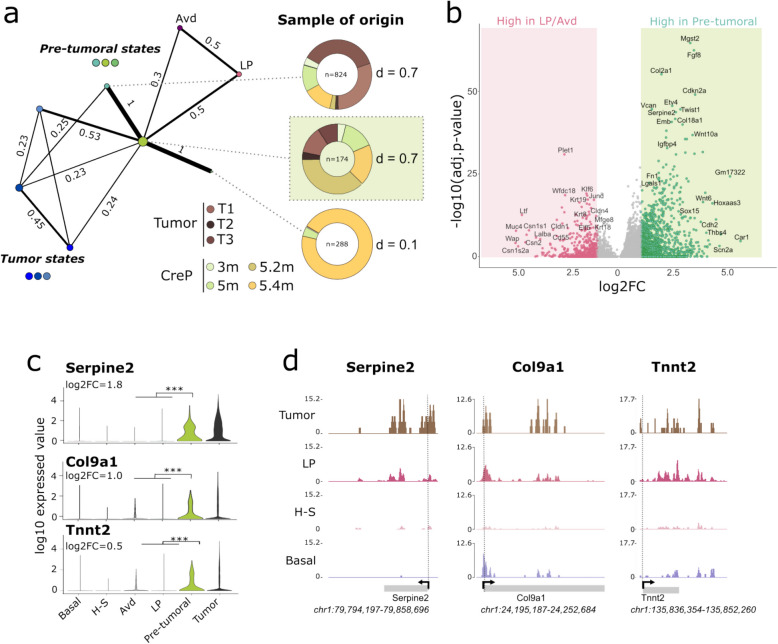
Fig. 2.
Identification of a primed pre-tumoral state in vivo. a Partition-based graph abstraction (PAGA) representation of scRNA-seq datasets, with cells from normal-like, pre-tumoral and tumor clusters. Nodes refer to clusters, and edge thickness is proportional to the transcriptional similarity between clusters. The sample of origin for the three pre-tumoral clusters is indicated, with the multi-sample pre-tumoral cluster (derived from multiple pre-tumoral mammary glands) highlighted with a light green background. b Volcano plot representing the log2 expression fold-change and log10 adjusted p-value comparing cells in pre-tumoral state and LP cells. c Violin representation of log10-normalized expression level of pre-tumoral marker genes Serpine2 and Tnnt2. *** corresponds to adjusted p-value < 10–3. d Snapshots of pseudobulk snH3 K4 me1 profiles for the Serpine2, Tnnt2 and Olfml3 genes in epithelial cells and tumor cells
Altogether, normal-like luminal progenitor cells deficient for Trp53 and Brca1 classified as normal-like cells based on their expression of physiological markers (Fig. 1b, Extended Data Supplementary Fig. 1 d), yet they displayed a drastic epigenomic disorder, potentially leading to the loss of cell lineage integrity observed here and by others [17].
Detection of a rare, epigenetically-primed pre-tumoral cell state
Next, we focused on the three pre-tumoral states we detected. Among these, one is sample specific, while two are multi-sample (Extended Data Supplementary Fig. 1e). Using the PAGA algorithm, we quantified cluster connectivity [18] and found that one of these pre-tumoral states served as a hub linking epithelial sub-clusters, with tumor cell states only accessible via this intermediate state (Fig. 2a). This cell state was found in multiple samples (Shannon index d = 0.70; Fig. 2a) and is composed primarily of cells from tumor-free CreP mice (75%), and to a lesser extent, from tumor samples (25%). We designed this cluster as "pre-tumoral" because tumor states were only reachable through this intermediate transcriptional state (Fig. 2a). Importantly, cells in the pre-tumoral state were overall very rare (0.9% of epithelial cells) yet detectable in 3-month-old CreP mice (0.4%); the fraction of these cells increased with age (1.8% at 5 months). In terms of cell identity, cells in a pre-tumoral state displayed a significant down-regulation of genes characteristic of the luminal compartments, as compared to LP and Avd cells (e.g. Krt8, Krt18, Krt19 and Csn2) (Fig. 2b, Extended Data Supplementary Fig. 1c and Supplementary Table 3). This result indicates that the loss of identity observed in some CreP LPs by 5 months becomes further exacerbated in the pre-tumoral state (Fig. 1d). Notably, while CreP LPs did not yet express pre-tumoral genes (Fig. 2c, e.g. Col9a1, Tnnt2 or Serpine2), we observed H3K4me1 enrichment at these loci within the LP compartment (Fig. 2d), suggesting an epigenetic priming of pre-tumoral genes in Brca1/Trp53-deficient LP cells.
To understand whether the pre-tumoral state originated from the expansion of isolated clones or was reached independently by multiple luminal cells, we evaluated the clonality of cells in the pre-tumoral state using inferred copy-number variation (CNV) profiles derived from scRNA-seq data, analyzed with inferCNV [19] (Extended Data Supplementary Fig. 3; see Methods). First, to understand when genomic alterations began accumulating in mammary glands of CreP mice, we quantified the percentage of genome with inferred CNVs in each cell as compared to reference basal cells. Both the proportion of cells with high CNV content and the average amount of CNV per cell increased with age (Extended Data Supplementary Fig. 3a). Notably, we detected cells with high CNV content as early as 3 months, prior to any observable tumor. This indicates that luminal cells can tolerate significant CNV accumulation without immediate tumor initiation. Luminal progenitors, in particular, exhibited the highest CNV burden, comparable to that observed in pre-tumoral and tumor cells (Extended Data Supplementary Supplementary Fig. 3b,c). Using consensus clustering to group inferred single-cell genomic profiles, we found that the LP compartment is multi-clonal, with a lack of stable partitioning into distinct clones and low pairwise correlation scores, while tumors appeared oligo-clonal, with only a few genetic sub-clones (Extended Data Supplementary Fig. 3 d,e). Notably, part of the pre-tumoral cell population (39%) was multi-clonal, suggesting that multiple luminal cells can switch to the pre-tumoral state. These findings suggest that non-genetic mechanisms can drive the transition of luminal progenitors to the pre-tumoral state.
Fig. 3.
Cell cycle defect and partial EMT in pre-tumoral state. a Barplot representation of the top hallmark pathways activated in pre-tumoral cells (in green) or LP/Avd (in gray); x-axis represents –log10 adjusted p-value. b Stack violin plot representation of expression of genes involved in EMT in epithelial clusters. ***p-value < 0.001, from Wilcoxon rank test comparing pre-tumoral cluster to LP/Avd clusters. c Pseudo-colored multiplex IHC staining for keratin 8 (red), keratin 5 (green), E-cadherin (magenta) and the mesenchymal markers N-cadherin (cyan) and vimentin (yellow). Scale bar, 50 µm. d Dot plot representation of the top 30 candidate TFs of the pre-tumoral expression program. TF ranks (x-axis) and scores (y-axis) were calculated using ChEA3. e Immunofluorescence staining for keratin 8 (red) and EMT-TF Snail (yellow) for CreN (control) or CreP mice and tumors. Scale bar, 50 µm. f Stack violin plot representation of the top markers of pre-tumoral cluster involved in the cell cycle regulation. ***p-value < 0.001, Wilcoxon rank test comparing pre-tumoral cluster to LP/Avd clusters. g Left: pseudo-colored multiplex IHC staining for identity markers keratin 5 (green) and keratin 8 (red), together with senescent marker p16 (cyan) and EMT markers E-cadherin (magenta) and vimentin (yellow). Scale bar, 50 µm. Right: Sunburst plot representation of the multiplex IHC staining for p16 + and p16– luminal cells from glands from 5-month-old CreP mice. Number of analyzed cells and mice are indicated
Altogether, thanks to the profiling of multiple animals nearing tumor initiation, we identified a continuum of cell states shared across several individuals. Luminal cells transition from an aberrant luminal state with compromised epigenetic and lineage integrity to a rare pre-tumoral state, demarked by a more pronounced loss of luminal identity, among other features. This pre-tumoral state serves as an intermediate stage before progressing to fully developed tumor cell states.
