Summary
Peripheral T cell lymphomas (PTCLs) comprise heterogeneous malignancies with limited therapeutic options. To uncover targetable vulnerabilities, we generate a collection of PTCL patient-derived tumor xenografts (PDXs) retaining histomorphology and molecular donor-tumor features over serial xenografting. PDX demonstrates remarkable heterogeneity, complex intratumor architecture, and stepwise trajectories mimicking primary evolutions. Combining functional transcriptional stratification and multiparametric imaging, we identify four distinct PTCL microenvironment subtypes with prognostic value. Mechanistically, we discover a subset of PTCLs expressing Epstein-Barr virus-specific T cell receptors and uncover the capacity of cancer-associated fibroblasts of counteracting treatments. PDXs’ pre-clinical testing captures individual vulnerabilities, mirrors donor patients’ clinical responses, and defines effective patient-tailored treatments. Ultimately, we assess the efficacy of CD5KO- and CD30- Chimeric Antigen Receptor T Cells (CD5KO-CART and CD30_CART, respectively), demonstrating their therapeutic potential and the synergistic role of immune checkpoint inhibitors for PTCL treatment. This repository represents a resource for discovering and validating intrinsic and extrinsic factors and improving the selection of drugs/combinations and immune-based therapies.
Keywords: T cell lymphoma, patient-derived tumor xenografts, clonal evolution, stratification, microenvironment, repository, precision medicine, CAR-T, drug screenings, pre-clinical trials
Graphical abstract

Highlights
-
•
We establish 102 patient-derived models covering common and rare PTCL entities
-
•
PTCL models genomically and transcriptionally recapitulate matched primary tumor
-
•
We identify pathogenic liabilities, effective combinations, and response predictors
-
•
We provide an innovative host-centric PTCL classification
Peripheral T cell lymphomas (PTCLs) are heterogeneous malignancies with limited therapeutic options. Fiore et al. introduce 102 patient-derived PTCL models to dissect pathogenetic mechanisms, identify intrinsic and host-mediated vulnerabilities, and implement innovative therapeutic approaches.
Introduction
In 2023, a total of 80,550 cases of lymphomas were diagnosed in the USA, with a staggering number of 20,180 deaths (https://seer.cancer.gov/statfacts/html/nhl.html). Peripheral T cell lymphomas (PTCLs) represent ∼15% of all lymphomas and comprise >30 different entities.1,2 PTCL patients display a remarkable clinical heterogeneity, with a 5-year overall survival (OS) ranging from 14% (adult T cell leukemia-lymphoma [ATLL]) to 70% (ALK+ anaplastic large cell lymphoma [ALCL]).3 Regrettably, chemotherapies (including anthracycline-containing regimens) have limited efficacy, and relapsed/refractory PTCLs experience a short OS (∼6 months),4 calling for effective agents and/or combinations.5 Improvements in PTCL classification, predictive biomarkers identification, and targeted agent development remain unmet medical needs.6
In recent years, multiple drugs were approved, and several are in clinical trials. Nevertheless, response rates remain disappointing (25%–29%), with progression-free survival < 4 months.6,7,8
This is mainly due to PTCL heterogeneity and rarity, as well as to the lack of informative models (only a few PTCL cell lines mainly corresponding to ATLL and ALCL subsets). Cell lines are unable to fully recapitulate the biology and therapeutic responsiveness of human cancers.9,10,11 Many of these limitations are shared by transgenic mice, including a few PTCL models.12,13,14,15,16
Patient-derived tumor xenografts (PDXs) can provide critical insight into overcoming treatment resistance, identifying targetable liabilities17,18,19,20,21,22 and microenvironment stimuli,23,24,25 and enabling pre-clinical trials.26,27 Despite limitations,28,29,30,31 PDXs are considered among the most informative tools to model human cancers.26,27,32,33 However, lymphoma PDXs remain poorly represented.17,34,35,36,37
Here, we describe an extensive library of PDXs corresponding to different PTCL entities. We show that these models (1) faithfully recapitulate the biological features and driver defects of their matched donor neoplasms, (2) allow the recognition of causative genetic defects and suitable dependencies, (3) underline the lymphoma-host dependencies and host-related refractory mechanisms, and (4) represent informative platforms to test established/innovative and cell-based therapeutic strategies. This repository will foster scientific discoveries and the development of therapeutic regimens tailored to molecularly defined subgroups, advancing personalized approaches for PTCL patients.
Results
Establishing a living PTCL PDX biorepository
We implanted 308 PTCLs from fresh or cryopreserved samples, yielding 102 patient-derived tumor models comprising 88 PDXs and 14 PDX-derived lines (PDX-Dlines; Figures 1A and 1B; Table S1) and 3,283 PDX cryopreserved seed samples (median passage: T4).
Figure 1.
Generation of PTCL PDX and PDX derivates
(A) Schematic representation of PDX generation and propagation strategies. Different primary sample sources and routes of implantation are annotated.
(B) Pie chart indicating the PTCL total and subtype-specific number of PDXs generated.
(C) PTCL PDX subtype-specific percentage of engraftment.
(D) Number of PDXs generated from naive or refractory patients for different PTCL subtypes.
(E) Time of engraftment (in days) of PDXs belonging to different PTCL subcategories along different rounds of propagation (T1 to T10). Error bars represent standard deviations.
(F) Representative MRI scanning of 4 different organs (lung, kidney, liver, and spleen) of an NSG mouse implanted with the different PDX. Arrows indicate lymphoma infiltration.
(G) Representative H&E staining of 4 different organs (lung, kidney, liver, and spleen) of NSG mice implanted with the AITL PDX model (magnification ×40).
(H) Flow cytometric analysis of EBV+ AITL PDX IL129A before (upper panels) and after CD19-ADC treatment (lower panels).
(I) Schematic representation of PDX-Dline generation strategies.
The biorepository represents the most common PTCL subtypes (Figure 1A). Higher engraftment rates were seen in relapsed/refractory PTCL (r/r, 62%; naive, 38%), with a 36% engraftment rate on average for the most common T/natural killer (NK) entities (PTCL not otherwise specified [PTCL-NOS], angioimmunoblastic T cell lymphoma [AITL], and ALCL). Also, rates were not linked to other parameters (Table S1). Rare entities engrafted less efficiently (Table S1,<15%) except Monomorphic epitheliotropic intestinal T cell lymphoma [MEITL] (Figures 1C and 1D). We also generated six PDXs from two longitudinal samples of the same patient. The engraftment time ranged from 3 weeks to 10 months (Figures 1E and S1A–S1H), remaining relatively stable along serial passages, except for AITL, which displayed progressively longer times over serial transfers (Figure S1B). Tumors implanted subcutaneously seldom homed to distant tissues (lungs, liver, and/or spleen), sometimes without a concomitant expansion at the implantation site (Figures 1F, 1G, and S1I). Seventy-three of 88 models propagated ≥2 serial passages without EBV+ (Epstein Barr virus) lymphoblastoid CD19+ B cells (EBV-LCLs). Some EBV+ LCLs (1%–90%) were detected (30 cases); primarily AITL (n = 10) (Table S1) and prominent expansions (>90%) were also seen (18 samples, 17% of all engraftments). EBV+ LCLs were present intratumorally in visceral tissues (i.e., kidney, spleen, liver, and lung) and, in most cases, expanded over serial passages (Figures 1H and S1J). EBV+ PDX and samples with no lymphoma expansion (6–9 months from injection) were failures (Figure S1J). Aiming to eradicate EBV+ LCLs, we treated six EBV+ PDXs with the anti-CD19 drug-conjugated loncastuximab tesirine (ADCT-402).38 The antibody successfully eradicated B cells, but only one PDX was established and serially propagated (interleukin [IL]129A AITL PDX, Figures 1H, S1K, and S1L). Similar data were obtained by treating EBV+ PDX in complement-proficient (Hc1) mice with rituximab (data not shown).
Lastly, we established fourteen continuous PDX-Dlines using cytokine-supplemented media (with/without IL-2 and IL15) or murine cancer-associated fibroblasts (CAFs; Figure 1I; Table S1).
PDXs maintain the immunophenotypic and TCR clonotype profiles of primary PTCLs
PTCL PDX histologically/cytologically resembled their matched donor samples (Figures 2A and S2A). In 29 cases, mostly AITL PDX, normal T and B cells (or EBV+ LCLs) were co-mingled with neoplastic elements, particularly in early passages (Figures 2B and 2C). PDX lineage fidelity and individual phenotypes were preserved, as demonstrated by immunohistochemistry and immunophenotyping (Figures 2D and S2B). Gene rearrangement analysis demonstrated that PDX and matched primaries share identical T cell receptor (TCR) DNA rearrangements (Tables S1 and S2; Figures 2E, S2C, and S2D). By total RNA sequencing (RNA-seq), primary and PDX displayed a rich TCR clonotype representation (Figures S2E–S2F). Explicit α and/or β TCR lymphoma clonotypes (>5% of the TCR) were documented in ∼50% of primary samples, with a higher clonal representation in PDX (Figures 2E and S2E–S2G).
Figure 2.
PDX faithfully mimics matched primary donor samples
(A) H&E staining of primary lymphomas (green frames) and matched PDX (yellow frames, magnification 40×).
(B) EBV in situ hybridization depicting EBV+ lymphoblastoid cells (EBER+) in AITL PDX.
(C) Multiparametric in situ imaging (MISI) of representative AITL lesions derived from diagnostic (lymph node) and patient-matched PDX (lung).
(D) Pie graph reporting the expression of 26 immune-histochemistry (IHC) markers in primary and PDX (T1 to T16) samples. Red: highly expressed, green: low expressed.
(E) α/β TCR clonal representation of primary and PDX, along serial passages.
(F) EBV positivity among PTCLs.
(G) TCR repertoire against EBV peptides and their mismatched sequences compare to known reference sequences.
(H) Prediction binding of EBV peptides to MHC class II determinants.
(I) In vitro competitive binding assay of EBV tetramers to recombinant DRB1.
Since antigen-driven TCR engagement facilitates cell growth and treatment resistance,39 we assessed the TCR usage and antigen specificity (https://vdjdb.cdr3.net/). We observed a relative over-representation of selected VDJ (TRA-V29DV5, V9-2, and V13-1), particularly in EBV+ AITL (Figure S2H). Notably, EBV transcripts were identified (Figure 2F), corresponding to both lytic and latent genes (Figure S2I) and, seldom, to HTLV-1 and HHV6 transcripts (Figure S2J).40 Remarkably, individual primary/PDX displayed dominant TCR clones expressing canonical or mismatched TCR CDR3 motifs known to bind EBV peptides (Figures 2G, S2K, and S2L; Table S2). To extend this prediction, we estimated the major histocompatibility complex (MHC) class II binding for multiple EBV peptides (Figure 2H). Next, we executed a competitive in vitro binding assay showing a high binding affinity of Epstein barr Nuclear Antigen (EBNA)-3B/4 peptides (IC50: 0.08 μM) to recombinant DRB1 (IL36, Figure 2I).
Lastly, we detected explicit immunoglobulin H (IgH) clones, at very low frequency in primary tumors (Figure S2M). Exceptions included rare primary AITL and EBV+ PDX (Figure S2N).
PDXs preserve the transcriptomic landscape of primary lymphomas
We first compared the transcriptomic profiles of primary (n = 79) and matched PDX (n = 140; Table S2) by principal-component analysis (PCA), demonstrating a partial overlap (Figure S3A), likely due to the tumor content (Figures S3B and S3C) and host human cells (Figures S3C–S3E). Hence, we performed a surrogate variable analysis (see STAR Methods) and established distinct clusters corresponding to ALK+ ALCL, ALK− ALCL, and PTCL-NOS/AITL, demonstrating a close correspondence of primary and PDX (Figures 3A, 3B, and S3F). PTCL-NOS and AITL were stratified using Gene Ontology (GO) enrichment, differential expression, pathway analyses, and publicly available signatures (Figures 3C and S3G–S3I), confirming known transcripts in different subtypes (Figure S3H). As selected biomarkers distinguish PTCL subtypes,41,42 we established PDX classifiers for each subgroup enriched by known differentially expressed genes (Figure S3J). The PDX-Dlines also showed close transcriptional signatures to matched donor PDX (Figure S3K).
Figure 3.
PDX maintains the inter- and intratumoral heterogeneity of matched lymphoma
(A) PCA of PDX and primary lymphoma-matched samples (AITL, PTCL-NOS, and ALCL) based on the bulk RNA expression levels excluding non-lymphoma reads in primary samples.
(B) Heatmap and unsupervised hierarchical clustering based on 1,000 top differentially expressed genes of PDX and primary lymphomas belonging to the main 4 PTCL subcategories (AITL, PTCL-NOS, ALK+ ALCL, and ALK- ALCL).
(C) Supervised hierarchical clustering of primary and PDX based on 12 known publicly available signatures stratifying different PTCL entities (A: PMC2817630_AITL, B: PMC2817630_ATLL, C; PMC4014836_TBX21/GATA3, D: PMC20159827_ALK+, E: PMC2817630_ALK+, F: PMC4014836_ALK+/−, G: PMC4014836_AITL, H: PMC4014836_ATLL, I; PMC4014836_ATLL, J: PMC2817630_CT_PTCL, K; PMC6161771_DUSP22, and L: PMC4014836_ENKTL).
(D) Uniform manifold approximation and projection (UMAP) clusters annotation based on single-cell RNA-seq expression of PTCL-NOS and AITL (IL-2 and IL138A) and ALCL (IL69, IL79 and IL89) PDX.
(E) Dot plot representation of top gene transcripts in each UMAP cluster of the PDX models sequenced by single-cell RNA-seq.
(F) UMAP cluster annotation based on single-cell RNA-seq expression (IL138A primary and T3 PDX model). Cell types have been annotated on the right part of the graph.
(G) Hallmark analysis of selected differentially expressed pathways among three tumor clusters of IL138A primary and PDX, based on single-cell RNA-seq expression data. Cluster 0 was present in both primary and PDX, while clusters 1 and 2 were enriched in IL138A PDX vs. the correspondent primary.
(H) Heatmap reporting fusions of primary and PDX samples belonging to different PTCLs. Only chimeras with a pathogenetic score ≥0.7 are depicted.
(I) Antitumoral effect of AZD-6244 in TO-ALCL-Belli PDX model (n = 8 mice/group). Error bars represent standard deviations.
(J) Circle pot depicting fusion landscapes of IL-2 and IL19 primary and PDX samples.
By single-cell RNA sequencing (scRNA-seq) (5 PDXs, Table S2), models were individually segregated at cluster resolution, even within the same entity (i.e., ALK+ ALCL, Figures 3D and 3E). Tumor clusters were common between primary and PDX (#1 and #2) (Figures 3F and 3G), and normal human stromal cells were depleted in PDX (#6 and #8, populated by normal CD8+ T cells and monocytes, respectively) (Figures 3F and S3L).
Fusion transcripts are common in PTCL and typical to histologic subtypes.2 We thus searched for chimeric transcripts (see Methods; Table S3) and identified 4,022 gene fusions across 206 samples. Previously identified fusions were annotated,43,44,45 as well as unknown putative tumorigenic fusions involving a variety of T cell genes (ACADVL-VAV1, MAZ-NF1, VASP-PPP2R1A, TET3-IMMT, MYL3-SETD2, TOX-MYBL1, IL17RA-RP11-363L24.3, and SMG1-NFATC3) (Table S3; Figure 3H). In ACADVL-VAV1, ACADVL was fused in-frame, while VAV1 translated to a truncated protein lacking its negative regulatory domain (Figure S3M), reminiscent of other oncogenic VAV1 fusion proteins.46,47 Since Ras signaling can contribute to PTCL pathogenesis,48,49,50,51 we validated the MAZ-NF1 fusion of an ALK− ALCL PDX. The predicted outcome was a truncated NF1 protein lacking activity (Figures S3N–S3P). Meanwhile, primary and PDX lost the second NF1 allele (Figure S3P), leading to the deregulated activation of Ras. Of note, we observed the loss of ERK1/2 phosphorylation (Figure S3Q) and the improved PDX outcome in vivo (Figure 3I) upon selumetinib treatment (AZD-6244, an MEK inhibitor), a compound with limited activity in other PDX-Dlines (Figure S3R).
Since fusion transcripts are informative biomarkers of disease identity52,53 and serve as surrogates for tracing clonal evolution,53 we explored their landscape in two models (IL-2 and IL19), derived from the same donor at different time points (Figures 3J, S3S, and S3T). PDX displayed several undetectable fusions in primary samples (IL-2, n = 62, and IL19, n = 68), some co-shared (n = 36). By quantitative reverse-transcription PCR (RT-qPCR - n = 9) and Sanger sequencing (n = 3, Figures S3U and S3V), we confirmed selected fusions in PDX and primary samples (Figure S3V). Remarkably, emerging fusions in the relapsed sample (primary IL19) and corresponding PDX suggested the occurrence of clonal trajectories in part shared and maintained along the diagnostic, PDX samples, and even PDX-Dlines (Figures S3U and S3V).