Cells in a pre-tumoral state display signs of cell cycle defects and undergo partial EMT
We then explored biological functions of cells in a pre-tumoral state. We first studied which biological pathways characterized the transition from luminal to the pre-tumoral state (Fig. 3a) by comparing pathway activity between cells from LP, Avd and pre-tumoral clusters. We identified that several hallmarks of cancer cells [20] were activated—namely, Myc signaling, the cell cycle (E2F target gene signature), the epithelial-to-mesenchymal transition (EMT) and angiogenesis—while the apoptosis pathway was repressed. These transcriptional signatures endorsed the pre-tumoral nature of this transition state.
To further understand what role the EMT plays in these cells, we investigated the marker genes of the pre-tumoral state. The Vim, Fn1 and Sparc genes were significantly up-regulated in cells in a pre-tumoral state (Fig. 3b and Supplementary Table 3), indicative of changes in cytoskeleton and extracellular matrix. Cdh1/E-cadherin and several claudin genes (Cldn4, -3 and -1) (Supplementary Table 3) were downregulated, indicative of the dissolution of adherens and tight junctions. Pre-tumoral state cells still expressed epithelial keratins, albeit to a lower level than their LP counterparts (Extended Data Supplementary Fig. 1c), suggesting that they reside in an intermediary epithelial and mesenchymal state [21]. We validated these findings using multiplex immunohistochemistry (IHC). We detected luminal cells in CreP mammary glands that expressed both vimentin and E-cadherin (n = 314 double positive cells, out of 2,079 luminal cells; P = 3.1e- 5 compared to CreN glands, Fisher’s extract test; Fig. 3c).
Predicting the transcription factors (TF) that potentially drive the transcriptomic changes from the luminal to the pre-tumoral state, we observed FOX family members among the top candidates as well as a series of EMT-associated TFs: Prrx1 and Prrx2 [22, 23], recently discovered to be EMT inducers, and the canonical EMT-associated TFs (Twist1, Twist2, Snail and Snail2) (Fig. 3d). To validate our finding that the EMT-associated TFs were expressed prior to tumor formation, we stained formalin-fixed, paraffin-embedded (FFPE) sections from mice at different ages for the EMT-related TFs Twist1 and Snail using immunofluorescence. In CreP mice, we detected luminal cells that expressed Snail prior to tumor formation, and the proportion of these cells increased with age; in turn, we found that Twist1 and Zeb1 were only expressed in full-grown tumors (Fig. 3e and Extended Data Supplementary Fig. 4a-c). These results are similar to previous findings of Snail expression leading up to tumor formation in mice [24]. We show here that EMT is one link of a series of state switches that occurs prior to tumor initiation.
Fig. 4.
Features of the pre-tumoral state are detected in human low grade basal-like tumors and BRCA1-deficient mammary glands. a Boxplot of the log10 normalized expression level of CDKN2A in the Pan Cancer breast dataset according to the tumor subtype, ***p-value < 0.001 from Wilcoxon rank test comparing CDNK2A expression score of basal tumors vs other samples. b Boxplot representation of the scores for the pre-tumoral signature according to the tumor subtype, ***p-value < 0.001 from Wilcoxon rank test comparing pre-tumoral score of basal tumors vs other samples. c Box plot representation of the scores for pre-tumoral signature according to the stage of the tumor, * p-value < 0.05 from Wilcoxon rank test comparing pre-tumoral score of stage I vs stage II/III tumors. d Kaplan–Meier disease-free survival curve for basal-like tumors, according to expression score of the human-derived pre-tumoral signature in the Pan Cancer breast dataset. e-f UMAP representation of the MERFISH datasets of 4 BRCA1m ± juxta-tumor human biopsies clustered by sample of origin (e) or cell identity (f). g Left: Merscope visualizer screenshots of two juxta-tumor breast BRCA1m human samples, analyzed using a custom 140 gene-panel (Supplementary Table 6). Scale bar, 1 mm. Middle: 2D spatial visualization of single cells from each experiment. Cells were labeled according to their corresponding cell type annotation. Right: 2D spatial visualization of the kernel-density score estimation of the co-expression of the top highly spatially variable pre-tumoral genes (CCND1, VIM, IGFBP4, AQP5) and the LP marker ELF5 positive cells. For a-c, *p-value < 0.05, ***p-value < 0.001, n.s: not significant, Wilcoxon's test comparing basal-like to each other breast cancer subtype or stage 1 to stage 2–3 cancers
We next focused on the top marker of the pre-tumoral state, Cdkn2a/p16 (Fig. 2b and Supplementary Table 3), which is a marker for cell cycle arrest and senescence [25–28]. We first tested for p16-positive cells in CreP tumor-free mammary gland and assessed the cell cycle status of these cells. Using both immunofluorescence and multiplex IHC, we showed that cells expressing p16 are specific to CreP mice and are mostly luminal cells (Extended Data Supplementary Fig. 5a-d). These cells were detectable starting at 3 months of age and displayed an increased fraction of Ki67 staining, as compared to luminal CreN cells, with mouse aging (Extended Data Supplementary Fig. 5e). Thus, luminal cells can activate p16 prior to tumor initiation and can apparently bypass the cell cycle arrest normally imposed by p16. Notably, other cell cycle related genes that together promote G1 to S transition—Cdk1, Cdk4 and Ccnd1— were overexpressed in cells in a pre-tumoral state (Fig. 3f and Supplementary Table 3). The overactivation of these genes could help cells bypass a p16 overexpression–induced cell cycle arrest [29].
Fig. 5.
Activation of FGF signaling pathway prior to tumor formation. a UMAP representation of all cells from CreP scRNA-seq datasets. b Circos plot representation of Fgfr1/Fgf8 communication between pre-tumoral cells and other cells. c Stack violin plot representation of log10 normalized expression values of top Fgf and Fgfr genes predicted to contribute to the FGF signaling between pre-tumoral cells and other cells. *** adjusted p-value < 0.001, n.s, non-significant for Wilcoxon rank test comparing expression scores in LP and pre-tumoral cells. d Immunofluorescence staining for keratin 8 (magenta), Krt5 (cyan) and pFGFR (yellow) of wild-type and BRCA1m prophylactic, juxta-tumoral and invasive carcinoma human samples. Scale bar, 100 µm. e Biplot illustrating the intensity of pFGFR staining per cell, together with the expression level of the luminal marker keratin 8 and the basal marker keratin 5
Next, we looked for traces of a present or past senescence-like phenomenon in CreP mammary glands. At the transcriptional level, the pre-tumoral state was significantly enriched for a senescence-related signature (Fridman_Senescence [30], adjusted P < 0.01) (Supplementary Table 3). In addition, cells in pre-tumoral state express the pro-senescence secreted factors, Igfbp4 and Igfbp7 (Supplementary Table 3), which can trigger senescence in neighboring cells [31]. We also screened CreP tissues for markers of senescence associated to p16 upregulation [18], including the presence of senescence-associated heterochromatin foci (SAHF) and senescence-associated-B-galactosidase (SABgal). We saw no SABgal staining within CreP mammary glands or tumor sections (Extended Data Supplementary Fig. 5 h); however, by immunofluorescence, we observed SAHF-like structures in tumor-free CreP glands, starting in 3-month-old mice (Extended Data Supplementary Fig. 5f,g). These results suggest that, in addition to p16 activation, cells in a pre-tumoral state might have undergone a senescent-like phenomenon.