PTCL PDXs retain pathogenetic drivers and inter- and intratumoral heterogeneity
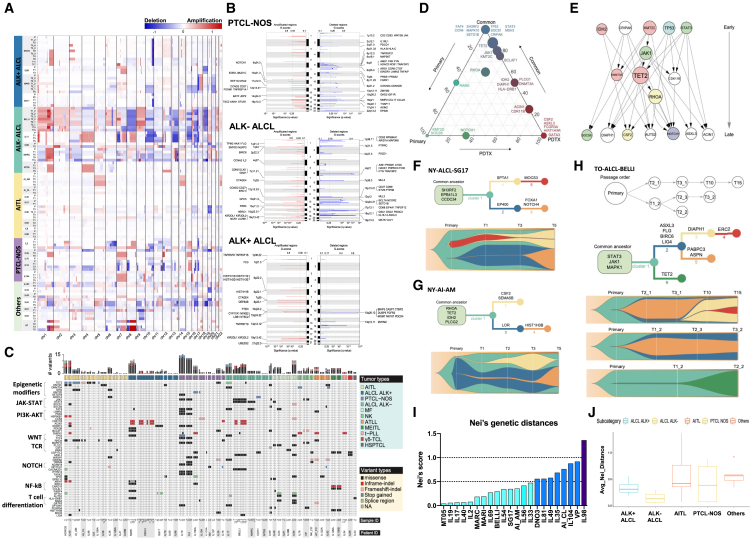
We performed whole-exome sequencing (WES) in 223 samples (34 primary, 29 normal, and 160 PDX samples and PDX-Dlines), derived from 49 different models (Table S2). Having assessed the contribution of human and mouse reads (Figure S4A), copy-number alteration (CNA) demonstrated significant overlaps between primary and corresponding PDX (Table S4; Figures 4A and S4B). Globally, PTCL-NOS and ALK− ALCL displayed a higher degree of DNA structural alterations (Figure 4B), including known abnormalities (e.g., 6q21 and 1q+ or 3q31.3+ in ALK− ALCL) and defects associated with pathogenic alterations (e.g., PRDM1 and MIR17HG), than other histology.54,55 ALCL displayed specific gains at 1p36.22 and 19q13.42, occurring in regions harboring pathogenetic TNFRSF8 (CD30) and KIR2FDL1/KIR3DL2 genes.56,57,58
Figure 4.
PTCL PDX models mutational landscape and clonal evolution
(A) Global copy-number variation (CNV) analysis of primary and PDX along propagation.
(B) Chromosome view of genes included in the recurrent deleted or amplified genomic regions in PTCL-NOS and ALCL (ALK+ and ALK−).
(C) Mutational landscape of PTCL primary and PDX samples assessed by WES. Variant sites with read depth lower than five are marked as NA. For the sample ID, “P” stands for primary tumors.
(D) Ternary plot of mutation frequency in recurrently mutated genes, comparing primary tumor-specific (left, green), PDX-specific (right, red), and shared (top, blue) alterations. The size of each node represents the mutation frequency.
(E) PDX tumor evolutionary directed graph of gene mutations. Arrows show the order in which mutations occur. The size of each node corresponds to the frequency of mutations.
(F–H) Tumor evolution models of NY-ALCL-SG, NY-AI-AM, and TO-ALCL-BELLI PDX models. Fish plots (bottom panels) show dynamic changes in CCF of each mutation cluster along serial passages, as depicted in the inferred phylogenetic trees (top panels).
(I) Nei’s genetic distance indicates the global evolution score of PDX models.
(J) Nei’s genetic distance indicates the global evolution score of PDX derived from different PTCL entities. Error bars represent standard deviations.
We found a high concordance of single-nucleotide variants (SNVs) and insertion or deletions (indels) between primary tumors and matched PDX (Figures S4C and S4D; Table S5) with a higher median variant allele frequency (VAF) in PDX (50% vs. 40%, respectively, p < 2.2e−16, t test; Figure S4E). Globally, 3,430 non-synonymous somatic variants were recognized (Table S5), mostly missense (77%), 11% splice site, 4% stop-gain, 4% in-frame, and 4% frameshift. A total of 1,582 were classified as pathogenetic SNVs. These involved chromatin modifiers, JAK-STAT, and TCR-associated genes (e.g., TET2, DNMT3A, JAK1, STAT3, RHOA, TP53, and NOTCH1).37,59,60,61,62 Previously undescribed putative tumorigenic variants (e.g., DIAPH1, FAT4, CRIPAK, SH3RF2, and BCLAF163,64,65,66,67) were detected (Figure 4C). Oncogenic drivers were enriched in AITL (TET2 and RHOA),68 ALK− ALCL (JAK1 and STAT3),69 and mycosis fungoides [MF] (PLCG1).70 Most mutations were faithfully shared between primary tumors and along PDX passages. Conversely, some were exclusive to either primary (e.g., KMT2D) or PDX (e.g., CSF2, GATA3, and ASXL3; Figure 4D) or emerged along serial PDX passages (AUTS2 and CSF2, Figure S4F). Higher mutational burdens were observed in ALK− ALCL, PTCL-NOS, and MF (Figure S4G). Lastly, we screened 104 normal/primary/PDX/PDX-Dline samples by deep sequencing and annotated 537 mutations in 450 pathogenetic genes (Table S2). When we compared WES and deep sequencing analyses (data not shown), we observed >90% concordance with an increased VAF of selected alterations in propagated PDX (Figure S4H).
To explore the evolutionary mutation trajectories, we built a tumor evolutionary directed graph71,72 showing that ancestor mutations usually occurred in STAT3, TP53, IDH2, and KMT2C, followed sequentially by those in JAK1, CDK11B, TET2, and RHOA. Mutations in ACIN1, NOTCH1, DIAPH1, and CSF2 were mostly acquired along PDX propagation (Figure 4E). We then implemented a clonal evolution analysis on 29/49 models using cancer cell fraction (CCF) estimation by ABSOLUTE.73 We classified a clonal mutation if the CCF was >0.85 with a probability >0.5 and subclonal otherwise. We identified 2,103 clonal and 1,544 subclonal mutations (Table S6) and constructed evolution models (e.g., T1-T3-T5 and, for selected cases, up to T15).74 All models showed a major cluster of co-shared mutations among primaries and PDX (Figures 4F–4H and S4I–S4K), with dominant clones preserved over propagations. Minor subclones branched and expanded along PDX propagations, with some clonal competition (Figures 4F–4H). Finally, we computed Nei’s genetic distances on 22/29 models to estimate clonal drifting along propagation (Figure 4I, 4J, and S4L)75 and defined three subgroups based on low/medium/high evolution rate (average Nei’s score): 14/22 (64%) low (<0.5), 7/22 (32%) median (>0.5 and <1), and only 1/22 (4%) high score (>1). This confirmed the overall stability of PDX compared to different systems (e.g., glioblastoma; Figure S4M).
In sum, PDXs maintain primary-matched pathogenetic drivers and degrees of lymphoma heterogeneity.76
PTCL PDXs recapitulate primary and host microenvironment interactions
To explore the nature of the PTCL tumor microenvironment (TME), we took advantage of our methodology24,77 to extract functional signatures from the crosstalk of TME with cancer cells (functional gene expression signatures [FGESs]) from bulk RNA-seq. We first used 24 FGESs to virtually reconstruct the TME of 845 PTCLs from 16 public datasets and our cohort. We separated them into four major clusters representing “lymphoma microenvironment” categories (Table S7; Figures 5A and S5A–S5E): “B cell rich” for the abundance of B cells and B cell trafficking FGES; “mesenchymal” for over-representation of FGES linked to stromal cells, extracellular matrix (ECM), and ECM remodeling; “inflammatory” for the presence of FGES related to macrophages and NK cells; and “depleted” that overall had the lowest representation of TME FGES. PTCL subgroups were distributed across the four TMEs without specific associations, although each group displayed distinct signatures and bore different genomic defects (Figure S5F). A survival analysis on two distinct PTCL patients’ cohorts (n = 253) showed that the “depleted” TME was associated with poorer prognosis (uncorrected p = 0.005) (Figure 5B). Remarkably, the only FGESs overrepresented in the “depleted” TME cases were related to “Th2-ILs and GATA3 activation” and “proliferation rate” (Figure 5A) and exhibited the highest proportion of tumor cells as determined by mutational load (Figure S5G, p < 0.01 vs. the other categories). Similar data were recently described in an independent PTCL cohort.78 Next, we performed a multiplex imaging analysis of primary PTCL using antibodies recognizing different subtypes of T cells, macrophages, and stromal elements, confirming the RNA deconvolution predictions (Figures 5C and S5H).
Figure 5.
The microenvironment of primary and PDX defines distinct subgroups of PTCLs
(A) Heatmap of the activity scores of 20 FGES and 4 signaling pathways (x axis) denoting four major TME clusters of primary PTCL (n = 845). In each dataset, signatures were median scaled using median and MAD (median absolute deviation) calculated only for samples with AITL or PTCL-NOS. MFP (microenvironment functional phenotype) portraits were predicted by Louvain clustering (with a threshold of closest points 0.25) within 20 signatures. Samples were sorted by MFP and by diagnosis and for each MFP and diagnosis by proliferation rate increasing. The bottom four molecular pathways were calculated by Progeny.
(B) Kaplan-Meier models of OS according to the PTCL TME category.
(C) TME annotation by multiplex analysis of PDX.
(D) Heatmap of the activity scores of 20 FGES (x axis) denoting four major TME clusters of PDX; signature scores (calculated by single sample Gene Set Enrichment Analysis - ssGSEA - algorithm) were median scaled for each biopsy site separately taking median and MAD only from AITL and PTCL-NOS samples. Oncoplot below the heatmap depicts mutations, ALK, and EBV status. Color palettes on the top indicate MFP, biopsy site, T-cell phenotype, and diagnosis for each sample.
(E) Left: Sankey plot showing changes in T differentiation throughout primary and three passages of PDX. Right: plot showing changes in MFP subtypes throughout primary and three passages of PDX.
(F) Proportion of macrophages M1 or M2 enriched in PDX by FGES. Error bars represent standard deviations (∗p < 0.05; ∗∗p < 0.001; ∗∗∗p < 0.0001).
(G) The proportion of myCAF enriched in selected PDX subtypes by FGES. Error bars represent standard deviations (∗p < 0.05; ∗∗p < 0.001; ∗∗∗p < 0.0001).
(H) The proportion of iCAF enriched in PDX by FGES. Error bars represent standard deviations (∗p < 0.05; ∗∗p < 0.001; ∗∗∗p < 0.0001).
(I) Barplot of apoptotic lymphoma cells cocultured with and without stromal cells (STCs). Data are representative of three replicates. Error bars represent standard deviations.
(J) Gene Ontology analysis indicates the biological processes enriched in educated vs. not-educated SCTs. Error bars represent standard deviations.
(K) Unsupervised hierarchical clustering of the top 100 differentially expressed genes in not-educated (cultured in vitro >3 days) and (re)educated (freshly isolated or co-cultured in vitro with PTCL cells >3 days) STCs isolated from PDX.
(L) Percentage of viable IL-2 PDX cells cultured in stress conditions alone (red bar) or cocultured with STCs isolated from different PDXs. Data are representative of three replicates. Error bars represent standard deviations.
(M) Percentage of viable MT05 PDX cells cultured in stress conditions alone (red bar) or cocultured with STCs isolated from different PDXs. Data are representative of three replicates. Error bars represent standard deviations.
(N) Barplots reporting the delta of the specific cell death of PDX-Dlines (IL-2 and IL142A) exposed to 40 drugs with or without STCs (72 h at 1 μM).
(O) Barplot showing viable PTCL PDX cells cocultured with STCs or cultured alone in the presence of targeting agents (72 h). Data are representative of three replicates. Error bars represent standard deviations.
We extended this approach to PDX models, using converted mouse FGES (mFGES), and showed the same four TME categories of primary tumors (Figures 5D and S5F–S5N). Using only human reads, we performed a clustering analysis of the PTCLs into 4 functional “intrinsic” T cell phenotypes: Th1 (mostly AITLs and PTCL-NOS), Tfh (AITL), Th2 (PTCL-NOS carrying JAK-STAT mutations), and cytotoxic (mainly ALCL) (Figure S5F). The Th1 and Tfh group frequently displayed TET2, RHOA mutations, and detectable EBV transcripts. The cytotoxic group had the lowest TCR signaling rate and few normal T cells (Figure S5K), in line with the ALCL low TCR signaling.79 Most samples with high TCR signaling displayed TET2/DNMT and/or RHOA/PCLG1 defects80; meanwhile, ∼50% of those lacking them were EBV+. TCR-negative samples were conversely enriched in JAK1/STAT3 mutants and/or clustered among ALCL.2,79 We next showed that TME cell populations were mostly conserved along PDX passages (Figure 5E), particularly in ALK+ ALCL, which maintained their cytotoxic phenotype, and for most Th2 PDX. Nevertheless, changes were observed, suggesting some plasticity (i.e., “mesenchymal” and “B cell-rich” TMEs; Figures 5E and S5I). A transition to a “mesenchymal” TME was seen in ALCLs whose tumor content rapidly increased after engraftment. Other PDXs displayed a progression to a “B cell-rich” TME (e.g., NY-PTCL-CR, IL33, and IL98), likely driven by an increased number of EBV-LCLs (Figures S5I and S5J).81,82
Additionally, we interrogated murine tumor-associated macrophages (TAMs) and CAFs, which increased along PDX passages (Figure S5M). Both M1-like and M2-like TAMs expanded in PDX, with an increased M2/M1 ratio (primary:1.53 vs. PDX: 2.27; Figure 5F). Regarding CAFs, signatures corresponding to “myCAF” and “iCAF” (Table S7) were also enriched (Figures 5G, 5H, and S5N).
As a functional validation, murine PDX stromal and tumor endothelial cells improved the survival of cocultured lymphoma cells (Figures 5I and S5O).83,84 Cocultured CAFs upregulated biogenesis, migration, cell mobility, and DNA replication pathways (Figure 5J), mimicking the phenotype of freshly isolated mesenchymal stromal cells (MSCs; Figure 5K), a phenotype partially lost when cultured alone. Remarkably, freshly isolated CAFs from matched tumors or in vitro re-educated-CAFs more efficiently rescued PDX under serum deprivation (Figures 5L and 5M). Finally, PDX-CAFs improved the viability of 2 models (IL-2 and IL142A) in co-culture drug screening platforms (Figures 5N and 5O), including compounds targeting PTCL driver pathways (Figure 5O). This effect was only seen with lymphoma-matched PDX-MSCs (Figure S5P), suggesting a lymphoma-specific education. We next identified putative pathways that mediated the host rescue, as depicted in Figure S5Q, where CAF rescue was partially abrogated with crizotinib (navitoclax/ABT263, belinostat, etc). Finally, we found that mouse mesenchymal cells (MS-5) protected ALCL cell lines from brentuximab-vedotin (BV)-induced death (Figure S5R).
Therapeutic responses are assessed in PTCL PDX
We first examined the therapeutic prediction of six PDXs (3 ALK+ ALCLs, 1 PTCL-NOS, and 2 ALK− ALCLs) performing a high-throughput in vitro drug screening (Figure S6A) targeting ∼634 proteins (Table S8; Figure 6A), demonstrating a high reproducibility among replicates (Figure S6B) and along serial propagations (Figures 6A–6C and S6C). Therapeutic responses differed according to the subtype (Figures 6B and 6C). Most compounds had little effect. However, a subgroup of 19 drugs showed higher efficacy across samples (Figure 6C). We then correlated the gene expression profiles with cell viability after drug exposure and identified predictive signatures to belinostat (Figures S6D and S6E) and ruxolitinib (R = 0.97, p = 0.033) (Figures 6D–6G, S6D, and S6F). Next, we expanded the screening to PDX-Dlines with 53 drugs, including the 30 most active compounds within the 433-drug library and 23 drugs from clinical trials (Table S9; Figure 6H), establishing dose-response curves (Figures 6I and S6G). Also, PDX-Dlines displayed individual patterns of responses to the ALK inhibitor (ALKi) crizotinib (TO-ALCL-DN03, ALK+ ALCL) and the JAK inhibitors (JAKi) ruxolitinib, tofacitinib, and cerdulatinib (IL-2, JAK1 mutant; Figure 6H), in line with their genetic alterations. Conversely, cytotoxic chemotherapeutics (daunorubicin, SN38, and vincristine), HDAC inhibitors (romidepsin, and panobinostat), a survivin inhibitor YM155, proteasome inhibitors (bortezomib and CEP18870), an aurora-kinase inhibitor (tozasertib), and a CDK9 inhibitor (AZD-4573) were pan-active, even at low concentrations.
Figure 6.
Ex vivo PDX drug responses
(A) Heatmap showing the magnitude of the cross-correlation of 6 PDX freshly isolated cells exposed to the drug library.
(B) Principal-component analysis (PCA) of 19 PDX freshly isolated cells based on the responses to 433 drugs. Circled dotted lines group together samples of PTCL subtype.
(C) Heatmap showing the responses of 6 PDX models (19 freshly isolated cell samples) to 433 drugs. Dendrograms on the left and bottom show unsupervised hierarchical clustering of drugs and PDX along the axis of maximum variation (ward) for the Euclidean distances. The dot plot denotes the average drug viabilities per PDX across 433 drugs (top). Dot plot shows the average sample viabilities per drug (right).
(D) Dot plots showing the correlation between the expression levels of JAK1 and JAK2 across PTCL subtypes with cell viability after ruxolitinib treatment (72 h, 1 μM). The correlation coefficients and p values are indicated.
(E) Heatmap and unsupervised clustering depicting the gene expression within the JAK-STAT pathway. Genes were selected based on the correlation between the expression and viability of samples treated with ruxolitinib (1 μM, 72 h). The viability values are indicated in the upper color bars.
(F) Heatmap and unsupervised clustering depicting the gene expression from a regression analysis obtained by modeling the cell viabilities as a function of the PTCL subtypes plus each gene expression.
(G) Dot plot showing the predicted vs. actual cell viabilities, with correlation and p value across PTCL subtypes. The prediction derives from the regression analysis in Figure 5F.
(H) Heatmap displaying the response of five PDX-Dlines to 40 compounds. Specific cell death is reported in percentage.
(I) IC50 assessment in five PDX-Dlines treated in vitro with increasing concentrations of compounds (day 3 and 6).
(J) Boxplot indicating the predicted synergy score by the DeepPTCL algorithm for the indicated drug combinations across PTCLs. Error bars represent standard deviations.
(K) Percentage of viable IL-2 and IL142A PDX-DLines cultured in the presence of the indicated concentrations of duvelisib and cerdulatinib (IL-2) or duvelisib and venetoclax (IL142A) for 72 h. Data are representative of three replicates. Error bars represent standard deviations.
Seeking drug combination candidates, we developed a deep learning-based algorithm named DeepPTCL (see Methods). Having demonstrated a consensus between the The Cancer Genome Atlas cell lines and PTCL (see Methods; Figure S6H), we screened 8 of the most effective drugs for synergies (irinotecan, romedepsin, duvelisib, pralatrexate, AZD-4573, cerdulatinib, azacytidine, and crizotinib) on PDX-Dlines (IL-2, IL89, IL142A, TO-ALCL-DN03, and TO-ALCL-Belli). DeepPTCL predicted 6 top synergy combinations with duvelisib (Figure 6J), some further validated in vitro (duvelisib/cerdulatinib in JAK1mut IL-2 PDX-Dline and duvelisib/venetoclax in IL-2 and IL142A PDX-Dlines, carrying PTEN and TP53 deletions) (Figure 6K; Figure S6I). Lastly, we linked responses of representative combinations to the mesenchymal TME within the FGES subtypes (Figure S6J).
PTCL PDX pre-clinical trials in vivo
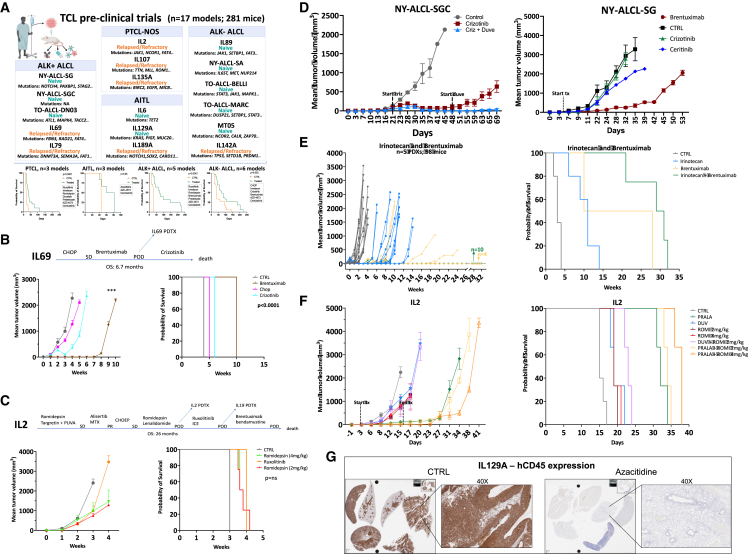
To assess responses to standard and innovative drugs/combinations, we selected 17 PTCL PDXs (10 naive and 7 refractory; Figure 7A; Table S9).
Figure 7.
The mouse hospital and pre-clinical trials
(A) PTCL pre-clinical trials overview and Kaplan-Meier plots representative of the overall survival of PDX models.
(B) Comparison of IL69 patient and matched PDX responses to CHOP, brentuximab, and crizotinib (n = 8–10 mice/group). Top panel: IL69 patient clinical history. Error bars represent standard deviations. p values were estimated with adjusted t test (∗p < 0.05; ∗∗p < 0.001; ∗∗∗p < 0.0001). Kaplan-Meier curve of the OS (right panel, log rank test, p < 0.0001).
(C) Comparison of IL-2 patient and matched PDX responses to ruxolitinib and romidepsin (n = 8–10 mice/group). Top panel: IL-2 patient clinical history. Error bars represent standard deviations. Kaplan-Meier curve of the OS (right panel, log rank test, p = ns: >0.05).
(D) Left panel: antitumoral effect of crizotinib alone or in combination with duvelisib in NY-ALCL-SGC PDX (n = 8–10 mice/group). Right panel: antitumoral effect of crizotinib, brentuximab, and ceritinib in NY-ALCL-SG PDX (n = 8–10 mice/group). Error bars represent standard deviations.
(E) Antitumoral effect of irinotecan, brentuximab, or combination in ALCL PDX (MT05: cutaneous ALCL - cALCL-, IL69, DN03; IL79: ALK+ ALCL; IL-2: PTCL-NOS). Kaplan-Meier curves of the OS (right panel, log rank test, p < 0.0001). Individual biological and technical replicates are depicted as single lines.
(F) Antitumoral effect of pralatrexate, duvelisib, and romidepsin or combinations in IL-2 PTCL-NOS PDX (n = 8–10 mice/group). Error bars represent standard deviations (∗p < 0.05; ∗∗p < 0.001; ∗∗∗p < 0.0001). Kaplan-Meier curves of the OS (right panel, log rank test, p < 0.0001).
(G) hCD45 IHC staining of IL129A PDX treated with vehicle or azacytidine. Left panels: mice organs (lungs, kidney, spleen, liver, and heart). Right panels: lungs (40x).
First, we tested whether PDX recapitulated patients’ responses (Table S10; Figures 7B, 7C, and S7A–S7C) to (1) CHOP, demonstrating that TO-ALCL-DN03 responded in line with the matched patient (partial response, Figures S7D–S7F) while MT05 and TO-ALCL-Marc were refractory as the donor patients, and (2) targeted agents. These latter experiments showed that JAK1mut IL-2 PDX was refractory to ruxolitinib and romidepsin, as its corresponding patient (Figure 7C), and the NPM-ALK+ IL69—derived from a patient refractory to CHOP, BV, and crizotinib—did not also show significant responses (Figure 7B). Similar responses were documented in the IL79 ALK+ ALCL model (Figure S7G). Conversely, the ALKi-naive TO-ALCL-DN03 was eradicated by crizotinib (Figure S7H), while NY-ALCL-SG showed little-to-no response to crizotinib and ceritinib (respectively) but was partially controlled by BV (Figure 7D). For the ALKi-naive NY-ALCL-SGC, we observed a significant response to crizotinib followed by a relapse; this latter phenotype was controlled by the duvelisib-crizotinib combination (Figure 7D).85 As STAT3 powers some ALK− ALCL,69 we treated a naive STAT3+ ALCL PDX with baricitinib, a JAK1/JAK2 inhibitor, using two different dosing schedules. Growth inhibition was partially achieved in BID-treated mice, demonstrating that prolonged and significant suppression of pSTAT3 is required for improved clinical outputs (Figure S7I).
Afterward, we harnessed multiple PDX to design precision-medicine-driven pre-clinical trials by integrating phenotypic, genomic, and drug screening data. We chose a debulking approach using irinotecan, which was effective in the in vitro screening (Figure 6C), followed by chemo-free approaches (Figures 7E and S7J). Irinotecan has some activity in PTCL patients86 and in relapsed/refractory non-Hodgkin patients.87,88,89 In detail, CD30+ ALCL PDX (IL69, IL79, MT05, and DN03) were challenged with irinotecan and BV (Figure 7E), while the IL-2 PTCL-NOS PDX was treated with irinotecan and ruxolitinib (Figure S7J). Combinations either eradicated and/or yielded improvement in survival. Finally, we tested effective in vitro drugs (Figure 6H), either as single agents or in combinations (Figures 7F, S7K, and S7L). Pralatrexate proved to be the most effective, improving OS and, in some cases, leading to lymphoma eradication as a single agent (IL142A, Figure S7K), or in combination with romidepsin (IL-2, Figure 7F) or duvelisib (IL107, Figure S7L). Conversely, azacitidine (Figures 7G and S7M)90,91 decreased lymphoma growth (Figures 7G and S7M–S7P) and prolonged survival (Figure S7P). We next combined CDK9 inhibitor (AZD-4573)92 with cerdulatinib,93 following a 1 × 1 × 1 pre-clinical design,26 in 9 PDXs (Figure 8A). As a single agent, cerdulatinib showed a modest effect in 3/9 models and significant readouts in only 2/9 models. AZD-4573 was more potent, with a modest effect in 2/9 and superior response in 4/9 models (Figures 8A–8C and S8A). Their combination yielded improved responses in 5/9 models, extending survival (pairwise log rank p = 0.019) (Figures 8C, S8A and S8B). By bulk RNA-seq, we observed that non-responders had an upregulation of TCR signaling, and conversely, responders were enriched in genes regulating migration, cytoskeleton, and cell interactions (Figure 8D).
Figure 8.
PDX pre-clinical trials support the implementation of drug combinations and immune-based regiments
(A) Swimmer plot of PDX models (n = 9 and 36 mice) treated with vehicle, cerdulatinib, AZD-4573, or combination.
(B) Barplot depicting PDX tumor size across time points (vehicle, cerdulatinib, AZD-4573, and combination). p values were calculated with one-way ANOVA with adjustment for multiple comparisons ∗: p < 0.05.
(C) Kaplan-Meier plots of the global OS of PDX models (n = 9 and 36 mice; log rank test, p = 0.017).
(D) Heatmap depicting the top differentially expressed genes in PDX model responders and not-responders to AZD-4573 in vivo treatment.
(E) Flow cytometry analysis of TO-ALCL-DN03 (above panels) and IL-2 (below panels) PDX-Dlines cocultured with CART30 cells at the indicated target (red dots)-to-effector (green dots) ratio.
(F) Antitumoral effect of CART5 cells alone or combinations with nivolumab in IL-2 PTCL-NOS PDX (n = 6–10 xenografts/group). Error bars represent standard deviations.
(G) Antitumoral effect of CART30 cells alone or combinations with nivolumab in NY-ALCL-SG ALK+ALCL PDX (n = 6–10 xenografts/group). Error bars represent standard deviations.
(H) Detection of untransduced - UTD -and CART30 within the peri-tumor and tumor masses (CART30 is depicted in green and NY-ALCL-SG cells in red).
(I) Multiparametric analysis demonstrates the positive PDL1 expression of NY-ALCL-SG (red color), and CD2 (green) and PD1 (low/partial white) of CART30 cells.
Lastly, we assessed the efficacy of CAR-T in PTCL PDX, a still largely unexplored field.94,95 We took advantage of two CART products specifically targeting CD30 or CD5 (CART30 and CART5, respectively), the latter engineered to lack endogenous CD5 expression, avoiding fratricide effects and with enhanced antitumor activity (see Methods96). Both products were effective in vitro (CART5 against IL-2, a PTCL-NOS, and CD5+/CD30− and CART30 against TO-ALCL-DN03, an ALK+ALCL, and CD30+/CD5−) (Figures 8E, S8C and S8D). In vivo, CART5 controlled lymphoma growth, especially in combination with nivolumab (Figure 8F). Meanwhile, CART30 or CART30/nivolumab combination showed a partial effect (Figure 8G). This was likely due to a defective intratumoral infiltration (Figures 8H and S8E) and disrupted crosstalk, as shown by multiplex imaging. Indeed, ALCL cells were strongly PDL1+, largely excluding PD1+ and EOMES+ CART at lymphoma periphery (Figure 8I) and distant locations (i.e., spleen; Figure S8F).
Discussion
This study establishes the largest available PDX biorepository for PTCL, offering a robust pre-clinical resource to study tumor evolution, drug resistance, and personalized therapies. PDXs faithfully replicate primary tumor characteristics, including histopathology, clonality, genomic, transcriptomic, and drug susceptibility, making them an invaluable resource for understanding PTCL pathogenesis.
PDX fidelity and drifting along propagation are still a matter of debate.30,33,97 Here, we proved that PDXs closely matched primary samples, displaying identical TCR rearrangements and retaining the same driver mutations/copy-number variation (CNV) and gene expression patterns. Nevertheless, distinct subclones were detectable with the acquisition of non-random defects in a stepwise fashion (i.e., loss of TET2 and DNMT3a) and the acquisition of mutations (i.e., RHOA, IDH2, or NOTCH1/4 in ALCL and AITL PDX). This supports the model of stepwise T cell transformation, defining preferential trajectories/pathways driven by intrinsic defects. These findings feature the relevance of PTCL PDX to inform the potential evolutionary trajectory of human tumors. Along the same lines, we discovered genomic aberrations (e.g., ACADVL-VAV1 and MAZ-NF1) converging on specific pathways and propelling T cell transformation. These findings support the implementation of agents selectively targeting downstream effectors (STAT3, IRF4 PROTACs, etc.). Strikingly, PTCL PDX and PDX-Dlines maintained a significant subclonal heterogeneity, a feature often lost by conventional cell lines, providing a better representation and higher predictive power.17,98 We took advantage of this predictive potential by comparing, via a deep learning model, PTCL PDX transcriptional signatures with those of the Cancer Cell Line Encyclopedia, to in silico predict and in vitro validate drug combinations.
Personalized treatments require cancer genomic stratification,99,100 providing a more granular landscape and pinpointing the role of the microenvironment.24,101 Here, we proved that PTCL and matched PDX can be stratified by microenvironment functional signatures (FGESs) derived by lymphoma-host cognate interactions. This stratification is of prognostic relevance, with the “Th2” subgroup (so-called Th2/GATA3, over-activating the PI3K pathway) bearing the most unfavorable outcome. Similar data were recently presented in an independent cohort.78 Strikingly, despite the paucity of the PDX microenvironment, the host elements’ compositions somehow recapitulated the PTCL landscapes, supporting a model predicting the education of the host by lymphoma elements.102,103,104,105 PDX cells could also instruct stromal remodeling in vitro, establishing prosurvival niches with drug-counteracting capabilities. This model allows the functional validation of host-mediated protumorigenic mechanisms and testing of ad hoc regimens strategies.106,107,108 Considering the limited array of intrinsic druggable liabilities, we believe that targeting the lymphoma microenvironment will become of pivotal importance in future studies.
Finally, PDXs are emerging as a powerful tool in clinical oncology for investigating rare tumors and neoplasms,109,110,111 such as PTCL.12,17 We believe that the future/systematic generation of PDX from patients enrolled in clinical trials will allow the direct comparison of PDX and patients’ responses, faster PDX-based predictions, and the possibility to assign/switch patients to the most effective therapeutic arms.32,111 Here, we performed pre-clinical trials using experimental drugs (mostly derived from drug screening approaches and/or in silico predictions) and proved their efficacy as single agents or combinations. Also, we proved that immune-based CAR-T strategies (CD5KO-CART5 and CART30) can be explored and validated in PTCL PDXs.
In summary, PDXs provide a compelling opportunity to foster the translation of drug and immune-based strategies from the bench to the bedside.24,112 Our biorepository provides a resource for PTCL research and serves as a pre-clinical platform for testing novel therapies. By integrating genomic, transcriptomic, and drug response data, this study advances precision medicine for PTCL patients.
Limitations of the study
Despite the significance of our findings, several limitations must be acknowledged.
Limited immune system representation
PDX models lack a functional immune system,25,111 limiting the evaluation of immunotherapies, including CAR-T and checkpoint inhibitors. Thus, studies should explore humanized mouse models to address this limitation.
Engraftment success varies
∼36% of PTCLs successfully engrafted, with lower rates for rare subtypes. We believe that tissue availability and technical (tissue amount/appropriateness) and biological features of rare lymphomas (individual and heterogeneous genotypes and host requirements) partially explain these failures.113,114 To improve success, we employed b2/MCH class I-class II knockout mice to lessen graft versus host disease (GVHD)-like reactions, which can jeopardize engraftments.115 Further optimization (e.g., IL15-NSG mice) did not improve NK/T PTCL engraftment, demonstrating that defined/multiple signals are required, hardly overcome by individual engineered models. We found liability in the emergence of EBV-transformed cells116 (especially in AITL), rarely controlled by anti-CD19-ADC (Antibody-Drug Conjugate) or rituximab in NSG Hc1 mice, in contrast to previous studies with solid cancers.117,118 This predicts that EBV+ and/or B cells may not be simply bystander elements but can contribute to the early stage of transformation and/or sustain lymphoma growth/survival.119 This model is in line with the EBV-peptide recognition (EBNA LMP1 etc.) by lymphoma/leukemia TCR/MHC class I complex and TCR triggering. These data extend the putative pathological role of B cells in the genesis and maintenance of AITL.120
Microenvironment changes over passages
While PDXs retain key tumor features, some human host components diminish over serial passages, affecting tumor-host interactions. Nevertheless, as for other PDXs,24,121,122, functional similarities between human and mouse TME and the protumorigenic role of CAFs were observed. Future work taking advantage of co-culture systems and engineered microenvironments is required to dissect the mechanisms of action.
Deep learning therapy prediction requires validation
The DeepPTCL algorithm successfully predicted effective drug combinations, fostering future (pre)clinical studies in patients before entering into clinics.
Clinical translation of PDX findings
While PDX drug responses nicely correlated with patient outcomes, the implementation of PDX in prospective clinical trials is needed for the translatability of our findings. We hope that new clinical trials will include the utilization of patients’ samples for the generation of PDX and, thus, the future design and validation of broader PTCL therapeutic strategies.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Giorgio Inghirami (ggi9001@med.cornell.edu).
Materials availability
Biobanked PDXs are cataloged by the Center for Technology Licensing (https://innovation.weill.cornell.edu) and can be requested. Distribution of PDX and derived cell lines to third (academic or commercial) parties requires completion of a material transfer agreement and must be authorized by the medical ethical committee of Weill Cornell Medicine (WCM) at request of the HUB to ensure the compliance with the Institutional Review Board-Research at the WCM research involving human subjects’ act. Use of PDX is subjected to patient consent; upon consent withdrawal, distributed PDX and any derived material will have to be promptly disposed of.