To spatially resolve the cell cycle and EMT associated changes in the mammary gland, we stained for epithelial (E-cadherin) and mesenchymal (N-cadherin, vimentin) markers as well as for p16, with multiplex IHC. p16 expression was significantly associated with the expression of vimentin in CreP mammary glands: both E-cadherin and vimentin were expressed in 43% of p16-positive luminal cells but only in 17% of p16-negative luminal cells (P = 3.3e- 6, Fisher’s exact test; Fig. 3g). These results indicate that alterations of cell cycle and a partial EMT can co-occur in luminal cells prior to tumor formation.
Features of pre-tumoral cell state are detected in early-stage breast cancers and in mammary glands of BRCA1m carriers
To first determine whether the pre-tumoral state is present in human tissues, we interrogated two large breast tumor cohorts [19, 32–34] and analysed the expression patterns of pre-tumoral genes, first focusing on the top marker of the pre-tumoral state, CDKN2A/p16. We found that CDKN2A was specifically over-expressed in almost all basal-like tumors (Fig. 4a). These findings mirrored our observations for Cdkn2a/p16 expression in mice (which was detectable prior to tumor initiation and maintained in tumor cells), suggesting that CDKN2A activation might be an early event in basal-like tumorigenesis. Next, we studied the expression pattern of the full pre-tumoral signature, defined as the top overexpressed genes in pre-tumoral cells versus LP and Avd cells (n = 50 human orthologs) (Supplementary Table 4). In the two cohorts, the pre-tumoral signature was significantly more expressed in basal-like tumors than in other tumors (Fig. 4b and Extended Data Fig. 6a). In addition, the signature was more expressed in low-stage than high-stage basal-like tumors and was associated with a longer disease-free survival (Fig. 4c,d and Extended Data Supplementary Fig. 6b,c). These results suggest that tumors with a pre-tumoral like expression program might be closer to an early-stage disease.
We next asked whether the pre-tumoral state can be detected in normal-like glands in humans near tumor initiation, by analysing mammary epithelium from women with BRCA1 germline deficiency. We reasoned that informative samples would be those where the epithelium had been exposed to the same intrinsic—here, BRCA1 deficiency—and extrinsic stresses as cells that already have initiated a tumor. This unique scenario, corresponding to juxta-tumoral tissue, enhances the likelihood of detecting pre-malignant molecular abnormalities. We used the spatial transcriptomics MERFISH [35] approach to probe the expression of genes of the pre-tumoral signature (Supplementary Tables 5 and 6), together with 20 marker genes to call cell identities in four juxta-tumoral tissues of BRCA1 m carriers (n = 49,970 cells; Extended Data Supplementary Fig. 6 d). We identified across four patients 20,021 epithelial cells, among which 11,263 LP cells. We detected the co-activation of several pre-tumoral genes – CCND1, VIM, AQP5 & IGFBP4— in patches of LP cells (Fig. 4e, Extended Supplementary Fig. 6e-f). Such observations mirror our findings in the mouse model, where pre-tumoral luminal cells acquire mesenchymal (Vim), senescence-related (Igfbp4) and aberrant cell cycle (Ccnd1) features. This analysis demonstrates that a fraction of LP cells displays a pre-tumoral signature in the tissues of BRCA1 m carriers that already had a tumor.
Luminal cells in pre-tumoral state activate immunosuppressive and FGF signaling
We then explored the way cells in pre-tumoral state communicate with one another and with other cell types from their microenvironment. To do so, we leveraged all cell types in our mouse scRNA-seq datasets and inferred cell–cell communication pathways using the CellChat algorithm [36] applied on all cells of CreP tumor-free mice (n = 8,855 cells; Fig. 5a). Pre-tumoral cells shared inward communication pathways with both LP and basal cells, expression genes coding for receptors for Notch, Kit and Epha signaling (Extended Data Supplementary Fig. 7a). Looking at the outward communications, we identified that pre-tumoral epithelial cells were predicted to send different signals than normal-like epithelial cells (Extended Data Supplementary Fig. 7b). Strikingly, pre-tumoral cells activated two signaling axes —MIF and SPP1— that were also found in tumor cells. Both pathways involved the emission of ligands from tumor cells to communicate with macrophages that bear the Cd44 receptor (Extended Data Supplementary Fig. 7c,d). The Spp1:Cd44 signaling axis has been previously reported to act as an immune checkpoint, inducing immune tolerance in tumors [13, 37, 38]. Pre-tumoral cells could thus be using such a signaling axis to evade immune surveillance during tumor initiation.
Cells in pre-tumoral state also transiently activated fibroblast growth factor (FGF) signaling (Fig. 5b and Extended Data Supplementary Fig. 7b,c) for autocrine and paracrine communication with epithelial cells and fibroblasts (Fig. 5b and Extended Data Supplementary Fig. 7 d). This signaling is not predicted to occur in tumor cells. Pre-tumoral cells displayed an over-expression of the receptor Fgfr1 as compared to LP cells (Fig. 5c) and were the only cells expressing the Fgf8 ligand. Expression of the ligand is lost in tumor cells, suggesting that the activation of the FGF signaling is transient and potentially necessary for early phases of transformation, but not for maintenance of tumor growth.
To assess the relevance of these findings in human tissues, we analysed the activation of the FGF signaling in human mammary glands. For this, we performed immunofluorescence staining for the activated phosphorylated form of all four isoforms of FGF receptors (pFGFR) in FFPE samples from n = 20 individuals (Supplementary Table 7). Samples included 8 healthy mammary glands from patients that never had a tumor (n = 4 BRCA1wt and n = 4 BRCA1m ‘prophylactic’), as well as normal-like mammary glands from BRCA1m carriers with a tumor (n = 9 BRCA1m juxta-tumoral) and tumors from BRCA1m carriers (n = 3, Fig. 5d). pFGFR levels were significantly elevated in specific regions of normal-like mammary glands of BRCA1 m carriers with and without a tumor, as compared to mammary glands of BRCA1wt individuals (Extended Data Supplementary Fig. 7e). Cells with high levels of pFGFR were specific to the luminal compartment (Fig. 5e). We did not detect high levels of pFGFR in any cells in the three tumors we studied, suggesting that activation of FGF signaling is not a prerequisite for tumor growth. These results show that FGF signaling is over-activated in BRCA1m tissues. Further studies will be needed to understand whether the number of FGF-high cells is associated to the timing of occurrence of tumors in BRCA1m carriers.
Discussion
In this study, we analysed human and mouse tissues to gain a non-genetic understanding of tumor initiation. We propose that an epigenomic disorder and aberrant activation of mesenchymal and cell cycle markers are manifestations that LP cells are undergoing oncogenic stress and can potentially engage in tumor initiation (Extended Data Supplementary Fig. 8). The general epigenomic disorder we observe is associated with a loss of cell lineage integrity. These findings echo studies in colon and pancreatic tumorigenesis, suggesting that improper maintenance of cell identity may precede tumor initiation [39, 40]. By leveraging single-cell histone modifications profiling, we showed that the LP compartment displays extensive non-genetic disorder prior to transformation with an extremely heterogeneous H3K4me1 epigenomes, which ranges from basal-like to basal and luminal H3K4me1 features. H3K4me1 is a proxy of enhancer activation and priming, suggesting that under oncogenic stress, LP cells either activate or prime enhancers in a disordered fashion, thereby losing their proper lineage integrity. H3K4me1 is only one histone modification among a wide variety of chromatin modifications that could encode relevant characteristics of early tumor evolution. Longitudinal studies of repressive and permissive chromatin modifications, alone or in combination [41], will be key to further mapping the epigenomic evolution of luminal cells towards transformation and to identifying additional recurrent, non-genetic features of tumor initiation.