Data and code availability
-
•
Raw sequencing data have been deposited to the Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra) and are publicly available. Accession numbers are listed in the key resources table (WES: PRJNA1198080; RNA-seq: PRJNA1214670; scRNA-seq: PRJNA1218277). Also, this paper analyzes existing, publicly available data, whose accession numbers are listed in the key resources table.
-
•
Codes used in this study are deposited at https://github.com/marchionniLab/ing-2023 (https://doi.org/10.5281/zenodo.14847458) and https://github.com/Mew233/DeepPTCL (https://doi.org/10.5281/zenodo.14845644) and listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
We thank all staff members of the Immunopathology Laboratory at Weill Cornell Medicine for their support. We are grateful to the Epigenomics and Genomics Cores of Weill Cornell Medicine for next-generation sequencing. We thank the members of the Weill Cornell Cell Sorting Core and Edward Meyer Cancer Center PDX Shared Resource. We thank Drs. Lorenzo Galluzzi and Shahin Rafii for intellectual discussions, feedback, and support. We thank the mouse facility operators and Drs. Kvin Lertpiriyapong and Rodolfo Ricart Arbona. We are grateful to Ani Arkur, Sofia Alayon, and Shefali B. Sha for their administrative support and financial management. G.I. is supported by CA229086, CA229100, CA195568, and LLS 7011-16; L.V.C. and R.F. were supported by the Italian Association for Cancer Research, Metastases 5x1000 Special Program, grant 21198; L.V.C. and G.Medico were supported by American-Italian Cancer Foundation Post-Doctoral Research Fellowship (AICF 2021-22 and 2023-24); D.F. was supported by the Rita-Levi Montalcini grant from the Italian Ministry of University and Research (MIUR); V.F. was supported by the Italian Association for Cancer Research, MFAG 2023 ID 28974; and D.M.W. was supported by NCI R35 CA231958, NCI P01 CA233412, and Leukemia and Lymphoma Society Specialized Center of Research 7026-21. R.R., A.W., and L.Z. were supported by NCI R35 CA253126, U01CA243073, and Stand Up to Cancer Convergence Program. L.Z. was supported by the NSFC 32170565 and the CAS Hundred Talents Program. W.C.C. was supported by the COH Cancer Center Support Grant P30, CA033572, and 1PO1CA229100.
Author contributions
Conceptualization: D.F., L.V.C., R.P., E.M., S.Y.N., A.M., Z.E., A.I., D.A., P.M., J.R., F.B., R.F., J.D.B., D.B., W.T., D.M.W., L.C., R.R., S.H., and G.I. Methodology and investigation: D.F., L.V.C., J.P., A.T., W.C., V.F., F.T., N.Z., C.K., G.Medico, R.P.P., M.G., R.M., G.A., M.T.C., G.Z., C.P., R.A.E., S.P., N.D.S., and I.K. Software and formal analysis: L.Z., N.K., P.G., M.S., P.Z., L.Y., A.W., X.H., A.N., O.K., D.Nikitin, D.T., E.P., A.B., A.K., E.B., V.S., K.T., N.D.S., S.D., G.Macari, L.C., S.G., L.S., J.O., C.Z., L.Q., O.E., and C.X. Resources: D.Novero, M.P., E.T., B.F., A.M., F.Z., J.S., E.D.S., Z.E., D.A., P.M., J.R., J.D.B., W.C.C., W.T., M.R., D.M.W., S.H., and G.I. Writing original draft: D.F., L.V.C., W.T., L.C., and G.I. Writing, reviewing, and editing: D.F., L.V.C., W.T., L.C., and G.I. Funding acquisition: D.F., S.H., D.M.W., R.F., and G.I. Project administration: G.I.
Declaration of interests
D.M.W. is an employee of Merck and has an equity interest in Ajax, Bantam, and Travera. F.B. receives institutional research funds from ADC Therapeutics, Bayer AG, Cellestia, Helsinn, HTG Molecular Diagnostics, ImmunoGen, iOnctura, Menarini Ricerche, NEOMED Therapeutics 1, Nordic Nanovector ASA, and Spexis AG; advisory board fees from Novartis; consultancy fee from Helsinn and Menarini; and travel grants from Amgen, AstraZeneca, and iOnctura and provided expert statements to HTG Molecular Diagnostics.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-CD3e conjugated to FITC (clone 17A2) | BD Bioscience | 349201; RRID: AB_395698 |
| anti-CD57 conjugated to PE (clone B3GAT1) | BD Bioscience | 560844; RRID:AB_2033965 |
| anti-CD5 conjugated to PerCP-Cy5.5 (clone L17F12) | BD Bioscience | 341099; RRID: AB_400220 |
| anti-CD4 conjugated to PE-Cy7 (clone SK3) | BD Bioscience | 348799; RRID: AB_400387 |
| anti-CD7 conjugated to APC (clone 124-1D1) | Invitrogen | 17-0079-42; RRID: AB_10671279 |
| anti-CD8 conjugated to APC-H7 (clone SK1) | BD Bioscience | 641409; RRID: AB_1645737 |
| anti-CD2 conjugated to BV421 (clone TS1/8) | BioLegend | 309216; RRID: AB_2073669 |
| anti-CD45 conjugated to V500C (clone 2D1) | BD Bioscience | 647450; RRID: AB_2814897 |
| anti-TCRa/b conjugated to FITC (clone WT31) | BD Bioscience | 340883; RRID: AB_400168 |
| anti-TCRg/g conjugated to PE (clone 111F2) | BD Bioscience | 347907; RRID: AB_400359 |
| anti-CD56 conjugated to PerCP-Cy5.5 (clone B159) | BD Bioscience | 560842; RRID: AB_2033964 |
| anti-CD3 conjugated to PE-Cy7 (clone SK7) | BD Bioscience | 341101; RRID: AB_400222 |
| anti-CD30 conjugated to APC (clone HRS4) | Beckman Coulter | A87939; RRID: N/A |
| anti-CD25 conjugated to APC-H7 (clone M-A251) | BD Bioscience | 560244; RRID: AB_1645472 |
| anti-CD16 conjugated to BV421 (clone 3G8) | Biolegend | 302032; RRID: AB_2104003 |
| anti-CD7 conjugated to FITC (clone M-T701) | BD Bioscience | 340699; RRID: AB_400100 |
| anti-CD185 conjugated to PE (clone J252D4) | Biolegend | 356904; RRID: AB_2561813 |
| anti-CD278 conjugated to PerCP-Cy5.5 (clone C398.4A) | Biolegend | 313518; RRID: AB_10641280 |
| anti-CD10 conjugated to APC (clone HI10A) | BD Bioscience | 340923; RRID: AB_400543 |
| anti-CD279 conjugated to BV421 (clone MIH4) | BD Bioscience | 565935; RRID:AB_2739399 |
| anti-CD5 conjugated to BV605 (clone UCHT2) | BD Bioscience | 563945; RRID: AB_2738500 |
| anti-Kappa conjugated to FITC (Rabbit anti-Human) | Agilent | FO434; RRID: N/A |
| anti-Lambda conjugated to PE (rabbit anti-Human) | Agilent | RO437; RRID: N/A |
| anti-CD23 conjugated to PE-Cy7 (clone M-L233) | BD Bioscience | 561167; RRID: AB_10611996 |
| anti-CD20 conjugated to APC-H7 (clone L27) | BD Bioscience | 641405; RRID: AB_1645729 |
| anti-CD19 conjugated to BV421 (clone HIB19) | BD Bioscience | 562440; RRID: AB_11153299 |
| anti-ALK-1 | Leica Biosystems | PA0831; RRID: AB_3073618 |
| anti-CD30 (clone BerH2) | Dako | GA602; RRID: AB_3675588 |
| anti-CD3 (clone PS1) | Leica Biosystems | NCL-CD3-PS1; RRID: AB_442061 |
| anti-CD2 (clone AB75) | Leica Biosystems | NCL-CD2-271; RRID: AB_442057 |
| anti-CD4 (clone 4B12) | Leica Biosystems | PA0371; RRID: AB_10554438 |
| anti-CD7 (clone LP15) | Novocastra | NCL-L-CD7-580 P; RRID: N/A |
| anti-CD5 (clone 4C7) | Novocastra | NCL-L-CD5-4C7; RRID: N/A |
| anti- TIA1 (clone 2G9A10F5) | Immunotech | IM2550; RRID: AB_131704 |
| anti-granzyme (clone GrB7) | Monosan | MON7029C; RRID: N/A |
| anti-perforin (clone 5B10) | Lab Vision | MS-1834-R7; RRID:AB_149381 |
| anti- TCRa/b (clone IP26) | Biolegend | 306718; RRID: AB_10612569 |
| anti-CD20 (clone L26) | Dako | M0755; RRID: AB_2282030 |
| anti-PAX5 (clone 24/PAX5) | BD Transduction Lab | 610863; RRID: AB_398182 |
| anti-CD25 (clone 4C9) | Leica Biosystems | PA0306; RRID: AB_10556556 |
| anti-CLA (rabbit polyclonal) | Dako | A 0423; RRID: AB_2335700 |
| anti-OCT2 (rabbit polyclonal) | Santa Cruz Biotechnology | sc-56822; RRID: AB_784955 |
| anti-CD33 (clone PWS44) | Leica Biosystems | PA0558; RRID:AB_10555285 |
| anti-MIB (clona MIB-1) | Agilent | GE020; RRID: N/A |
| anti-GAS1 (clone C-17) | Santa Cruz Biotechnology | N/A |
| anti-pSTAT3 (clone M9C6) | Cell Signaling Technology | 4113; RRID: AB_2198588 |
| anti-C/EBP (C19) | Santa Cruz Biotechnology | sc-150; RRID: AB_2260363 |
| anti-NFATC2 (clone M20) | Santa Cruz Biotechnology | sc-1151; RRID: AB_632026 |
| anti-pJAK2 (clone E132) | abcam | ab219728; RRID: N/A |
| anti-pJAK3 (rabbit polyclonal) | Santa Cruz Biotechnology | sc-16567; RRID:AB_2128682 |
| anti-p44/42 MAPK (Erk1/2) | Cell Signaling Technology | 9102; RRID: AB_330744 |
| anti-GAPDH | Cell Signaling Technology | 5174s; RRID: AB_10622025 |
| anti-CD2 | Leica Biosystems | AB75; RRID: AB_2528815 |
| anti-CD30 | Agilent | BERH2; RRID: AB_10670808 |
| anti-PDL1 | Roche | SP263; RRID: AB_2819099 |
| anti-EOMES | eBioscience/Invitrogen/Thermo | WD1928; RRID: AB_2572615 |
| anti-CD3 (clone SP7) | Lab Vision | RM-9107-S; RRID: AB_149922 |
| anti-CD68 (clone KP1) | Agilent | IR609; RRID: N/A |
| anti-CD163 (clone 10D6) | Leica Biosystems | NCL-L-CD163, RRID:AB_2756375 |
| anti-SMA (clone 1A4) | Agilent | M0851; RRID: AB_2223500 |
| anti-CD20 (clone L26) | Leica Biosystems | NCL-L-CD20-L26; RRID:AB_563521 |
| DAPI | Cell Signaling Technology | 4083S |
| Biological samples | ||
| PTCL Patient Samples | Weill Cornell Medicine, New York PresbyterianHospital (NY), Memorial Sloan Kettering Cancer Center (NY), City of Hope (CA), The Tisch Cancer Institute at Mount Sinai, University of Torino (IT), San Raffaele (IT). | WCM IRB: 1302013582, 0107004999, 1410015560; MSKCC IRB: 13–014, 09–141, 12–245, 06–107; University of Torino: 0081521 |
| Healthy Peripheral blood mononuclear cells | New York Blood Bank | Custom Order |
| Tissue Blocks of human PTCL | as described in Crescenzo et al., 2015 | 10.1016/j.ccell.2015.03.006 |
| Chemicals, peptides, and recombinant proteins | ||
| 17AAG | Selleckchem | S1141 |
| ABT-199 | Selleckchem | S8048 |
| ABT263 | Selleckchem | S1001 |
| AUY922 | Selleckchem | S1069 |
| Azacytidine | Selleckchem | S1782 |
| AZD4573 | Selleckchem | S8719 |
| Belinostat | Selleckchem | S1085 |
| Bortezomib | Selleckchem | S1013 |
| Brentuximab vedoton | Boc Science | 914088-09-8 |
| CC220 | Selleckchem | S8760 |
| CEP18870 | Selleckchem | S1157 |
| Cerdulatinib | Selleckchem | S3566 |
| Ceritinib | Selleckchem | S4967 |
| Chidamide | Selleckchem | S8567 |
| CHIR124 | Selleckchem | S2683 |
| Crenolanib | Selleckchem | S2730 |
| Crizotinib | Selleckchem | S1068 |
| Cyclophosphamide | Selleckchem | S2057 |
| Daunorubicin | Selleckchem | S3035 |
| Decitabine | Selleckchem | S1200 |
| Dexamethason | Selleckchem | S5956 |
| Doxorubicin | Selleckchem | S1208 |
| Duvelisib | Selleckchem | S7028 |
| Enzastaurin | Selleckchem | S1055 |
| Ganetespib | Selleckchem | S1159 |
| GDC-0077 | Selleckchem | S8668 |
| Idelalisib | Selleckchem | S2226 |
| Irinotecan | Selleckchem | S1198 |
| KPT330 | Selleckchem | S7252 |
| Lenalidomide | Selleckchem | S1029 |
| MK1775 | Selleckchem | S1525 |
| MLN2238 | Selleckchem | S2180 |
| NSC319726 | Selleckchem | S7149 |
| NVP742 | Selleckchem | S1088 |
| Ouabain | Selleckchem | S4016 |
| Panobinostat | Selleckchem | S1030 |
| PomalidomideS1567 | Selleckchem | S1567 |
| Prednisone | MedChemExpress | HY-B0214 |
| Pralatrexate | Selleckchem | S1497 |
| PU-H71 | Selleckchem | S8039 |
| RGFP-966 | Selleckchem | S7229 |
| RO4929097 | Selleckchem | S1575 |
| Romidepsin | Selleckchem | S3020 |
| Ruxolitinib | Selleckchem | S1378 |
| SC144 | Selleckchem | S7124 |
| Selumetinib | Selleckchem | S1008 |
| Semagacestat | Selleckchem | S1594 |
| SN38 | Selleckchem | S4908 |
| Stattic | Selleckchem | S7024 |
| Tozasertib | Selleckchem | S1048 |
| TGR1202 | Selleckchem | S8194 |
| Tofacitinib | Selleckchem | S2789 |
| TSA | Selleckchem | S1045 |
| Valemetostat | Selleckchem | S8926 |
| Vincristine | Selleckchem | S9555 |
| YKL-5-124 | Selleckchem | S8863 |
| YM155 | Selleckchem | S1130 |
| Ficoll-Paque PLUS | Cytiva | 17144003 |
| Trypan Blue | Sigma Aldrich | T10282 |
| RPMI 1640 Medium | Gibco™ | 11875093 |
| DMEM Medium | ThermoFisher Scientific | C11965092 |
| Phosphate Buffered Saline (1X) | ThermoFisher Scientific | 20021–027 |
| Fetal Bovine Serum; Heat inactivated | Corning | 35-011-CV |
| Ethylenediaminetetraacetic acid (EDTA) 0.5M | VWR | E522-100ML |
| Penicillin-Streptomycin-Glutamine (100X) | Gibco/Invitrogen | 15140–122 |
| Normocin | Invivogen | ant-nr-1 |
| Gemcitabine | Selleck Chemicals | S1714 |
| Mitomycin C | Sigma-Aldrich | M4287 |
| 0.25% Trypsin-EDTA | Gibco™ | 25200056 |
| Collagenase Type IV | Sigma-Aldrich | C5138-5G |
| Accutase cell dissociation reagent | ThermoFisher Scientific | A1110501 |
| Cell strainer, 100μm, filter | Corning | 431742 |
| Cell strainer, 70μm, filter | Corning | 431751 |
| Cell strainer, 40μm, filter | Falcon, Fisher Scientific | C352340 |
| Recombinant Human IL-2 | R&D systems | 202-IL |
| Recombinant Human IL-7 | R&D systems | 207-IL-025 |
| Recombinant Human IL-15 | R&D systems | 247-ILB |
| DNase I | Worthington Biochemical | LS002007 |
| LB agar Lennox | Gibco | 244520 |
| Trizol | ThermoFisher | 15596018 |
| SYBR™ Select Master Mix | Applied Biosystems | 4472897 |
| EBER probes | Leica | ISH5687-A |
| CellTrace™ CFSE Cell Proliferation Kit | Invitrogen | C34554 |
| CellTrace™ Violet | Invitrogen | C34557 |
| Critical commercial assays | ||
| SureSelect Strand-Specific RNA Library Agilent Preparation Kit | Agilent | G9691A |
| TruSeq® Stranded Total RNA Library Prep Human/Mouse/Rat | Illumina | 20020597 |
| SureSelectXT Human All Exon 50 Mb v4 Kit | Agilent | 5190–4632 |
| SureSelectXT Human All Exon 50 Mb v5 Kit | Agilent | 5190–6209 |
| DNeasy Blood & Tissue Kit | Qiagen | 69504 |
| RNeasy Mini kit | Qiagen | 74106 |
| Qubit dsDNA HS and BR Assay Kits | Thermo Fisher Scientific | Q32851 |
| Bio-Rad protein assay kit | Bio-Rad Laboratories | 5000001 |
| PCR Mycoplasma Detection Kit - Quantity: 100 Reactions | Applied Biological material | G238 |
| CD3 MicroBeads, human | Miltenyi Biotec | 130-097-043 |
| CD19 MicroBeads, human | Miltenyi Biotec | 130-050-301 |
| TCRB Gene Clonality Assay | Invivoscribe | 12050011 |
| IGH + IGK B-Cell Clonality Assay | Invivoscribe | 11000031 |
| Chromium™ Next GEM Single Cell 5′ Library and Gel Bead Kit v1.1 | 10x Genomics | 1000165 |
| Chromium™ Single Cell 5′ Library Construction Kit | 10x Genomics | 1000020 |
| Deposited data | ||
| Raw Data Files from RNA-seq | This paper - SRA | SRA: PRJNA1214670 |
| Raw Data Files from WES | This paper - SRA | SRA: PRJNA1198080 |
| Raw Data Files from scRNASeq | This paper - SRA | SRA: PRJNA1218277 |
| WES data | Nat Genet Da Silva Almeida 2015 | https://www.nature.com/articles/ng.3442#Sec17 |