Using a mouse model played a crucial role in investigating how luminal progenitor cells can give rise to fully developed tumors. Based on multiple sampling at different stages of tumorigenesis—and in particular, on profiling tumor-free animals that are litter mates of mice with tumors, we demonstrated that luminal cells consistently pass through a pre-tumoral state before progressing to fully-developed tumors. This state is characterized by a loss of luminal identity, activation of a partial mesenchymal phenotype and immunosuppressive and FGF signaling. Additionally, this pre-tumoral state retains signs of previous cell cycle arrests; indeed, our data suggest that p16 activation could be a very early event in basal-like tumorigenesis. Further studies will be needed to understand precisely how luminal cells escaped such an arrest. Based on the observation that p16 and vimentin expression are correlated prior to tumor formation, we propose that partial EMT could be one of the mechanisms that enables cells to escape the cell cycle arrest imposed by oncogenic stress. EMT in early transformation had been observed by others in mammary glands in vitro [42–44] and in vivo [24, 45]. In addition, transcriptional signatures of EMT have been detected in pancreatic [40] and prostate tumorigenesis [46], advocating for a general occurrence of EMT during early transformation.
We observed a significant delay—typically of several months—between tumor suppressor gene deletion and tumor appearance. In addition, we and others detect CNVs prior to tumor initiation in luminal progenitors [47, 48]. In concordance with other studies [25, 26], this supports the fact that genetic alterations are not sufficient to launch tumor initiation in luminal cells. This is in line with the accumulation of somatic mutations in normal tissues prior to tumor formation [49–51]. Collectively, these findings suggest that non-genetic processes involved in tumor evolution, such as clonal selection or epigenomic mechanisms, could be the detonators needed to achieve tumor formation. In this line, a recent study [52] proposed that loss of one Brca1 allele induces cancer-associated epigenomic changes that prime cells for subsequent transformation.
Early events in human BRCA1-tumorigenesis have so far been mostly studied using prophylactic surgeries performed on BRCA1m carriers. By employing a range of techniques from cytometry to immunofluorescence and single-cell approaches [7–10], studies have revealed a series of malfunctions within the epithelial compartment in BRCA1m carriers, including replicative stress and homologous recombination defect [53], accumulation of DNA breaks [54], altered homeostasis of cell populations [6, 8, 10, 53] and an increased proportion of cells that exhibit both luminal and basal characteristics [8, 10] and exist physiologically in the mammary gland. Nee et al. [10] also observed pro-tumorigenic changes in the microenvironment in BRCA1m carriers with the identification of pre-cancer associated fibroblasts. Here we leveraged a mouse model to first identify a signature of recurrent pre-tumoral events. We then showed that we can actually detect these features in tissues from BRCA1 m carriers that already had a tumor. We identified luminal progenitor cells with aberrant expression of senescence, mesenchymal and cell cycle markers. Further studies will be needed to understand whether the amount of cells with pre-tumoral features could relate to the timing of occurrence of cancer in patients, and could serve as predictive tools or as potential targets to delay tumor initiation. Our work paves the way to early detection of pre-tumoral events in humans in individuals with high cancer risk.
Methods
Experimental methods
Animal models
The generation of Brca1fl/fl and Trp53fl/fl mice has been previously described [55, 56]. Blg-Cre transgenic mice were purchased from The Jackson Laboratory. Mice strains were crossed to obtain Blg-Cre Trp53fl/fl Brca1fl/fl animals. Genotypes were determined by PCR (primers Cre: 3’ CGAGTGATGAGGTTCGCAAG 5’—3’ TGAGTGAACGAACCTGGTCG 5’; primer Brca1: 3’TATCACCACTGAATCTCTACC 5’—3’ GACCTCAAACTCTGAGATCCAC 5’; Trp53: 3’ AAGGGGTATGAGGGACAAGG 5’—3’ GAAGACAGAAAAGGGGAGGG 5’). Mice were sacrificed by cervical dislocation. For each sample (gland or tumor), one piece was fixed in 4% paraformaldehyde (15,710, Euromedex) for histological analysis, one piece was snap frozen in dry ice and stored at − 80 °C and one piece was kept fresh for the desired experimentation.
Ethics statement
All procedures used in the animal experimentations are in accordance with the European Community Directive (2010/63/EU) for the protection of vertebrate animals. The project has been approved by the ethics committee n°02265.02. We followed the international recommendations on containment, replacement and reduction proposed by the Guide for the Care and Use of Laboratory Animals (NRC 2011). We used as few animals as possible and minimized their suffering, no painful procedures were performed. The breeding, care and maintenance of the animals were performed by the Institut Curie animal facility (facility license #C75 - 05–18). Patients (n = 4 BRCA1m carriers for MERFISH analysis, Supplementary Table 6; n = 20 patients for anti-phospho FGFR staining, Supplementary Table 7) gave informed consent for the use of their tissue in the study.
Immunofluorescence
Freshly dissected mammary glands or tumors were fixed overnight in 4% neutral-buffered paraformaldehyde at 4 °C, paraffin-embedded and sectioned at 3 µm thickness. Tissue sections were deparaffinized and rehydrated through a series of xylene and ethanol washes, and subjected to antigen retrieval in boiling citrate buffer pH6 (C9999) for 20 min. Permeabilization was obtained with 0.3% Triton X- 100; non-specific antibody binding was blocked with 5% FBS and 2% BSA (2 h at RT), and then sequentially incubated with primary (overnight at 4 °C, following by 3 washes in PBS 10 min) and secondary antibodies (2 h at RT). Primary antibodies used: chicken Keratin 5 (BioLegend 905,901, 1:500), rat Keratin 8 (Sigma MABT329, 1:500), rabbit Snail (PA5 - 115,940, 1:100), rabbit Zeb1 (Abcam 155,249, 1:100), rabbit Twist1 (Cell Signaling 31,174, 1:400), mouse IgG3 H3 K27 me3 (Abcam 6002, 1:200), rabbit p16 (Abcam 211,542 1:100), rabbit phospho-FGFR (Cell Signaling 3471S, 1:100), chicken GFP (Thermo Fisher PA1 - 9533). Fluorochrome-conjugated secondary antibodies included AlexaFluor 488-conjugated anti-chicken IgG, A488-conjugated anti-mouse IgG3, Cy3-conjugated anti-rabbit IgG, Cy5-conjugated anti-rat. All secondary antibodies were used at 1:1000 dilutions, sections were counterstained with DAPI (1 mg/mL; Sigma). After 3 wash in PBS 10 min, sections were mounted in Aquapoly mount media.
Image acquisition and analysis of immunofluorescence data
Image acquisition of stained sections were done using a laser scanning confocal microscope (LSM780, Carl Zeiss) with a LD LCI PLAN-APO × 40 or × 65/08 NA oil objective. The acquisition parameters were: zoom 0.6; pixel size xy 554 nm; spectral emission filters (bandwidth): 414–485 nm, 490–508 nm, 588–615 nm, 641–735 nm; laser wavelengths: 405, 488, 561 and 633 nm. At least 3 independent images were acquired for each biological replicate. Image processing was performed using Fiji Software, version 1.0. A quantitative score (“signal-to-noise”) was computed for interpretation of transcription factors expression, by measuring the mean nuclear fluorescence values for each keratin 8 + cell and dividing these values by the mean of 6 independent negative nuclei from the same image. The counting of µ-HF was done in Fiji with a custom macro, for each nucleus, we selected the most representative Z, then the counting was done automatically with the AutoThreshold MaxEntropy.
Senescence-associated-beta-galactosidase staining
Straight away after harvesting, samples were embedded in optimal cutting temperature (OCT) medium (23–730–751) in molds and cooled on a metal support previously cooled on dry ice. The samples were stored at –80 °C before being cut in a cryostat at –30 °C into 10 µm sections. Slides were stored at –80 °C before use. For the staining, the slides were removed from − 80 °C storage and dried at room temperature for 30 min. Samples were fixed in a fixation solution of PFA 2% [Electron Microscopy Sciences #15714] and glutaraldehyde 0.2% [Sigma #49629] in PBS at room temperature for 15 min. After three PBS washes of 5 min, samples were incubated 6 h at 30 °C in a SAβGal solution containing 40 mM of citrate buffer pH 6.0 [VWR #28,027.295; Sigma #C7129]; 5 mM of K3 Fe(CN)6 [Sigma #P8131]; 5 mM of K4 Fe(CN)6 [Sigma #P9387]; 2 mM of MgCl2 [Sigma #M8266]; 150 mM of NaCl [Sigma #31434]; 0.1% of NP40 [Sigma #I8896]; 0.5 mg/mL of X-gal [Euromedex #EU0012] and ultrapure Milli-Q water. After incubation, samples were washed in PBS, post-fixed in PFA 4% [Electron Microscopy Sciences #15714] for 15 min and washed again on PBS. Tissue sections were then counterstained with Nuclear FastRed [Vector #H- 3403] for 2 to 5 min (checked under the microscope for a good contrast), dehydrated by one 5 min bath of EtOH 95% and two 5 min in 100% EtOH baths before mounted with Eukitt [Sigma Aldrich #49629].
Multiplex histological staining (multiplex IHC)
Multiplexed IHC was performed according to the protocol developed by [57], with some adjustment. Tissues were baked at 60 °C for 1 h, deparaffinized in Xylene (Fisher Scientific, 10,467,270) and rehydrated. The heat-induced epitope retrieval was done with pH 6.1 citrate buffer (Dako, S169984 - 2) or pH 9 EDTA buffer (Dako, S236784 - 2) in a 95 °C water bath for 30 min for the first staining (otherwise 15 min) followed by incubation in REAL peroxydase blocking solution (Agilent Dako, S202386 - 2) for 10 min. If the primary antibody was the same species as any antibody used in prior stains, another blocking step was added with Fab Fragment, only for anti-rabbit (Jackson ImmunoResearch Europe Ltd, 711–007 - 003) for 20 min. Protein block serum free (Agilent Dako, X090930 - 2) was added for 10 min. Primary antibody was incubated for 1 or 2 h at room temperature or overnight at 4 °C. The primary antibody was detected using a secondary antibody directed against the first one, conjugated with horseradish peroxydase (Anti-rabbit: Agilent Dako, K400311 - 2) (Anti-rat: BioTechne, VC005 - 050) followed by chromogenic revelation with 3-amino- 9-ethylcarabazole (AEC) (Agilent Dako, K3468). Slides were counterstained with hematoxylin (Thermo Scientific, 6,765,001) and mounted with Glycergel aqueous mounting medium (Dako, C056330 - 2). After scanning (Philips Ultra Fast Scanner 1.6 RA), tissues were bleached with ethanol baths, and another cycle was performed starting with heat-induced epitope retrieval.
Overlay of multiplex IHC stainings
Histological analysis was performed using the open-source image analysis QuPath software (QuPath- 0.3.2, http://qupath.github.io/) [58] and ImageJ/Fiji. We created a new QuPath project containing all scans of each slide which allow us to crop and export (BioFormats plugin) and then overlay the images using Fiji script following these different steps: 1. color deconvolution (separation of hematoxylin and AEC signal); 2. alignment on hematoxylin images; 3. creation of transformation matrix on AEC images; 4. For some of the staining (Edac, Vim, Ki67), an automatic threshold using MaxEntropy was done to remove background; for the rest (p16, Krt5, Krt8, Ncad), a different threshold was determined using control cell signal. Each staining was colored as desired. For further analysis, the composite image was transferred back to QuPath. The different structures of the gland/tumors were annotated (duct, stroma, juxta-tumoral duct, mm-tumor, tumor) by hand. The ‘cell detection’ function based on hematoxylin nucleus staining was used to identify all cells, and then the ‘show detection measurement’ function was used to export the annotation and the intensity signals for all stainings for each cell, which were analyzed in R.
Quantification of multiplex IHC images
The resulting measurements were exported and analyzed in R (4.1.1). Briefly, high signal channels, corresponding to Ki67, Vim were thresholded by the Maximum Entropy algorithm, whereas the remaining channel markers were subjected to a custom thresholding approach. To identify true positive cells for each marker, mean “Cell” signal values were binarized as follows:—non-zero values of the max entropy thresholded markers were set to 1, whereas zero values were set to 0. To determine positive cells for p16, Ncad and Krt5, the local minimum after the highest peak was fitted on the density distribution of the merged cells from all the samples corresponding to each marker. Different thresholds were defined for each sample for the following markers: Krt8 and Ecad. Briefly, the “approxfunc” R interpolation function was applied on the density distribution of each marker on each sample, followed by an optimization step using the “optimize” R function to retrieve the local minimum within the interval of the density function. Higher values as compared to each threshold were set to 1, whereas smaller values were set to 0. Basic R functions were used to calculate the percentages of positive cells for each marker and the ggplot package was used for graphical representations. Stromal regions were excluded from the analyses based on manual curation of images, based in H&E and Vimentin stainings.
Mammary gland & tumor dissociation and flow cytometry
Samples were cut roughly with dissecting scissors and then with two scalpels for approximatively 10 min. Single-cell dissociation was done by enzymatic digestion with 3 mg/ml collagenase I (Roche, 11,088,793,001) and 100 U/ml hyaluronidase (Sigma-Aldrich, H3506) in complete media (HBSS 24020117; 5% SVF) during 1 h 30 min under agitation at 170 rpm at 37 °C. Cells were then dissociated in PBS 0.25% trypsin-versen (Thermo Fisher Scientific, 15,040–033) pre-warmed at 37 °C for 1 min 30 s with pipetting for 45 s. The cell suspension was then treated with dispase 5 mg/ml (Sigma-Aldrich, D4693) and DNase 0.1 mg/ml (Roche, 11,284,932,001) in complete media for 5 min at 37 °C. A treatment with red blood cell lysis buffer (Thermo Fisher Scientific, 00–4333 - 57) was carried out then the suspension was filtered at 40 µM before counting and FACS staining. Cell suspensions were stained 20 min in dark at 4 °C with anti-CD45-APC 1:100 (BioLegend, 103,112), anti-CD31-APC 1:100 (BioLegend, 102,510), anti-CD24-BV421 1:50 (BioLegend, 101,826), and anti-CD49f-PE 1:50 (BioLegend, 313,622). Cells were resuspended in cytometry media (PBS, BSA, EDTA). For mammary gland samples, either the total epithelium was recovered, or the luminal and basal cells populations were recovered separately.
scRNA-seq data generation
In accordance with the protocol of 10X Chromium, the cells were resuspended in PBS 0.04% BSA (Sigma, A8577). Depending on the samples, approximately 3000 or 4000 cells were loaded on the Chromium Single Cell Controller Instrument (Chromium single cell 3’ v3 or 3’ NextGem, 10X Genomics, PN- 1000075) in accordance with the manufacturer's protocol. Libraries were prepared according to the same protocol. scRNA-seq libraries were sequenced on a NovaSeq 6000 (Illumina).