| WES data | Nat Genet Sakata-Yanagimoto 2014 | https://www.nature.com/articles/ng.2872#Sec24 |
| WES data | Nat Genet Choi 2015 | https://www.nature.com/articles/ng.3356#Sec34 |
| WES data | Mod Pathol 2020 Laginestra | https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6994417/ |
| WES data | Nat Genet Kataoka 2015 | https://www.nature.com/articles/ng.3415#Sec45 |
| WES data | Frontiers in Oncology Mirza 2020 | https://www.frontiersin.org/articles/10.3389/fonc.2020.00514/full |
| WES data | Nat Genet Jiang 2015 | https://www.nature.com/articles/ng.3358 |
| WES data | Cancer Cell Crescenzo 2015 | https://www.cell.com/cancer-cell/fulltext/S1535-6108(15)00094-X#secsectitle0015 |
| WES data | Palomero, Nat Genet. 2014 | https://www.nature.com/articles/ng.2873#Sec26 |
| Targeted genomic | Schatz, Hortwitz, Weinstock Leukemia 2014 | https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4286477/table/tbl1/?report=objectonly |
| Targeted genomic | Yoshida, Weinstock Blood 2020 | https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7180081/ |
| Targeted genomic | Nat Genet Sakata-Yanagimoto 2014 | https://www.nature.com/articles/ng.2872#Sec24 |
| Targeted genomic | Kataoka, Nat Genet. 2015 | https://www.nature.com/articles/ng.3415#Sec44 |
| Targeted genomic | Nat Genet Jiang 2015 | https://www.nature.com/articles/ng.3358 |
| Targeted genomic | Cancer Cell Crescenzo 2015 | https://www.cell.com/cancer-cell/fulltext/S1535-6108(15)00094-X#secsectitle0015 |
| RNA_Sequencing data | EGA | EGA: EGAS00001001296 |
| RNA_Sequencing data | dbGaP | dbGaP: phs000689 |
| RNA_Sequencing data | SRA | SRA: SRP049695 |
| RNA_Sequencing data | SRA | SRA: SRP029591 |
| RNA_Sequencing data | SRA | SRA: SRP099016 |
| RNA_Targeted data | NCBI GEO | GEO: GSE58445 |
| RNA_Targeted data | NCBI GEO | GEO: GSE45712 |
| RNA_Targeted data | NCBI GEO | GEO: GSE19069 |
| RNA_Targeted data | NCBI GEO | GEO: GSE90597 |
| RNA_Targeted data | NCBI GEO | GEO: GSE6338 |
| RNA_Targeted data | NCBI GEO | GEO: GSE36172 |
| RNA_Targeted data | EMBL-EBI | EBI: E-TABM-783 |
| RNA_Targeted data | NCBI GEO | GEO: GSE65823 |
| RNA_Targeted data | NCBI GEO | GEO: GSE118623 |
| RNA_Targeted data | EMBL-EBI | EBI: E-TABM-702 |
| RNA_Targeted data | NCBI GEO | GEO: GSE78513 |
| RNA_Targeted data | NCBI GEO | GEO: GSE51521 |
| RNA_Targeted data | NCBI GEO | GEO: GSE14317 |
| RNA_Targeted data | NCBI GEO | GEO: GSE80631 |
| RNA_Targeted data | NCBI GEO | GEO: GSE19067 |
| RNA_Targeted data | NCBI GEO | GEO: GSE20874 |
| Experimental models: Cell lines | ||
| TO-ALCL-Belli PDX-Dline | This paper | N/A |
| TO-ALCL-DN03 PDX-Dline | This paper | N/A |
| TO-ALCL-MARI PDX-Dline | This paper | N/A |
| IL2 PDX-Dline | This paper | N/A |
| IL69 PDX-Dline | This paper | N/A |
| IL79 PDX-Dline | This paper | N/A |
| IL86 PDX-Dline | This paper | N/A |
| IL89 PDX-Dline | Fiore, Cappelli et al. Cancers 2020123 | https://pubmed.ncbi.nlm.nih.gov/32560455/ |
| IL104 PDX-Dline | This paper | N/A |
| IL135A PDX-Dline | This paper | N/A |
| IL142A PDX-Dline | This paper | N/A |
| IL 223B PDX-Dline | This paper | N/A |
| IL 228 PDX-Dline | This paper | N/A |
| COH1 PDX-Dline | This paper | N/A |
| MS-5 cell line | DSMZ | ACC 441 |
| SUPM2 cell line | DSMZ | ACC 509 |
| L82 cell line | DSMZ | ACC 597 |
| MAC1 cell line | Expasy | CVCL_H631 |
| TLBR1 cell line | DSMZ | ACC 904 |
| Experimental models: Organisms/strains | ||
| NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ | The Jackson Laboratory | 5557 |
| NOD.Cg-B2mtm1Unc Prkdcscid Il2rgtm1Wjl/SzJ | The Jackson Laboratory | 10636 |
| NOD.Cg-Prkdcscid H2-K1b-tm1Bpe H2-Ab1g7-em1Mvw H2-D1b-tm1Bpe Il2rgtm1Wjl/SzJ | The Jackson Laboratory | 025216 |
| NOD.Cg-Hc1 Prkdcscid Il2rgtm1Wjl/SzJ | The Jackson Laboratory | 030511 |
| Patient-derived xenografts (PDX) | This paper | Table S1 |
| Oligonucleotides | ||
| Primers for RTqPCR | This paper | Table S12 |
| Software and algorithms | ||
| Excel 2016 | Microsoft | https://www.office.com/ |
| GSEA (4.1.0) | Subramanian et al. 2005 | https://www.gsea-msigdb.org/gsea/index.jsp |
| Halo | Indica labs | https://www.indicalab.com |
| STARTRAC | Zhang et al., 2018b | https://github.com/Japrin/STARTRAC |
| STAR aligner v2.6.1 | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| DiVa V8.0.1 | BD Biosciences | https://www.bdbiosciences.com/en-eu/products/software/instrument-software/bd-facsdiva-software |
| FlowJo v10.7.1 | FlowJo, LLC | N/A |
| R v4.3.1 | R Core Team, 2014124 | https://www.r-project.org |
| CIBERSORT | Newman et al., 2015 | https://cibersort.stanford.edu/ |
| Seurat v.4.0.3 | Stuart et al., 2019 | https://satijalab.org/seurat/ |
| BLAST | Altschul (1990) | ftp.ncbi.nlm.nih.gov/blast/executables/blast=/LATEST |
| ImageJ Software | Open source | N/A |
| GraphPad Prism software version 9.3.1 | GraphPad Software, Inc. | https://www.graphpad.com/ |
| IGV | Robinson et al., 2011 | http://software.broadinstitute.org/software/igv/ |
| Kassandra code | Zaitsev et al.77 | https://github.com/BostonGene/Kassandra |
| Shiny v1.7.4 | R Studio Partners, R Core Team 2019125 | https://www.r-project.org/nosvn/pandoc/shiny.html |
| Tidyverse v1.3.9 | vignettes/paper.Rmd126 | https://www.tidyverse.org/packages/ |
| Bioconductor v3.17 | Huber et al. 2015127 | https://www.bioconductor.org |
| Paper code #1 | This paper | https://github.com/marchionniLab/ing-2023 - https://doi.org/10.5281/zenodo.14847458 |
| Paper code #2 | This paper | https://github.com/Mew233/DeepPTCL - https://doi.org/10.5281/zenodo.14845644 |
| Other | ||
| Clinical Annotations | This paper | Table S1 |
| BD FACSCanto™ II | BD Biosciences | N/A |
| BD LSR Fortessa | BD Biosciences | N/A |
| LSRII | BD Biosciences | N/A |
| BD FACSAriaTM III Cell Sorter | BD Biosciences | N/A |
| BD FACSCelestaTM Cell Analyzer | BD Biosciences | N/A |
| NovaSeq 6000 | Illumina | N/A |
| TissueLyser II | Qiagen | 85300 |
| Leica Bond-III | Leica instruments | N/A |
| Leica Bond-RX | Leica instruments | N/A |
| Rodent diet | PicoLab Rodent Diet 20 | 5053 |
| Ventilated cages | N/A | N/A |
| Mouse hair removal kit | 3 M | 9667L |
| Gauze Sponges | Thermo Fisher Scientific | 13-761-52 |
| VWR® Dissecting Scissors, Sharp Tip, 41/2″ | VWR | 82027–578 |
| VWR Dissecting Forceps | VWR | 89259–944 |
| Isoflurane chamber with nose cone | N/A | N/A |
| Wound Clip Complete Kit | Thermo Fisher Scientific | BD427638 |
| 1 mL syringe | VWR | 76124–644 |
| 5 mL syringe | VWR | 76163–596 |
| Animal Ear Punch, Plier-Style | VWR | N 10806-290 |
| Freezing container | VWR | 55710–200 |
| 50mL Falcon Tubes | VWR | CA21008-940 |
| Sterile Petri Dish | VWR | 25384–342 |
| Scalpel with blade no. 10 | VWR-Miltex | 21909–654 |
| Sterile razor | VWR | 55411–050 |
| 2mL Serological Pipette | Thermo Fisher Scientific | 170365 |
| 5mL Serological Pipette | Thermo Fisher Scientific | 170355 |
| 10mL Serological Pipette | Thermo Fisher Scientific | 170367 |
| 25mL Serological Pipette | Thermo Fisher Scientific | 170357 |
| Alcohol pad | VWR | 720–2586 |
| Puralube® Ophthalmic Ointment | Patterson veterinary | 211–38 |
| Sutures: Dermalon Suture, Blue, Size 5/0, 18″, CE-4 Needle | Medline | D-G175621 |
| Insulin Syringes | VWR | BD328438 |
| RAM Scientific Safe-T-Fill™ Capillary Blood Collection Systems: Lithium Heparin | ThermoFisher | 14-915-65 |
| Lab animal scale | N/A | N/A |
| Digital caliper | VWR | 36934–152 |
| Carbon Dioxide (CO2) | N/A | N/A |
| Isoflurane | N/A | N/A |
| Feeding gavage needles | braintreescientific | N-VP 22G-15S |
| Mouse Tail Illuminator Restrainer | braintreescientific | MSPP-MTISTD |
| Matrigel | Corning | 354234 |
| Trocar for mouse surgery | braintreescientific | TRO 14MS |
| Surgical Scrub Betadine | Purdue Products LP | 6904214–40890 |
| Betadine Iodine Solution | Purdue Products LP | 158348 |
| Meloxicam | Boeringer Ingelheim | L20805A-42 |
| Tear gel | Optixcare Eye Lube | BP231-1 |
| Ethyl Alcohol Anhydrous | Commercial Alcohols | PO16EAAN |
Experimental model and study participant details
Human study
The collection of PTCL patient data and tissue for the generation and distribution of PDX and derivatives were performed according to the guidelines of the Institutional Review Board-Research at the Weill Cornell Medicine, Memorial Sloan Kettering’s Institutional Review Board (IRB)/Privacy Board and the Comitato Etico Interaziendale, AOU San Giovanni Battista di Torino and CTO Maria Adelaide di Torino. All patients participating in the study signed informed consent forms approved by the authority responsible (see above). In all cases, patients can withdraw their consent at any time, leading to the prompt disposal of their tissue and any derived material. Biobanked Patient-Derived models can be requested at https://innovation.weill.cornell.edu. Clinical information is available in Table S1. Additional data i.e., age, gender, genetics, therapy, etc. of subjects can be inquired through https://innovation.weill.cornell.edu.
Pathological samples most frequently from diagnostic tissue samples (76/88), and some from bone marrow (3/88), pleural effusions (2/88), or peripheral blood (7/88) were collected at the Weill Cornell Medicine (WCM) of New York, University of Torino, and Memorial Sloan Kettering Cancer Center (MSKCC). Both fresh (n = 54) and viably cryopreserved tissue samples were implanted (n = 34). Diagnoses were assigned according to the WHO classification by expert pathologists. De-identified patients’ samples (323) were obtained with informed consent under WCM (78), Torino (6), MSKCC (234), S. Raffaele at Milan (1), City of Hope (3), and Mount Sinai (1) Institutional Review Boards (IRB)-approved protocols, according to the Declaration of Helsinki.
Mice models
NOD Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG), NOD.Cg-B2mtm1Unc Prkdcscid Il2rgtm1Wjl/SzJ (NSG B2m), NOD.Cg-Prkdcscid H2-K1b-tm1Bpe H2-Ab1g7-em1Mvw H2-D1b-tm1Bpe Il2rgtm1Wjl/SzJ (NSG-MHC I/II DKO), and NOD.Cg-Hc1 Prkdcscid Il2rgtm1Wjl/SzJ (NSG-Hc1) mice were originally purchased from Jackson Laboratories and then bred in-house and handled according to WCM Institutional Animal Care and Use Committee (protocol #2014-24).
Primary PTCL samples were implanted subcutaneous (sc, 2 fragments, 1mm3 each) or via intravenous (iv; 1 × 106 cells, 150 μL of DPBS) or intra bone (ib; 1 × 106 cells, 10–20 μL of DPBS) routes, in 4-6-week-old (male/female ratio: 1:1) NSG B2m/NSG-MHC I/II DKO mice.128 PDX-Dlines were s.c. implanted in Matrigel (25%, 1x106 cells, 150 μL of DPBS). Engraftment was monitored every week by visual inspection (s.c.) and/or multicolor flow cytometry on peripheral blood. Mice were sacrificed at the earlier sign of distress. All tissues were collected for histology, immunohistochemistry, and additional ancillary studies. Viable and dry samples were cryopreserved for PDX transplantation/biobanking and genomic/functional studies. Tumors were then propagated along multiple generations corresponding to serial passages (T).
Cell culture
PDX-Dlines, PDX derived stromal cells, PTCL continuous cell lines (SUPM2, L82, MAC1 and TLBR1), and MS-5 stromal cell line were cultured in RPMI (Sigma) supplemented with 20% FBS (Corning), 100 U/ml glutammine (Sigma), Normocin 1:500 (InVivoGen) and 100μg/ml streptomycin (Sigma) and maintained at 37°C in a humidified 5% CO2 atmosphere. IL2 and TO-ALCL-BELLI PDX-Dlines were supplemented with exogenous interleukin-2 (50U/ml) and interleukin15 (10μg/ml) (R&D). Cells were analyzed by flow cytometry using a panel of monoclonal antibodies against human T cell surface markers twice per year.
Method details
Isolation of viable PDX-derived tumor cells
Tumor tissue was finely dry minced using sterile blades and digested for 30–45 min at 37°C. Digestion media was composed of RPMI1640 (Sigma) and digestion buffer (4:1). The digestion buffer was prepared according to the following: 140nM NaCl (Sigma), 5mM KCl (Sigma), 2.5mM Phosphate buffer ph7.4 (prepared by solving 3.1g of NaH2PO4-H2O and 10.9g of Na2HPO4 anhydrous in 1 L of sterile cell culture grade water), 10mM HEPES (Sigma), 2mM CaCl2 (Sigma), 1.3mM MgCl2 (Sigma), 25 mg/ml of Collagenase A (Roche), 25 mg/ml Dispase II (Sigma), 250 mg/ml DNAase (Roche). The digested tissue was then passed through 70μm nylon filters (Corning) and the resulting cell suspension was washed twice with PBS (Sigma). Cells were resuspended in RPMI1640 (Sigma) plus 20% FBS (Gibco) and seeded at 1 million/ml in T150 flasks (Corning) overnight. The day after, floating T-Cells were separated from stromal cells attached to the flasks and centrifuged on a Ficoll Paque (Sigma) gradient to remove dead cells, red blood cells, and debris, to isolate a pure (>95%) and viable (>95%) T cell population. Stromal adherent cells were cultured (RPMI1640 20%FBS - Sigma) and used for experimental purposes if needed. T cell suspensions were cultured (RPMI1640 20%FBS – Sigma/Corning) and analyzed by flow cytometry using a panel of monoclonal antibodies against human T cell surface markers.
Histopathological analyses
Tissues were recovered within 0.5-1h after mouse sacrifice, fixed in 10% neutral buffered formalin (Sigma), and processed for histology and immunohistochemistry. Immunohistochemistry was performed on 4μm Formalin Fixed Paraffin Embedded (FFPE) sections of multiple organs (lungs, spleen, liver, heart, kidney, and tumor mass). Deparaffinization, rehydration, and antigen retrieval were performed by BERS2 (prediluted; pH 9.0) antigen retrieval solution performed on the Bond-III Leica automated slide stainer for 20 min at 100°C. Specimens were incubated with primary antibodies (Table S11 and key resources table) followed by visualization with the Leica Bond detection kit (BOND Polymer Refine Detection) for 15 min at room temperature. For dual optical staining, sections were then incubated with a second primary antibody and developed using a BOND Polymer Refine Red Detection, as described above. The specimens were then counterstained with hematoxylin and the cover slipped. Each IHC run contained external positive controls.
All the antibodies used (Table S11 and key resources table) were diluted in PBS (Sigma).
Multicolor flow cytometry
Flow cytometry was performed by staining lymphoma cells with a mix of antibodies diluted 1:100 in PBS and recognizing T- and B-cell restricted markers. The antibodies used are summarized in the supplementary material and were purchased from BD Biosciences. Briefly, lymphoma cells were identified after gating on human CD45+ cells, and the selected markers were analyzed inside the human CD45-positive cell population. At least 10000 events were acquired. Samples were run on the BD FACSCanto and analyzed with the BD FACSDiva software.
Dual-color fluorescence in situ hybridization (FISH)
BAC clones spanning the gene loci were obtained from BACPAC Resources at http://bacpac.chori.org. Commercial FITC centromeric probes were used as well (Abbott Park, Illinois, U.S.A). DNA was labeled by nick-translation using spectrum green or Spectrum red-dUTP fluorochromes (Abbott). FISH was performed by standard methods and at least 20 metaphase spreads or 200 interphase nuclei on DAPI-stained slides were scored.
Multiplex immunofluorescence tissue staining
Multiplexed immunofluorescence (mIF) was performed using the Opal system (Akoya Biosciences) and SignalStar by staining 4 micron-thick Bouin-fixed, paraffin-embedded whole-tissue sections from diagnostic biopsy specimens in a Bond RX automated tissue stainer (Leica Biosystems, Buffalo Grove, IL).129 Alternatively, we used a combination of oligonucleotides and fluorophores to amplify the antibody signal, allowing for the detection of targets, at low expression levels (https://www.cellsignal.com/applications/signalstar-multiplex-ihc-overview). Whole slide scans were subsequently obtained at 20× magnification using the Vectra Polaris Automated Quantitative Pathology Imaging System (Akoya Biosciences) to generate a collection of tiled images, which were subsequently spectrally unmixed in InForm (v2.4.8, Akoya Biosciences). Unmixed tiles were finally fused in HALO (v3.3.2541.231, Indica Labs) to generate a single multi-layered TIFF image file for each sample, which was used in downstream analyses. The analysis was performed using QuPath 0.5.1 (https://qupath.github.io) where we identified each phenotype based on the median intensity of independent marker histograms.
The main phenotypes of the TME validation were classified as T cells (CD3 marker), Macrophages (CD68 and CD163), Fibroblast (SMA), B cells (CD20), and Others (DAPI without any other marker).
DNA and RNA extraction
Total RNA extraction from (dry frozen) tissues or cells was performed using TRIZOL (Invitrogen) according to the manufacturer’s protocols. RNA quality was checked on an Agilent Bioanalizer (Agilent Technologies). Samples with an RNA integrity number (RIN) > 7 were selected for further analysis. RTqPCR was executed using the iQ SYBR Green Real-Time PCR Supermix (BioRad). The list of primers is reported in the Table S12. Genomic DNA was extracted from (dry frozen) tissues or cells using phenol/chloroform (Sigma). Quality and quantity were checked using the Agilent Tapestation (Agilent) and Qubit (Invitrogen).
Identification of clonal antigen receptor gene rearrangements
Clonal rearrangements of BCR (IgH and IgK) and TCR genes were determined using the Invivoscribe kit based on the BIOMED-2 assay130 on extracted DNA. PCR products are analyzed by capillary electrophoresis (CE) using the ABI 3500 Genetic Analyzer.
Total RNA and whole-exome sequencing
Total RNA was used for cDNA library preparation using the TruSeq-Stranded Total RNA sample preparation (HS protocol) following the manufacturer’s instructions (Illumina). DNA1000 Kit (Agilent) was used to size and quantify the library preparation on an Agilent 2100 Bioanalyzer. Sequencing data were aligned to the human reference genome (hg38) using the STAR v2.3.5 aligner(Dobin, Davis et al. 2013) after human-mouse read disambiguation via BBsplit v37.76. Gene counts were calculated by using featureCounts(Liao, Smyth et al. 2014) v1.4.6 concerning Gencode v19 annotations.
TCR signaling activity was determined from RNA-sequencing using validated signatures available at MSigDB (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp).
For whole-exome sequencing, genomic DNA was used to prepare the libraries using the SureSelect 6.0 kit (Agilent), according to the manufacturer’s protocol. Whole exome sequencing data were aligned to the human reference genome (hg38) using mem from BWA v-0.7.12(Li 2013). Duplicate reads were removed using the MarkDuplicates command from Picard v1.124 (http://broadinstitute.github.io/picard/), and local realignment around indels was performed using ABRA v0.92(Mose, Wilkerson et al. 2014). Somatic mutations were called with MuTect v1.1.5. FACETS v0.9.7–13 was employed for copy number analysis (Shen and Seshan 2016), and the segmentation was visualized in R with the plotAberration function from the copy number package (Nilsen, Liestol et al. 2012). Both RNA and DNA libraries were sequenced on an Illumina HiSeq 4000 (paired-end, 100bp or 50bp).
RNAseq data filtering and surrogate variable analysis
To analyze RNAseq data on PDX samples, mouse reads were filtered out using bbsplit.sh from the BBMap v37.76 package (https://github.com/BioInfoTools/BBMap/tree/master). We then only focused on human reads to perform subsequent expression analyses.
To overcome the heterogeneity in gene expression of different samples (e.g., primary and PDX) we constructed “surrogate variables” to assess so-called latent co-variables (variables that are not related to the factor of interest) using the svaseq package.131,132 This method provides a sample-wise matrix of numeric vectors representing surrogate variables which capture sources of variation in the data that are not of primary interest. When incorporated into downstream analyses, e.g., differential expression analysis, these variables are accounted for in the determination of differentially expressed genes. These adjustments resulted in a PCA plot (Figure 3A) defining the separation by subtype and intermingling between primary and PDX samples.
Binding affinity prediction
NetMHCIIpan-4.1 algorithm was used to Predict Binding Affinity of EBV Peptides.133 Each peptide’s binding affinity was predicted for MHCII alleles found in PTCL primary and patient-derived models. To improve binding affinity, the original peptide sequences were extended followed by performing predictions for the prevalent MHCII alleles, which are common in over 90% of the population.134 For the synthesis, peptides with a predicted binding affinity of <1000 nM for the haplotypes found in the patient-derived models were chosen. The peptides were synthesized by Genscript Biotech, NJ, USA.
Single-cell RNA-seq and data analyses
The 10x Genomics Chromium v.2 protocol was carried out according to the manufacturer’s recommendations (10x Genomics, Pleasanton, CA). A total of 2000–5000 cells per sample were processed using Cell Ranger version 2.1.0 with default parameters. Reads were aligned to the human reference sequence hg38. Reads from the PDX tissues were additionally aligned to the mouse reference sequence mm10. Downstream analysis was performed using the Seurat package (version 3.2.2) in R (Butler et al., 2018). Low-quality cells (e.g., cells with >200 or <5000 unique molecular identifiers (UMIs) per cell and cells with >10% mitochondrial gene percentage) were removed. Doublets were identified using DoubletFinder135 and removed. Normalization, variance stabilization, and integration were performed using SCTransform workflow.136 Uniform manifold approximation and projection (UMAP) dimensionality reduction was used to visualize cell clusters. Cells were then annotated using the SingleR package,137 which performs reference-based cell annotation. Two cell-type reference datasets from the celldex package137 were utilized for cell annotation: the human primary cell atlas138 and the Blueprint/ENCODE ref. 139
Targeted deep sequencing and mutation calling
A targeted sequencing gene panel including coding exons and splice sites of 538 genes (target region: ∼3.2 Mb) that are recurrently mutated (>2) in mature T cell neoplasms as well as genomic regions corresponding to recurrent translocations were designed to investigate the genomic profile of the primary and PDX tumors. Using an input of genomic DNA of at least 100 ng isolated from frozen tissues, the next-generation sequencing (NGS) libraries were constructed using the KAPA Hyperplus Kit (Roche), and hybrid selection was performed with the Twist Library Prep Kit (Twist Biosciences), according to manufacturer’s protocols. Multiplexed libraries were sequenced using 150-bp paired end on Hiseq4000 sequencers (Illumina).
Paired sequencing fastq files were aligned to the human genome assembly (hg19) using the Burrows–Wheeler Aligner (version 0.7.1).140 SAMtools (version 1.2)141 were used to sort, index the reads. Then, duplicated reads were marked with Picard (version 1). Somatic mutations were called by the variance calling software SAVI2, which is based on the empirical Bayesian method.142 Somatic mutations that annotated as a synonymous variant, intragenic variant, or intron variant; annotated as a common SNP (dbSnp138); with variant allele frequency ≤ 5% in the tumor sample; with altered reads ≥2 in the matched normal control; with overall mismatch rate >0.02 (https://github.com/genome/bam-readcount) were removed. For PDX samples, reads originated from the mouse genome were removed by filtering out reads with 2 or less unmatched bases compared to the mouse reference genome (GRCm38). For those tumors without normal control, somatic mutations detected in any normal samples in the cohort were additionally filtered out. To annotate and predict the putative pathogenicity of the variants, we used the Annovar dbNSFP variants annotation and set a threshold of at least 3 concordant entries.59 Mutation signature analysis was performed using R package SomaticSignatures.143 Next, we used R package to generate the Ternary plot.144 To infer the mutation order of somatic variants in PTCL, we followed the strategy in Wang et al.71 for the reconstruction of tumor evolutionary directed graph. We selected the recurrently mutated genes to build patient specific evolutionary networks. A mutation observed in both the primary tumor and the PDX was defined as clonal events, whereas subclonal events were mutations only observed in one of these samples. Then, we pooled multiple sequential networks from different patients to construct the mutation order during tumor progression.
Copy number calling, tumor purity estimation, CCF estimation and phylogenetic tree reconstruction
CNVkit145 was used to generate estimated copy number variations (CNV) in a tumor specimen compared with its matching normal. GISTIC 2.0 was used to detect recurrent copy number variations (CNV).146 ABSOLUTE was used to infer tumor purity and cancer cell fraction (CCF) for each WES sample by integrating variant allele frequencies (VAF) and copy number variations (CNV). Then, the ABSOLUTE-annotated MAF files were utilized by PhylogicNDT147 to infer the clonal structure, phylogenetic relationship between clones and evolution among different time points. Fish plots were generated by R package fishplot.148
TCR clonality and prediction of antigen binding
We used MiXCR (http://mixcr.milaboratory.com/and https://github.com/milaboratory/mixcr/), a universal framework that processes big immunome data from raw sequences to quantitated clonotypes.149 We defined explicit clones, populations expressing defined TCRs with a read coverage greater than 20, and the most abundant clone(s) with a significantly higher coverage compared to a second-most covered clonotype. We next interrogated the predicted amino acid sequences of the CDR3 using a curated database of T cell receptor (TCR) sequences with known antigen specificities (https://vdjdb.cdr3.net/search)150 Both TCR alpha and beta chains were evaluated, and mismatched were scored. We then correlated putative binding peptides and their corresponding amminoacidic sequences with HLA-A or HLA-B alleles. T cell Receptor Gamma Gene Rearrangement was performed using a commercial Kit (Gamma TCR clonality was determined by Assay 2.0 - ABI Fluorescence).
Functional experiments
Cell number and viability were assessed by Trypan blue exclusion count (Invitrogen). Cell metabolism was evaluated using luminescence of CTG-tagged ATP kit (cell titer glo Promega kit), the plates were analyzed on a plate reader (Synergy 4, Biotek). Apoptosis was detected using Annexin V-7AAD Apoptosis Detection Kit I (BD Pharmingen), while cell death with propidium iodide staining (PI). Cells were analyzed by flow cytometry (BD LSR-II).
Protein isolation and Western blotting
Cells were lysed in JST buffer (Tris-HCl 20mM Ph7.5, 150 mM NaCl, 1% Triton X-100, 5 mM EDTA, 1mM Na3VO4, 1mM PMSF, 10mM NaF, and 1X protease inhibitor cocktail, Sigma). Protein concentration was determined with the DC protein assay (BioRad) using bovine serum albumin (Sigma) as the standard, and equal amounts of protein were analyzed by SDS-PAGE (12% acrylamide). Gels were electroblotted into nitrocellulose membranes (G & E Healthcare). Membranes were blocked for 1 h with 5% non-fat dry milk (Sigma) in PBS plus 0.1% Tween 20 and incubated at 4°C overnight with the primary antibody. Detection was performed with peroxidase-conjugated secondary antibodies, using the enhanced chemiluminescence system (Thermo Scientific). Primary antibodies used were anti-phospho-p44/42 (Cell Signaling #9101, 1:10 dilution), anti-p44/42 (Cell Signaling #9102, 1:10 dilution) and anti-GAPDH (Cell Signaling #5174, 1:100 dilution).
DeepPTCL
DeepPTCL, a deep learning model designed to predict drug synergy on cell lines from the Cancer Genome Atlas. Drugs represented as SMILES were converted using RDKit into a PyG graph, with atoms represented as nodes and bonds represented as edges. The features of the cell line were gathered from the Genomics of Drug Sensitivity in Cancer. From the normalized expression levels of 17,737 genes, we selected 908 landmark genes. To develop the model, we used DrugComb (the largest database of high-throughput combination screening data) as a training dataset with 2,174 drugs on 164 cell lines, altogether a total of 16,3816 drug-drug-cell triplets. We then trained the model with a 5-fold cross-validation (each colored line represents a run). We designed two challenging tasks for training to improve model generalizability. In the leave-drug-out setup, we excluded drugs seen from training from the test set. For the leave-combination-out setup, the drug pairs from training were removed from the test set, although the same individual drugs might appear in both datasets. The performance of the model was evaluated using AUROC and PRAUC metrics. The higher AUROC and PRAUC value quantifies the better performance of the model. We show DeepPTCL, achieved an AUROC score of 0.806 (CI = 0.801–0.819) and a PRAUC score of 0.54(CI = 0.518–0.576) on the held-out test data of 32,000 drug combinations.
Drug screenings and response prediction
The HTS drug-screening library, composed of 433 targeted-compounds, was purchased from SelleckChem, and consists of a subset of SelleckChem’s ‘Targeted Selective inhibitory Library’. Drugs were selected based on current clinical applications (FDA approved), selectivity (target of canonical signaling pathways [JAK/STAT, Ras/ERK, PI3K/ATK, β-catenin, chromatin-modifiers, anti-apoptotic etc.]) and redundancy (multiple drugs targeting the same pathways). Collectively, a total of 634 proteins were targeted. Drug screening plates were prepared at a concentration of 1μM spanning 2x 384 well plates using the Tecan Freedom EVO 150 (Tecan, CH) in the High throughout and Spectroscopy facility at Rockefeller University. ∼33,000 PTCL cells were added per well, having 150μL of total volume (drug solution + cells), and incubated at 37°C for 72 h. After drug incubation, cell viability was evaluated based on luminescence of CTG-tagged ATP (cell titer glo Promega kit), and assessed using a plate reader (Synergy 4, Biotek), and the data was processed, analyzed, and plotted using MATLAB (Mathworks, MA). To determine compound activity, each data point was normalized to its corresponding in-plate vehicle control (16 wells of vehicle controls per plate), and then linearized to transform the response-matrix (16x33) into a 433x1 drug-response vector. To assess the degree of concordance, sample-replicates were plotted analyzed using principal component analysis (PCA).
For the flow cytometry-based 53 drugs screening, the following conditions were applied: PTCL cells were labeled with Cell Tracer Violet (1μM, Invitrogen), washed and plated (83,000 cells/well) in 96-well plates and challenged with the drug library (1μM) in duplicate/triplicate. After 72 h, all cells were collected and stained with propidium iodide (Sigma). In selected cases, 10000 stromal cells stained with CSFE (1μM, Invitrogen) were previously added and allowed to attach to the plate overnight. Cell viability was assessed by HTS flow cytometry (BD Celesta, Fortessa and Symphony). At least 10,000 events were recorded per well. Flow data were analyzed by FCSExpress 7 (DeNovo Software) and Prism 9 (GraphPad Software, Inc.). When two drugs were screened in combination, compounds were delivered simultaneously at the indicated concentrations on PTCL cells plated as above. Readouts were collected and analyzed as described for the flow cytometry-based 53 drugs screening.
To perform prediction models using viability data and RNAseq data, we used the R caret (Classification And REgression Training) package as previously described, adopting a splitting data procedure with 70% training to 30% test ratio as validation.11
For each drug, we explored gene signatures included in the drug-related KEGG pathway database and focused on gene sets predicting cell viability (“feature selection”) using recursive feature elimination or Gam scoring.151,152
TME FGES
We previously developed a method for TME cell deconvolution and machine learning algorithm to re-construct the TME using bulk expression data.24 This method is based on the presence and association of Functional Gene Expression Signatures (FGES) covering distinct cellular subtypes. Twenty FGES relevant to TME (e.g., cancer-associated fibroblasts and tumor-infiltrating lymphocytes), noncellular components of the tumor microenvironment (e.g., immune-suppressive cytokines and extracellular matrix), biological processes (e.g., secretion and proliferation) were curated from the original FGES publication24 and four canonical pathways activation were calculated using PROGENy (Pathway RespOnsive GENes153). Twenty-four FGES (including four signaling pathways) were developed (Table S7). Selected cellular FGES were also validated using publicly available single-cell RNA-seq data.24
TME clustering
FGES signatures were used to identify microenvironmental patterns among PTCL samples by unsupervised dense clustering using the Louvain method for community detection (63). FGES intensities were median-transformed within each cohort. Non-PTCL samples were also transformed using PTCL samples’ median and MAD values. Inter sample similarity was calculated using Pearson correlation. The resulting distance matrix was converted into a graph where each sample formed a node, and two nodes formed an edge with weight equal to the pair’s Pearson correlation. Edges with weight lower than specified thresholds were removed and the Louvain community detection algorithm was applied to calculate graph partitioning into clusters. To mathematically determine the optimum threshold for observed clusters, we used minimum David Bolduin, maximum Calinski Harabasz and Silhouette scores excluding separations with low-populated clusters (<5% of samples).