snCUT&Tag data generation
snCUT&Tag was adapted from [16, 59]. All washes were made with 500 µL unless otherwise stated and all centrifugations were done using a swinging bucket centrifuge at 1300 g for 4 min at 4 °C for nuclei preparation, or 600 g for 8 min at 4 °C for the subsequent steps. Nuclei were extracted from 1–2 million cells for 10 min on ice in 6 mL ice-cold NE1 buffer (20 mM HEPES pH7.2, KCl 10 mM, spermidine 0.5 mM, glycerol 20%, BSA 1%, NP- 40 0.1%, digitonin 0.01%, proteases inhibitor 1x). Nuclei were filtered with a 30 uM cell strainer, washed in 6 mL PBS + BSA 1% and resuspended in Dig-Wash buffer (20 mM HEPES pH7.2, NaCl 150 mM, spermidine 0.5 mM, BSA 1%, NP- 40 0.01%, digitonin 0.01%, proteases inhibitor 1x), checked under microscope and counted with 4,6-diamidino- 2-phenylindole (DAPI) staining. 50,000 to 100,000 nuclei were resuspended in 50uL antibody buffer (EDTA 2 mM, 20 mM HEPES pH7.2, NaCl 150 mM, spermidine 0.5 mM, BSA 1%, NP- 40 0.01%, digitonin 0.01%, proteases inhibitor 1x) with 1:50 antibody (Anti-H3K4me1 #5326 D1 A9, Cell Signaling) and incubated overnight at 4 °C with rotation. Next day, nuclei were washed with Dig-Wash buffer and resuspended in 100uL Dig- 300 buffer (20 mM HEPES pH7.2, NaCl 300 mM, spermidine 0.5 mM, BSA 1%, NP- 40 0.01%, digitonin 0.01) with the proteinA-Tn5 fusion (Diagenode, #C01070001, 1:250) and incubated for 1 h at room temperature with rotation. Then, nuclei were washed three times with Dig- 300 buffer, resuspended in 300 µL Tag-Buffer (20 mM HEPES pH7.2, NaCl 300 mM, spermidine 0.5 mM, BSA 1%, NP- 40 0.01%, digitonin 0.01%, MgCl2 10 mM) and incubated for 1 h at 37 °C. Tagmentation was stopped by addition of one volume of 1 × Diluted Nuclei Buffer (DNB, 10X Genomics) supplemented with 2% BSA, 12.5 mM EDTA. The nuclei were then centrifuged at 1300 g, 4 min at 4 °C and washed twice with 200 µL 1 × DNB supplemented with 2% BSA. The nuclei were resuspended in 10–70 µl of DNB + 2% BSA. If the sample did not show nuclei aggregates, nuclei were loaded on a 10 × Chromium system using 10 × Chromium Single Cell ATAC–Seq kit v2 (10X Genomics) as described [16]. Final library amplification with 15 PCR cycles was performed according to the Chromium Single Cell ATAC Library kit manual. snCUT&Tag libraries were sequenced on a NovaSeq 6000 (Illumina) in PE50 mode.
MERFISH data generation
A custom multipurpose 140-gene panel was developed by our group balancing genes for cell type annotation and identification of abnormal luminal cells using our previously defined pre-tumoral signature from the mouse dataset (Supplementary Table 5). Frozen tissue sections of 10 µm thickness from 4 juxta-tumor breast biopsies were adhered to the MERSCOPE beaded slide, and stored in 70% ethanol at 4 °C for a maximum of 1 month. Adhesion was optimized to be able to fit two biological samples on the same coverslip. Standard sample preparation for frozen tissues was performed following the Vizgen instruction manual, without any specific adaptations. In short, a cell boundary staining was applied, followed by hybridization with a custom-made 140 gene panel. The section was then embedded and cleared. Slides were imaged with the MERSCOPE machine, and processed with the software version 233.
Computational methods
Code related to the following sections can be found at https://github.com/vallotlab/BRCA1_Tumorigenesis
Processing of scRNA-seq datasets
Demultiplexed FASTQ files from the raw single cell RNA-seq reads were aligned using the pre-built reference mm10 genome proposed by 10X Genomics toolkit, and gene counts were obtained using the Cell Ranger “count” function. Empty droplets were filtered out using the DropletUtils package, implemented in the Cell Ranger suite, at an FDR of 0.01. Doublets were detected using the DoubletDetection algorithm (https://zenodo.org/record/6349517) with default parameters. Singlet expression matrices were processed individually in R (v4.0.2) mainly using Seurat (v3) R package [60]. We kept cells with a coverage in between 1000–8000 genes, 1000–60000 UMIs and < 30% of mitochondrial reads. Filtered gene-cell barcodes matrices were normalized individually using the “SCTransform''method, and filtered gene-nuclei barcodes were normalized using the “NormalizeData” function using the “LogNormalize” method.
Clustering, UMAP reduction and cell annotation
Normalized expression matrices were merged without data integration using Seurat package (v3.9). Coarse-grained clustering (resolution = 0.8) was applied on the merged scRNA-seq datasets after PCA projection (n = 50 PCs), and canonical markers were used to annotate cell types (Epithelial, Immune, Fibroblast, Endothelial). To annotate epithelial subtypes, we created a Seurat object containing these cells only, reran Variable Features selection, PCA, clustering at a higher resolution (res = 1.2) and UMAP projection. Differential Gene Expression (DGE) between the epithelial subtypes was run using FindAllMarkers (only.pos = TRUE, log2FC = 0.05, adjusted P < 0.05). The resulting clusters were manually annotated using canonical markers from 37. If a given cluster is mainly composed of either mmT or tumor cells (> 50%), the cluster name is a concatenated form of the sample of origin and a given index (of integer type). UMAP projections were plotted using “uwot” as umap.method, n.neighbours = 30, distance metric = “cosine”, min.dist = 0.3) and “random.state = 42”.
Partition-based graph abstraction (PAGA)
PAGA was performed using “scanpy” Python library loaded on RStudio using “reticulate” R package; using default parameters and a threshold of 0.1 to keep highly connected nodes. Connectivity scores were extracted from the PAGA output, along with the nodes and edges connections. Centrality scores (number of edges) were computed by counting the number of edges for each cell cluster (node).
Copy number variation (CNV) inference from scRNA-seq data
CNVs were inferred using inferCNV (https://github.com/broadinstitute/infercnv) with default parameters, taking as reference the CreP basal cells for mouse scRNA-seq. The “observation” and “reference” transformed matrices were extracted and quantified for each nucleus the number of genomic regions, with an absolute inferCNV score (from the “observations” transformed matrix) above the 95 th percentile of the reference dataset (from the “references” transformed matrix). This score was expressed as a percentage of the total number of regions in the genome; this metric is referred to as “the percentage of genome with CNVs”.
Pathway enrichment and signature analysis
PEA was performed using the “enricher” function (ClusterProfiler R package) on the top overexpressed genes by the pre-tumoral as compared to the LP/Avd clusters (avg_log2FC > 0.5, adjusted P < 0.05), using the Hallmark collection from the Molecular Signature Database (MSigDB) 21 available through the msigdbr R package. Transcriptional signatures were quantified using the “UCell” [61] package. We computed the gene signatures using the wrapper function “AddModuleScore_UCell”, giving as input a list of features, along with the seurat object. The “FindMarkers” Seurat function was used to define DEG between the pre-tumoral and LP/Avd cell clusters (avg_log2FC ≥ 1 and adjusted P < 0.05, only.pos = TRUE). This mouse gene list (n = 53 genes) was converted to human orthologs using the “gorth” function from the “Gprofiler” R package, obtaining a list of n = 50 human genes (Supplementary Table 4).
Analysis of snCUT&Tag datasets_Preprocessing
Fastq files were processed using an in-house pipeline https://github.com/vallotlab/scCUT-Tag_10X to generate fragment files, count matrices and pseudo-bulk bigwig files. The unique number of fragments per nuclei, and the fraction of reads that fall within peaks (FRiP), were computed. Genomic regions were binned using the “createArrowFiles” function from the ArchR suite [62], taking as input the list of fragment files corresponding to each sample, with minFrags = 100, maxFrags = 3000, TileMatParams: binsize = 20,000 bp, binarize = FALSE, addGeneScoreMatrix = TRUE, using the mm10 reference genome, excluding the Y and mitochondrial chromosomes. Doublets were identified using the “addDoubletScores” ArchR function with k = 20, n_neighbors = 40, corCutOff = 0.75. Nuclei with a DoubletEnrichment scores higher than 30 were annotated as “Doublets” and were discarded for the downstream steps. The filtered 20,000 bp TileMatrix was further processed by performing TF-IDF normalization and 5 iteration steps of Latent Semantic Indexing (LSI) dimension reduction. LSI components with a correlation score > 0.8 with the “total_fragments” were removed. Two-dimensional visualizations were performed using the “addUMAP” ArchR function, taking as input the LSI-based dimension reduction. Louvain graph-based clustering was performed on the LSI-based matrix using the “addClusters” ArchR function with resolution = 0.8, n_neighbors = 30.
Analysis of snCUT&Tag datasets_cluster annotation
A manual annotation step was performed on the GeneScoreMatrix assay of the ArchR project to annotate the clusters: Differential Gene Signal (DGS) between the clusters was run using getMarkerFeatures(testMethod = ”wilcoxon”, bias = ”nFrags”, useMatrix = ”GeneScoreMatrix). The top markers for each cluster (log2 FC > 2 & FDR < 0.05) were intersected with the previously used canonical markers to annotate the scRNAseq data. The annotation accuracy was manually checked by visualizing each canonical marker signal on IGV.
Analysis of snCUT&Tag datasets_2D basal-LP plots
A 1-vs-all wilcoxon test was performed between the epithelial subtypes of the creN sample using the GeneScoreMatrix assay. Top 200 most-enriched gene signals (log2 FC > 4 & FDR < 0.05) per subtype were considered as “canonical” H3 K4 me1 signal genes. creP epithelial cells were scored by the three canonical signatures using the “AddModuleScore” function. Two-dimensional plots of the Basal-LP signatures were plotted for the three histological categories: creP, creN and Tumor. For the sake of clarity, “geom_jitter” was used to split the nuclei and increase the plot resolution.
Analysis of PanCancer breast and CPTAC datasets
Expression matrix were downloaded from cbioPortal web-portal (https://www.cbioportal.org/study/summary?id=brca_tcga_pan_can_atlas_2018) and normalized CPTAC Breast datasets (https://www.linkedomics.org/data_download/TCGA-BRCA/). Metadata tables (including stage and breast tumor subtype) were also downloaded from the two websites. PanCancer expression matrix was normalized using the DESeq2 R package. CDKN2 A/p16 log-normalized expression level was compared between basal-like and other tumor subtypes (HER2, LumA, LumB) and normal-like samples using a two-sided Wilcoxon test. In both datasets, UCell package was used to compute pre-tumoral signature scores on each sample, and compare the scores distribution between the basal-like and the remaining breast subtypes using the two-sided Wilcoxon rank-sum test. Within the basal-like subtype only, these same pre-tumoral scores were compared between early (stage1) and late (stage 2 or more) stage samples. Survival metrics, including overall survival from the CPTAC (included in the metadata) and Progression-free survival from the PanCancer (also included in the metadata), were compared between samples with high or low signature scores using the Survfit and Survival R packages.
Transcription factor (TF) enrichment analysis
TF enrichment was performed on the overexpressed genes by the pre-tumoral cluster as compared to the LP/Avd clusters (avg_log2FC > 0.5, adjusted p-value < 0.05), using the Chea3 R package49 with default parameters. Top 30 enriched TFs were further represented on the figures.
Preprocessing of MERFISH datasets
Cell segmentation was performed based on DAPI and membrane staining, using the CellPose algorithm as part of the Merscope workflow. Data was inspected using the visualizer software, and subsequently analyzed using a custom pipeline. The output cell gene count and cell coordinate matrices were loaded individually into Seurat (v5). Only regions with a raw number of cells > 4,000 were kept for further analysis. To ensure consistency in cell annotation across the 4 samples, the individual expression matrices were merged without considering their spatial coordinates. Cells with nUMI > 20 and nGenes detected > 10 were kept for further steps. Count matrices were processed using NormalizeData (normalization.method ="LogNormalize", scale.factor = 10,000), ScaleData, RunPCA (npcs = 30), RunUMAP(reduction ="pca", dims = 1:20). Graph-based clustering was performed using the FindNeighbors(reduction ="pca", dims = 1:20) and FindClusters(resolution = 0.8) functions in Seurat. Clusters were annotated using the same procedure as for the scRNAseq (see above). Two-dimensional plots of the individual samples were represented using the “DimPlot” function, with “reduction = ”spatial”.
Analysis of MERFISH datasets & identification of expression patterns of the pre-tumoral signature
We focused on the LP compartment in each sample and kept the spatial coordinates of each LP cell. Neighbor relationships between LPs in each sample were computed using the “getSpatialNeighbors” function from the MERINGUE R package, setting the minimal distance between LPs to consider as neighbors to dist = 15. Significantly spatially auto-correlated genes were obtained by computing the MORAN’s I test, taking as input the normalized-gene expression matrix and the weighted adjacency matrix computed before. These steps were run independently on each LP compartment from each sample. The top 95 th quantile highly spatially variable genes from the four samples were kept as “TRUE_spatially variable”. From this list of genes, only those part in the pre-tumoral signature were considered; namely: VIM, CCND1, AQP5, IGFBP4. The coexpression of the four genes retained from the previous steps was assessed on each sample using a kernel-density of co-expression from the Nebulosa (https://github.com/powellgenomicslab/Nebulosa) R package.
Supplementary Information
Acknowledgements
We thank Dr. Alain Puisieux and Dr. Josh Waterfall for providing critical discussion. We also thank the animal facility, the sequencing and imaging platforms from Institut Curie. We thank Dr. J. Jonkers for providing mouse strains.
Authors’ contributions
C.L. and M.L. performed experiments throughout the manuscripts. A.D and A. T. performed single-cell epigenomic experiments. M.S and P.P performed computational analysis. R.L, K.B performed MERFISH experiments. M.F. performed mouse work. A.C. performed senescence associated experiments. A.V.S. selected patient samples. J.M. and H. S. performed and analysed multiplex IHC experiments. C.V designed and led the project. C.V., C.L., M.S. and M.L. made figures and wrote the manuscript with input from all authors.
Funding
This work was supported by the ATIP Avenir program, by Plan Cancer, by the SiRIC-Curie program SiRIC Grants #INCa-DGOS- 4654 and #INCa-DGOS-Inserm_12554, support from Bettencourt-Schueller Foundation and by a starting ERC grant from the H2020 program #948528-ChromTrace (C.V.). High-throughput sequencing was performed by the ICGex NGS platform of the Institut Curie supported by the grants Equipex #ANR- 10-EQPX- 03, by the France Genomique Consortium from the Agence Nationale de la Recherche #ANR- 10-INBS- 09–08 ("Investissements d’Avenir"program), by the ITMO-Cancer Aviesan—Plan Cancer III and by the SiRIC-Curie program SiRIC Grant #INCa-DGOS- 4654.
Data availability
The datasets described in this study have been deposited in the GEO repository, SuperSeries GSE200444.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Camille Landragin, Melissa Saichi and Marthe Laisné contributed equally to this work.