TME heterogeneity and pseudotime analysis
Shannon diversity indexes were calculated from the TME cell deconvolution profiles. To calculate the TME heterogeneity index for each tumor, we performed cell deconvolution on the bulk RNA-seq samples from tumor tissue. From this prediction, we calculated the estimated proportion (p) of cells that belong to each distinct cell type. The subpopulation diversity index was then calculated as Shannon Index: DI = −Σi(pi × lnpi), with larger values representing higher TME heterogeneity within the lymphoma. The monocle 2.0 R package154 was implemented for dimensionality reduction and the construction of pseudotime.
Tumor clonality
To process immunome data from RNA sequences into quantitated clonotypes, we applied MiXCR v2.1.7.149,155 Single clonotypes were grouped into clones with specific VDJ combination and identical CDR3 nucleotide sequences. For T cells, the clones were further aggregated into clone groups if the VDJ combination was the same and CDR3 nucleotide sequences differed no more than 1 nucleotide. The biggest clone group was assigned as tumor if the absolute clonotype counts >20; the relative clonotype counts >5%; the ratio of the second biggest group to the first <0.6 and the group contains an enriched clone >25%. The TCRa/b chains were called if there was an enriched clonotype in one of the TCR chains. In cases with an enriched clone in both chains, the biggest by absolute counts was selected.
Viral reads identification
Viral reads identification was based on GATK Pathseq software kit,142 quantitative assessment expressed in VRM (viral read per million human reads). Viral status and serotype verification were performed using the VIRTUS pipeline.143 Threshold for determining viral status as "positive" was chosen at 2 VRM (viral reads per million host reads). At this threshold, the number of raw reads makes it possible to evaluate the expression of viral transcripts. Mapping, score, and quantification of viral transcripts was also analyzed by ViGEN.144
Analysis of murine TME signatures
We used Kallisto v.0.42.4156 to align RNA-sequencing reads to the transcriptome reference GRCm38.p6. Transcript annotation (protein coding and noncoding), transcript to gene mapping, and annotation to human homologs for murine genes were retrieved from the Ensembl database.157For the PDX sample xengsort algorithm146 was used to split reads into human, mouse or neither origin. Gene expression was calculated by combining host and graft raw Kallisto output and refactoring TPM values. Only protein-coding genes with human homologs were used in subsequent analysis. To measure the similarity between the human lymphoma sample microenvironment and a particular murine lymphoma phenotype, we developed the microenvironment similarity (MES) metric, which is a reversed Euclidean distance between estimated percentages of different cell types in murine and human samples as follows: MES (P, s) = 1i(phumani − pmousei)2, where P is a murine lymphoma phenotype, s is a human lymphoma sample, phumani is the estimated cell percentage of cell type i in the sample s, pmousei is the median estimated cell percentage of cell type i among murine samples with phenotype P.
Kassandra deconvolution
To get more quantitative insights into the human and PDX TME we utilized previously developed algorithm Kassandra. It allows to estimate percentages of the major populations of cells using bulk RNA-sequencing.77
CART generation and production
Human T cells obtained from the University of Pennsylvania Human Immunology Core were combined in a 1:1 ratio of CD4+ and CD8+ cells. For cells subjected to electroporation (CD5 KO CART5 cells), a mixture of 5 μg sgRNA and 10 μg of TrueCut Cas9 Protein v2 (Invitrogen; Cat# A36499) was prepared and incubated at room temperature for 10 min to form a ribonucleoprotein (RNP) complex before electroporation. The CRISPR-Cas9 sgRNA was chemically synthesized (Integrated DNA Technologies). A total of 10 × 106 T cells in 100 μL of the buffer provided with P3 Primary Cell 4D-Nucleofector X Kit L (Lonza; Cat# V4XP-3024) were mixed with the RNP complex and electroporated using the pulse code EO-115 in a 4D-Nucleofector (Lonza; Cat# AAF-1002B). Mock KO UTD cells underwent the same electroporation procedure described above without the presence of an RNP complex. Following electroporation, T cells were incubated at 37°C for 24 h and subsequently activated using CD3/CD28 Dynabeads (Gibco; Cat# 40203D) at a ratio of 3 beads per cell. For cells not subjected to electroporation (UTD and CART30), the expansion process began at the CD3/CD28 Dynabead activation step. The following day, CAR lentiviral vectors (CAR5 or CAR30) were introduced to the stimulated cultures at a multiplicity of infections ranging between 1 and 3. Beads were removed between days 6–8 post-stimulation, and cell counting was performed every other day using a Moxi GO II (Orflo) until growth kinetics and cell size indicated they had rested from stimulation. Initially, all electroporated T cells were cultured with 20 ng/mL of supplemental cytokines IL-7 and IL-15, which were gradually reduced to 0 ng/mL by the end of the expansion process.
For the in vitro cytotoxic assays, PDX-Dline cells (5x105/well) were cocultured with CAR T cells at the indicated E:T ratios in 96 well round bottom for 24-48-72 h. Cells were harvested, washed and stained with an amine-reactive viability dye for dead cell detection and surface T cell associated/restricted antigens. The samples were analyzed by flow cytometry (FCM). Percent of viable cells, and target cells (PDX-Dline cells of CART) were enumerated. Specific lysis was calculated using the following formula: % specific lysis = [(experimental lysis – spontaneous lysis)/(maximum lysis – spontaneous lysis)] × 100.
For the in vivo CART adaptive therapies 6–10-week-old NOD-SCID-γc−/− (NSG) mice were injected on day 7–14 after s.c. tumor delivery, 1 × 106 T cells (control or CAR engineered) were injected via tail vein in 0.2mL sterile PBS. Animals were monitored for signs of disease progression and overt toxicity, such as xenogeneic graft-versus-host disease, as evidenced by >10% loss in body weight, loss of fur, diarrhea, conjunctivitis, and disease-related hindlimb paralysis.
Preclinical in vivo studies
For in vivo experiments, xenografts were surgically implanted in 4–8 weeks old adult NSG mice and numbers, age, and gender of the mice were equally distributed among arms. Compounds were administered according to Table S10. Mice were closely followed for symptoms of tumor progression until moribund. Tumor burden was evaluated by digital caliper twice a week on s.c. masses or MRI where specified. Bodyweight was used as a surrogate for drug toxicity. Statistics and graphic representation were generated using Prism 9 (GraphPad Software, Inc.).
Quantification and Statistical analysis
Statistics and software
Statistical significance was calculated by two-tailed Student’s t test and Mantel–Cox test with GraphPad Prism software. GraphPad Prism software was used to calculate the IC50 values. Statistically significant differences are indicated with asterisks in the figures, accompanied by p values in the figure legends. Error bars indicate standard error of the mean (SEM) for the number of replicates, as indicated in the figure legends. Data visualization and plots were generated in R. Schematic visualizations were produced at https://biorender.com.
Published: March 26, 2025
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2025.102029.
Supplemental information
A: PDX and PDX-Dlines list, B: patient clinical annotation, C: primary-PDX identity, D: all implanted samples source
A: WES, B: bulk RNA-seq, C: scRNA-seq, D: DeepSeq, E: TCR_data, F: BCR_data, G: EBV status, H: TCR_EBV, I: EBV match TCR, J: MHC12_AITL
A: Fusions detected in the TCL PDX cohort
A: Copy-number aberrations estimated from WES data in the TCL PDX cohort
A: NY-AI-AM, B: NY-AI-CL, C: NY-AT-FC, D: TO-ALCL-BELLI, E: COH1, F: TO-ALCL-DN03, G: IL114A, H: IL-2, I: IL6, J: IL17, K: IL19, L: IL33, M: IL34, N: IL35, O: IL36, P: IL38B, Q: IL40, R: IL41, S: IL49, T: IL54, U: IL56, V: IL65, W: IL69, X: IL76, Y: IL78, Z: IL79, AA: IL81, AB: IL86, AC: IL88, AD: IL89, AE: IL92, AF: IL94, AG: IL98, AH: IL104, AI: IL107, AJ: IL122A, AK: IL123, AL: IL126A, AM: IL126B, AN: IL129A, AO: IL135A, AP: IL138A, AQ: TO-ALCL-MARC, AR: TO-ALCL-MARI, AS: MT05, AT: NY-NK-CD, AU: NY-PTCL-RV, AV: NY-ALCL-SG, AW: NY-NK-VP
A: clonal evolution analysis reporting cancer cell fraction estimates and mutation clonality
A: datasets utilized to generate microenvironment signatures, B: TME subtype of 70 primary PTCL
A: list of 433 compounds utilized in the high-throughput (HTS) drug screening
References
- 1.Swerdlow S.H., Campo E., Arber D.A., Cazzola M., Cook J.R., Döhner H., Dreyling M., Hasserjian R.P., Jaffe E.S., Orazi A., et al. Response to "The WHO classification of haematolymphoid tumours" (Editorial) Leukemia. 2022;36:2748–2749. doi: 10.1038/s41375-022-01689-9. [DOI] [PubMed] [Google Scholar]
- 2.Fiore D., Cappelli L.V., Broccoli A., Zinzani P.L., Chan W.C., Inghirami G. Peripheral T cell lymphomas: from the bench to the clinic. Nat. Rev. Cancer. 2020;20:323–342. doi: 10.1038/s41568-020-0247-0. [DOI] [PubMed] [Google Scholar]
- 3.Vose J., Armitage J., Weisenburger D., International T-Cell Lymphoma Project International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J. Clin. Oncol. 2008;26:4124–4130. doi: 10.1200/JCO.2008.16.4558. [DOI] [PubMed] [Google Scholar]
- 4.Mak V., Hamm J., Chhanabhai M., Shenkier T., Klasa R., Sehn L.H., Villa D., Gascoyne R.D., Connors J.M., Savage K.J. Survival of patients with peripheral T-cell lymphoma after first relapse or progression: spectrum of disease and rare long-term survivors. J. Clin. Oncol. 2013;31:1970–1976. doi: 10.1200/JCO.2012.44.7524. [DOI] [PubMed] [Google Scholar]
- 5.Khan N., Cheson B.D. PTCL therapies: a review of treatment and outline of novel therapies. Am. J. Therapeut. 2013;20:524–533. doi: 10.1097/MJT.0b013e31828232a3. [DOI] [PubMed] [Google Scholar]
- 6.Moskowitz A.J., Stuver R.N., Horwitz S.M. Current and upcoming treatment approaches to common subtypes of PTCL (PTCL NOS, ALCL, TFHs) Blood. 2024;144:1887–1897. doi: 10.1182/blood.2023021789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broccoli A., Argnani L., Zinzani P.L. Peripheral T-cell lymphomas: Focusing on novel agents in relapsed and refractory disease. Cancer Treat Rev. 2017;60:120–129. doi: 10.1016/j.ctrv.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Ma H., Marchi E., O'Connor O.A. The peripheral T-cell lymphomas: an unusual path to cure. Lancet. Haematol. 2020;7:e765–e771. doi: 10.1016/S2352-3026(20)30207-6. [DOI] [PubMed] [Google Scholar]
- 9.Jack J., Rotroff D., Motsinger-Reif A. Lymphoblastoid cell lines models of drug response: successes and lessons from this pharmacogenomic model. Curr. Mol. Med. 2014;14:833–840. doi: 10.2174/1566524014666140811113946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan R., Chen S., Wang Y. Computational Prediction of Drug Responses in Cancer Cell Lines From Cancer Omics and Detection of Drug Effectiveness Related Methylation Sites. Front. Genet. 2020;11:917. doi: 10.3389/fgene.2020.00917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurilov R., Haibe-Kains B., Brors B. Assessment of modelling strategies for drug response prediction in cell lines and xenografts. Sci. Rep. 2020;10:2849. doi: 10.1038/s41598-020-59656-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cutucache C.E., Herek T.A. Burrowing through the Heterogeneity: Review of Mouse Models of PTCL-NOS. Front. Oncol. 2016;6:206. doi: 10.3389/fonc.2016.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiarle R., Gong J.Z., Guasparri I., Pesci A., Cai J., Liu J., Simmons W.J., Dhall G., Howes J., Piva R., Inghirami G. NPM-ALK transgenic mice spontaneously develop T-cell lymphomas and plasma cell tumors. Blood. 2003;101:1919–1927. doi: 10.1182/blood-2002-05-1343. [DOI] [PubMed] [Google Scholar]
- 14.Dierks C., Adrian F., Fisch P., Ma H., Maurer H., Herchenbach D., Forster C.U., Sprissler C., Liu G., Rottmann S., et al. The ITK-SYK fusion oncogene induces a T-cell lymphoproliferative disease in mice mimicking human disease. Cancer Res. 2010;70:6193–6204. doi: 10.1158/0008-5472.CAN-08-3719. [DOI] [PubMed] [Google Scholar]
- 15.Mhaidly R., Krug A., Gaulard P., Lemonnier F., Ricci J.E., Verhoeyen E. New preclinical models for angioimmunoblastic T-cell lymphoma: filling the GAP. Oncogenesis. 2020;9:73. doi: 10.1038/s41389-020-00259-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Debackere K., Marcelis L., Demeyer S., Vanden Bempt M., Mentens N., Gielen O., Jacobs K., Broux M., Verhoef G., Michaux L., et al. Fusion transcripts FYN-TRAF3IP2 and KHDRBS1-LCK hijack T cell receptor signaling in peripheral T-cell lymphoma, not otherwise specified. Nat. Commun. 2021;12:3705. doi: 10.1038/s41467-021-24037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng S.Y., Yoshida N., Christie A.L., Ghandi M., Dharia N.V., Dempster J., Murakami M., Shigemori K., Morrow S.N., Van Scoyk A., et al. Targetable vulnerabilities in T- and NK-cell lymphomas identified through preclinical models. Nat. Commun. 2018;9:2024. doi: 10.1038/s41467-018-04356-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mer A.S., Ba-Alawi W., Smirnov P., Wang Y.X., Brew B., Ortmann J., Tsao M.S., Cescon D.W., Goldenberg A., Haibe-Kains B. Integrative Pharmacogenomics Analysis of Patient-Derived Xenografts. Cancer Res. 2019;79:4539–4550. doi: 10.1158/0008-5472.CAN-19-0349. [DOI] [PubMed] [Google Scholar]
- 19.Ireson C.R., Alavijeh M.S., Palmer A.M., Fowler E.R., Jones H.J. The role of mouse tumour models in the discovery and development of anticancer drugs. Br. J. Cancer. 2019;121:101–108. doi: 10.1038/s41416-019-0495-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Tunon I., Hernandez-Sanchez M., Ordonez J.L., Alonso-Perez V., Alamo-Quijada M., Benito R., Guerrero C., Hernandez-Rivas J.M., Sanchez-Martin M. The CRISPR/Cas9 system efficiently reverts the tumorigenic ability of BCR/ABL in vitro and in a xenograft model of chronic myeloid leukemia. Oncotarget. 2017;8:26027–26040. doi: 10.18632/oncotarget.15215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei X., Yang J., Adair S.J., Ozturk H., Kuscu C., Lee K.Y., Kane W.J., O'Hara P.E., Liu D., Demirlenk Y.M., et al. Targeted CRISPR screening identifies PRMT5 as synthetic lethality combinatorial target with gemcitabine in pancreatic cancer cells. Proc. Natl. Acad. Sci. USA. 2020;117:28068–28079. doi: 10.1073/pnas.2009899117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szlachta K., Kuscu C., Tufan T., Adair S.J., Shang S., Michaels A.D., Mullen M.G., Fischer N.L., Yang J., Liu L., et al. CRISPR knockout screening identifies combinatorial drug targets in pancreatic cancer and models cellular drug response. Nat. Commun. 2018;9:4275. doi: 10.1038/s41467-018-06676-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maykel J., Liu J.H., Li H., Shultz L.D., Greiner D.L., Houghton J. NOD-scidIl2rg (tm1Wjl) and NOD-Rag1 (null) Il2rg (tm1Wjl) : a model for stromal cell-tumor cell interaction for human colon cancer. Dig. Dis. Sci. 2014;59:1169–1179. doi: 10.1007/s10620-014-3168-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotlov N., Bagaev A., Revuelta M.V., Phillip J.M., Cacciapuoti M.T., Antysheva Z., Svekolkin V., Tikhonova E., Miheecheva N., Kuzkina N., et al. Clinical and Biological Subtypes of B-cell Lymphoma Revealed by Microenvironmental Signatures. Cancer Discov. 2021;11:1468–1489. doi: 10.1158/2159-8290.CD-20-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdolahi S., Ghazvinian Z., Muhammadnejad S., Saleh M., Asadzadeh Aghdaei H., Baghaei K. Patient-derived xenograft (PDX) models, applications and challenges in cancer research. J. Transl. Med. 2022;20:206. doi: 10.1186/s12967-022-03405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao H., Korn J.M., Ferretti S., Monahan J.E., Wang Y., Singh M., Zhang C., Schnell C., Yang G., Zhang Y., et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 2015;21:1318–1325. doi: 10.1038/nm.3954. [DOI] [PubMed] [Google Scholar]