References
- 1.Martincorena I, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015). [DOI] [PMC free article] [PubMed]
- 2.Martincorena I, et al. Somatic mutant clones colonize the human esophagus with age. Science. 2018;362:911–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yizhak K, et al. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science 364, (2019). [DOI] [PMC free article] [PubMed]
- 4.Frank TS, et al. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol. 2002;20:1480–90. [DOI] [PubMed] [Google Scholar]
- 5.Molyneux G, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7:403–17. [DOI] [PubMed] [Google Scholar]
- 6.Lim E, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–13. [DOI] [PubMed] [Google Scholar]
- 7.Pal B, et al. A single-cell RNA expression atlas of normal, preneoplastic and tumorigenic states in the human breast. EMBO J. 2021;40: e107333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shalabi SF, et al. Evidence for accelerated aging in mammary epithelia of women carrying germline BRCA1 or BRCA2 mutations. Nat Aging. 2021;1:838–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar T, et al. A spatially resolved single-cell genomic atlas of the adult human breast. Nature. 2023;620:181–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nee K, et al. Preneoplastic stromal cells promote BRCA1-mediated breast tumorigenesis. Nat Genet. 2023;55:595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guy CT, et al. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci U S A. 1992;89:10578–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Keymeulen A, et al. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature. 2015;525:119–23. [DOI] [PubMed] [Google Scholar]
- 13.Bach K, et al. Time-resolved single-cell analysis of Brca1 associated mammary tumourigenesis reveals aberrant differentiation of luminal progenitors. Nat Commun. 2021;12:1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–8. [DOI] [PubMed] [Google Scholar]
- 15.Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49:825–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartosovic M, Kabbe M, Castelo-Branco G. Single-cell CUT&Tag profiles histone modifications and transcription factors in complex tissues. Nat Biotechnol. 2021;39:825–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langille E, et al. Loss of Epigenetic Regulation Disrupts Lineage Integrity, Induces Aberrant Alveogenesis, and Promotes Breast Cancer. Cancer Discov. 2022;12:2930–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolf FA, et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 2019;20:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao R, et al. Delineating copy number and clonal substructure in human tumors from single-cell transcriptomes. Nat Biotechnol. 2021;39:599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2020;21:341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fazilaty H, et al. A gene regulatory network to control EMT programs in development and disease. Nat Commun. 2019;10:5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lv Z-D, et al. Silencing of Prrx2 Inhibits the Invasion and Metastasis of Breast Cancer both In Vitro and In Vivo by Reversing Epithelial-Mesenchymal Transition. Cell Physiol Biochem. 2017;42:1847–56. [DOI] [PubMed] [Google Scholar]
- 24.Ye X, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525:256–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erickson A, et al. Spatially resolved clonal copy number alterations in benign and malignant tissue. Nature. 2022;608:360–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jakubek YA, et al. Large-scale analysis of acquired chromosomal alterations in non-tumor samples from patients with cancer. Nat Biotechnol. 2020;38:90–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao T, et al. A pan-tissue survey of mosaic chromosomal alterations in 948 individuals. Nat Genet. 2023;55:1901–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karaayvaz-Yildirim M, et al. Aneuploidy and a deregulated DNA damage response suggest haploinsufficiency in breast tissues of BRCA2 mutation carriers. Sci Adv 6, eaay2611 (2020). [DOI] [PMC free article] [PubMed]
- 29.Lee H-S, Park J-H, Kim S-J, Kwon S-J, Kwon J. A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 2010;29:1434–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fridman AL, Tainsky MA. Critical pathways in cellular senescence and immortalization revealed by gene expression profiling. Oncogene. 2008;27:5975–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collado M, Serrano M. Senescence in tumours: evidence from mice and humans. Nat Rev Cancer. 2010;10:51–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whiteaker JR, et al. CPTAC Assay Portal: a repository of targeted proteomic assays. Nat Methods. 2014;11:703–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Durante MA, et al. Single-cell analysis reveals new evolutionary complexity in uveal melanoma. Nat Commun. 2020;11:496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015). [DOI] [PMC free article] [PubMed]
- 36.Jin S, et al. Inference and analysis of cell-cell communication using Cell Chat. Nat Commun. 2021;12:1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shurin MR. Osteopontin controls immunosuppression in the tumor microenvironment. J Clin Invest. 2018;128:5209–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klement JD, et al. An osteopontin/CD44 immune checkpoint controls CD8+ T cell activation and tumor immune evasion. J Clin Invest. 2018;128:5549–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bala P, et al. Aberrant cell state plasticity mediated by developmental reprogramming precedes colorectal cancer initiation. Sci Adv 9, eadf0927 (2023). [DOI] [PMC free article] [PubMed]
- 40.Burdziak C, et al. Epigenetic plasticity cooperates with cell-cell interactions to direct pancreatic tumorigenesis. Science 380, eadd5327 (2023). [DOI] [PMC free article] [PubMed]
- 41.Bartosovic M, Castelo-Branco G. Multimodal chromatin profiling using nanobody-based single-cell CUT&Tag. Nat Biotechnol. 2023;41:794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ansieau S, et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14:79–89. [DOI] [PubMed] [Google Scholar]
- 43.Morel A-P, et al. A stemness-related ZEB1–MSRB3 axis governs cellular pliancy and breast cancer genome stability. Nat Med. 2017;23:568–78. [DOI] [PubMed] [Google Scholar]
- 44.De Blander H, et al. Cooperative pro-tumorigenic adaptation to oncogenic RAS through epithelial-to-mesenchymal plasticity. Sci Adv 10, eadi1736 (2024). [DOI] [PMC free article] [PubMed]
- 45.Harper KL, et al. Mechanism of early dissemination and metastasis in Her2+ mammary cancer. Nature. 2016;540:588–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan JM, et al. Lineage plasticity in prostate cancer depends on JAK/STAT inflammatory signaling. Science. 2022;377:1180–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin Y, et al. Normal breast tissues harbour rare populations of aneuploid epithelial cells. Nature. 2024. 10.1038/s41586-024-08129-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williams MJ, et al. Luminal breast epithelial cells of BRCA1 or BRCA2 mutation carriers and noncarriers harbor common breast cancer copy number alterations. Nat Genet. 2024;56:2753–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li R, et al. A body map of somatic mutagenesis in morphologically normal human tissues. Nature. 2021;597:398–403. [DOI] [PubMed] [Google Scholar]
- 50.Lee-Six H, et al. The landscape of somatic mutation in normal colorectal epithelial cells. Nature. 2019;574:532–7. [DOI] [PubMed] [Google Scholar]
- 51.Fowler JC, et al. Selection of Oncogenic Mutant Clones in Normal Human Skin Varies with Body Site. Cancer Discov. 2021;11:340–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li CM-C, et al. Brca1 haploinsufficiency promotes early tumor onset and epigenetic alterations in a mouse model of hereditary breast cancer. Nat Genet. 2024;56:2763–75. [DOI] [PubMed] [Google Scholar]
- 53.Pathania S, et al. BRCA1 haploinsufficiency for replication stress suppression in primary cells. Nat Commun. 2014;5:5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Network CGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jonkers J, et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–25. [DOI] [PubMed] [Google Scholar]
- 56.Liu X, et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci U S A. 2007;104:12111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blom S, et al. Systems pathology by multiplexed immunohistochemistry and whole-slide digital image analysis. Sci Rep. 2017;7:15580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bankhead P, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep. 2017;7:16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaya-Okur HS, et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun. 2019;10:1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888-1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andreatta M, Carmona SJ. UCell: Robust and scalable single-cell gene signature scoring. Comput Struct Biotechnol J. 2021;19:3796–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Granja JM, et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat Genet. 2021;53:403–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets described in this study have been deposited in the GEO repository, SuperSeries GSE200444.