- 27.Sun H., Cao S., Mashl R.J., Mo C.K., Zaccaria S., Wendl M.C., Davies S.R., Bailey M.H., Primeau T.M., Hoog J., et al. Author Correction: Comprehensive characterization of 536 patient-derived xenograft models prioritizes candidates for targeted treatment. Nat. Commun. 2022;13:294. doi: 10.1038/s41467-021-27678-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aparicio S., Hidalgo M., Kung A.L. Examining the utility of patient-derived xenograft mouse models. Nat. Rev. Cancer. 2015;15:311–316. doi: 10.1038/nrc3944. [DOI] [PubMed] [Google Scholar]
- 29.Klco J.M., Spencer D.H., Miller C.A., Griffith M., Lamprecht T.L., O'Laughlin M., Fronick C., Magrini V., Demeter R.T., Fulton R.S., et al. Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell. 2014;25:379–392. doi: 10.1016/j.ccr.2014.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ben-David U., Ha G., Tseng Y.Y., Greenwald N.F., Oh C., Shih J., McFarland J.M., Wong B., Boehm J.S., Beroukhim R., Golub T.R. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat. Genet. 2017;49:1567–1575. doi: 10.1038/ng.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willyard C. The mice with human tumours: Growing pains for a popular cancer model. Nature. 2018;560:156–157. doi: 10.1038/d41586-018-05890-8. [DOI] [PubMed] [Google Scholar]
- 32.Hidalgo M., Amant F., Biankin A.V., Budinská E., Byrne A.T., Caldas C., Clarke R.B., de Jong S., Jonkers J., Mælandsmo G.M., et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 2014;4:998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woo X.Y., Giordano J., Srivastava A., Zhao Z.M., Lloyd M.W., de Bruijn R., Suh Y.S., Patidar R., Chen L., Scherer S., et al. Conservation of copy number profiles during engraftment and passaging of patient-derived cancer xenografts. Nat. Genet. 2021;53:86–99. doi: 10.1038/s41588-020-00750-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chapuy B., Cheng H., Watahiki A., Ducar M.D., Tan Y., Chen L., Roemer M.G.M., Ouyang J., Christie A.L., Zhang L., et al. Diffuse large B-cell lymphoma patient-derived xenograft models capture the molecular and biologic heterogeneity of the disease. Blood. 2016;127:2203–2213. doi: 10.1182/blood-2015-09-672352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Townsend E.C., Murakami M.A., Christodoulou A., Christie A.L., Köster J., DeSouza T.A., Morgan E.A., Kallgren S.P., Liu H., Wu S.C., et al. The Public Repository of Xenografts Enables Discovery and Randomized Phase II-like Trials in Mice. Cancer Cell. 2016;29:574–586. doi: 10.1016/j.ccell.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L., Nomie K., Zhang H., Bell T., Pham L., Kadri S., Segal J., Li S., Zhou S., Santos D., et al. B-Cell Lymphoma Patient-Derived Xenograft Models Enable Drug Discovery and Are a Platform for Personalized Therapy. Clin. Cancer Res. 2017;23:4212–4223. doi: 10.1158/1078-0432.CCR-16-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng S.Y., Brown L., Stevenson K., deSouza T., Aster J.C., Louissaint A., Jr., Weinstock D.M. RhoA G17V is sufficient to induce autoimmunity and promotes T-cell lymphomagenesis in mice. Blood. 2018;132:935–947. doi: 10.1182/blood-2017-11-818617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zammarchi F., Corbett S., Adams L., Tyrer P.C., Kiakos K., Janghra N., Marafioti T., Britten C.E., Havenith C.E.G., Chivers S., et al. ADCT-402, a PBD dimer-containing antibody drug conjugate targeting CD19-expressing malignancies. Blood. 2018;131:1094–1105. doi: 10.1182/blood-2017-10-813493. [DOI] [PubMed] [Google Scholar]
- 39.Wang T., Lu Y., Polk A., Chowdhury P., Murga-Zamalloa C., Fujiwara H., Suemori K., Beyersdorf N., Hristov A.C., Lim M.S., et al. T-cell Receptor Signaling Activates an ITK/NF-kappaB/GATA-3 axis in T-cell Lymphomas Facilitating Resistance to Chemotherapy. Clin. Cancer Res. 2017;23:2506–2515. doi: 10.1158/1078-0432.CCR-16-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakhoul H., Lin Z., Wang X., Roberts C., Dong Y., Flemington E. High-Throughput Sequence Analysis of Peripheral T-Cell Lymphomas Indicates Subtype-Specific Viral Gene Expression Patterns and Immune Cell Microenvironments. mSphere. 2019;4 doi: 10.1128/mSphere.00248-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iqbal J., Wright G., Wang C., Rosenwald A., Gascoyne R.D., Weisenburger D.D., Greiner T.C., Smith L., Guo S., Wilcox R.A., et al. Gene expression signatures delineate biological and prognostic subgroups in peripheral T-cell lymphoma. Blood. 2014;123:2915–2923. doi: 10.1182/blood-2013-11-536359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amador C., Greiner T.C., Heavican T.B., Smith L.M., Galvis K.T., Lone W., Bouska A., D'Amore F., Pedersen M.B., Pileri S., et al. Reproducing the molecular subclassification of peripheral T-cell lymphoma-NOS by immunohistochemistry. Blood. 2019;134:2159–2170. doi: 10.1182/blood.2019000779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott D.W., Mungall K.L., Ben-Neriah S., Rogic S., Morin R.D., Slack G.W., Tan K.L., Chan F.C., Lim R.S., Connors J.M., et al. TBL1XR1/TP63: a novel recurrent gene fusion in B-cell non-Hodgkin lymphoma. Blood. 2012;119:4949–4952. doi: 10.1182/blood-2012-02-414441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Della-Valle V., Roos-Weil D., Scourzic L., Mouly E., Aid Z., Darwiche W., Lecluse Y., Damm F., Mémet S., Mercher T., et al. Nfkbie-deficiency leads to increased susceptibility to develop B-cell lymphoproliferative disorders in aged mice. Blood Cancer J. 2020;10:38. doi: 10.1038/s41408-020-0305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramis-Zaldivar J.E., Gonzalez-Farré B., Balagué O., Celis V., Nadeu F., Salmerón-Villalobos J., Andrés M., Martin-Guerrero I., Garrido-Pontnou M., Gaafar A., et al. Distinct molecular profile of IRF4-rearranged large B-cell lymphoma. Blood. 2020;135:274–286. doi: 10.1182/blood.2019002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abate F., da Silva-Almeida A.C., Zairis S., Robles-Valero J., Couronne L., Khiabanian H., Quinn S.A., Kim M.Y., Laginestra M.A., Kim C., et al. Activating mutations and translocations in the guanine exchange factor VAV1 in peripheral T-cell lymphomas. Proc. Natl. Acad. Sci. USA. 2017;114:764–769. doi: 10.1073/pnas.1608839114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujisawa M., Sakata-Yanagimoto M., Nishizawa S., Komori D., Gershon P., Kiryu M., Tanzima S., Fukumoto K., Enami T., Muratani M., et al. Activation of RHOA-VAV1 signaling in angioimmunoblastic T-cell lymphoma. Leukemia. 2018;32:694–702. doi: 10.1038/leu.2017.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turner S.D., Yeung D., Hadfield K., Cook S.J., Alexander D.R. The NPM-ALK tyrosine kinase mimics TCR signalling pathways, inducing NFAT and AP-1 by RAS-dependent mechanisms. Cell. Signal. 2007;19:740–747. doi: 10.1016/j.cellsig.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 49.Chiarle R., Martinengo C., Mastini C., Ambrogio C., D'Escamard V., Forni G., Inghirami G. The anaplastic lymphoma kinase is an effective oncoantigen for lymphoma vaccination. Nat. Med. 2008;14:676–680. doi: 10.1038/nm1769. [DOI] [PubMed] [Google Scholar]
- 50.Nicolae A., Xi L., Pham T.H., Pham T.A., Navarro W., Meeker H.G., Pittaluga S., Jaffe E.S., Raffeld M. Mutations in the JAK/STAT and RAS signaling pathways are common in intestinal T-cell lymphomas. Leukemia. 2016;30:2245–2247. doi: 10.1038/leu.2016.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boddicker R.L., Razidlo G.L., Feldman A.L. Genetic alterations affecting GTPases and T-cell receptor signaling in peripheral T-cell lymphomas. Small GTPases. 2019;10:33–39. doi: 10.1080/21541248.2016.1263718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stengel A., Shahswar R., Haferlach T., Walter W., Hutter S., Meggendorfer M., Kern W., Haferlach C. Whole transcriptome sequencing detects a large number of novel fusion transcripts in patients with AML and MDS. Blood Adv. 2020;4:5393–5401. doi: 10.1182/bloodadvances.2020003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Foltz S.M., Gao Q., Yoon C.J., Sun H., Yao L., Li Y., Jayasinghe R.G., Cao S., King J., Kohnen D.R., et al. Evolution and structure of clinically relevant gene fusions in multiple myeloma. Nat. Commun. 2020;11:2666. doi: 10.1038/s41467-020-16434-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boi M., Rinaldi A., Kwee I., Bonetti P., Todaro M., Tabbò F., Piva R., Rancoita P.M.V., Matolcsy A., Timar B., et al. PRDM1/BLIMP1 is commonly inactivated in anaplastic large T-cell lymphoma. Blood. 2013;122:2683–2693. doi: 10.1182/blood-2013-04-497933. [DOI] [PubMed] [Google Scholar]
- 55.de Mel S., Soon G.S.T., Mok Y., Chung T.H., Jeyasekharan A.D., Chng W.J., Ng S.B. The Genomics and Molecular Biology of Natural Killer/T-Cell Lymphoma: Opportunities for Translation. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19071931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lone W., Bouska A., Sharma S., Amador C., Saumyaranjan M., Herek T.A., Heavican T.B., Yu J., Lim S.T., Ong C.K., et al. Genome-Wide miRNA Expression Profiling of Molecular Subgroups of Peripheral T-cell Lymphoma. Clin. Cancer Res. 2021;27:6039–6053. doi: 10.1158/1078-0432.CCR-21-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gasparini P., Cascione L., Landi L., Carasi S., Lovat F., Tibaldi C., Alì G., D'Incecco A., Minuti G., Chella A., et al. microRNA classifiers are powerful diagnostic/prognostic tools in ALK-EGFR-and KRAS-driven lung cancers. Proc. Natl. Acad. Sci. USA. 2015;112:14924–14929. doi: 10.1073/pnas.1520329112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bagot M., Porcu P., Marie-Cardine A., Battistella M., William B.M., Vermeer M., Whittaker S., Rotolo F., Ram-Wolff C., Khodadoust M.S., et al. IPH4102, a first-in-class anti-KIR3DL2 monoclonal antibody, in patients with relapsed or refractory cutaneous T-cell lymphoma: an international, first-in-human, open-label, phase 1 trial. Lancet Oncol. 2019;20:1160–1170. doi: 10.1016/S1470-2045(19)30320-1. [DOI] [PubMed] [Google Scholar]
- 59.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38 doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vallois D., Dobay M.P.D., Morin R.D., Lemonnier F., Missiaglia E., Juilland M., Iwaszkiewicz J., Fataccioli V., Bisig B., Roberti A., et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell-derived lymphomas. Blood. 2016;128:1490–1502. doi: 10.1182/blood-2016-02-698977. [DOI] [PubMed] [Google Scholar]
- 61.Lemonnier F., Couronné L., Parrens M., Jaïs J.P., Travert M., Lamant L., Tournillac O., Rousset T., Fabiani B., Cairns R.A., et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood. 2012;120:1466–1469. doi: 10.1182/blood-2012-02-408542. [DOI] [PubMed] [Google Scholar]
- 62.Waldmann T.A., Chen J. Disorders of the JAK/STAT Pathway in T Cell Lymphoma Pathogenesis: Implications for Immunotherapy. Annu. Rev. Immunol. 2017;35:533–550. doi: 10.1146/annurev-immunol-110416-120628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaustio M., Nayebzadeh N., Hinttala R., Tapiainen T., Åström P., Mamia K., Pernaa N., Lehtonen J., Glumoff V., Rahikkala E., et al. Loss of DIAPH1 causes SCBMS, combined immunodeficiency, and mitochondrial dysfunction. J. Allergy Clin. Immunol. 2021;148:599–611. doi: 10.1016/j.jaci.2020.12.656. [DOI] [PubMed] [Google Scholar]
- 64.Dobashi A., Tsuyama N., Asaka R., Togashi Y., Ueda K., Sakata S., Baba S., Sakamoto K., Hatake K., Takeuchi K. Frequent BCOR aberrations in extranodal NK/T-Cell lymphoma, nasal type. Genes Chromosomes Cancer. 2016;55:460–471. doi: 10.1002/gcc.22348. [DOI] [PubMed] [Google Scholar]
- 65.Li D.Q., Nair S.S., Ohshiro K., Kumar A., Nair V.S., Pakala S.B., Reddy S.D.N., Gajula R.P., Eswaran J., Aravind L., Kumar R. MORC2 signaling integrates phosphorylation-dependent, ATPase-coupled chromatin remodeling during the DNA damage response. Cell Rep. 2012;2:1657–1669. doi: 10.1016/j.celrep.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim T.W., Kang Y.K., Park Z.Y., Kim Y.H., Hong S.W., Oh S.J., Sohn H.A., Yang S.J., Jang Y.J., Lee D.C., et al. SH3RF2 functions as an oncogene by mediating PAK4 protein stability. Carcinogenesis. 2014;35:624–634. doi: 10.1093/carcin/bgt338. [DOI] [PubMed] [Google Scholar]
- 67.Zhou X., Wen Y., Tian Y., He M., Ke X., Huang Z., He Y., Liu L., Scharf A., Lu M., et al. Heat Shock Protein 90alpha-Dependent B-Cell-2-Associated Transcription Factor 1 Promotes Hepatocellular Carcinoma Proliferation by Regulating MYC Proto-Oncogene c-MYC mRNA Stability. Hepatology. 2019;69:1564–1581. doi: 10.1002/hep.30172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sakata-Yanagimoto M., Enami T., Yoshida K., Shiraishi Y., Ishii R., Miyake Y., Muto H., Tsuyama N., Sato-Otsubo A., Okuno Y., et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat. Genet. 2014;46:171–175. doi: 10.1038/ng.2872. [DOI] [PubMed] [Google Scholar]
- 69.Crescenzo R., Abate F., Lasorsa E., Tabbo F., Gaudiano M., Chiesa N., Di Giacomo F., Spaccarotella E., Barbarossa L., Ercole E., et al. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell. 2015;27:516–532. doi: 10.1016/j.ccell.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perez C., Mondejar R., Garcia-Diaz N., Cereceda L., Leon A., Montes S., Duran Vian C., Perez Paredes M.G., Gonzalez-Moran A., Alegre de Miguel V., et al. Advanced-stage mycosis fungoides: role of the signal transducer and activator of transcription 3, nuclear factor-kappaB and nuclear factor of activated T cells pathways. Br. J. Dermatol. 2020;182:147–155. doi: 10.1111/bjd.18098. [DOI] [PubMed] [Google Scholar]
- 71.Wang J., Khiabanian H., Rossi D., Fabbri G., Gattei V., Forconi F., Laurenti L., Marasca R., Del Poeta G., Foà R., et al. Tumor evolutionary directed graphs and the history of chronic lymphocytic leukemia. Elife. 2014;3 doi: 10.7554/eLife.02869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang J., Cazzato E., Ladewig E., Frattini V., Rosenbloom D.I.S., Zairis S., Abate F., Liu Z., Elliott O., Shin Y.J., et al. Clonal evolution of glioblastoma under therapy. Nat. Genet. 2016;48:768–776. doi: 10.1038/ng.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carter S.L., Cibulskis K., Helman E., McKenna A., Shen H., Zack T., Laird P.W., Onofrio R.C., Winckler W., Weir B.A., et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brastianos P.K., Carter S.L., Santagata S., Cahill D.P., Taylor-Weiner A., Jones R.T., Van Allen E.M., Lawrence M.S., Horowitz P.M., Cibulskis K., et al. Genomic Characterization of Brain Metastases Reveals Branched Evolution and Potential Therapeutic Targets. Cancer Discov. 2015;5:1164–1177. doi: 10.1158/2159-8290.CD-15-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee J.K., Wang J., Sa J.K., Ladewig E., Lee H.O., Lee I.H., Kang H.J., Rosenbloom D.S., Camara P.G., Liu Z., et al. Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat. Genet. 2017;49:594–599. doi: 10.1038/ng.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schurch C.M., Federmann B., Quintanilla-Martinez L., Fend F. Tumor Heterogeneity in Lymphomas: A Different Breed. Pathobiology. 2018;85:130–145. doi: 10.1159/000475530. [DOI] [PubMed] [Google Scholar]
- 77.Zaitsev A., Chelushkin M., Dyikanov D., Cheremushkin I., Shpak B., Nomie K., Zyrin V., Nuzhdina E., Lozinsky Y., Zotova A., et al. Precise reconstruction of the TME using bulk RNA-seq and a machine learning algorithm trained on artificial transcriptomes. Cancer Cell. 2022;40:879–894. doi: 10.1016/j.ccell.2022.07.006. [DOI] [PubMed] [Google Scholar]
- 78.Huang Y.H., Qiu Y.R., Zhang Q.L., Cai M.C., Yu H., Zhang J.M., Jiang L., Ji M.M., Xu P.P., Wang L., et al. Genomic and transcriptomic profiling of peripheral T cell lymphoma reveals distinct molecular and microenvironment subtypes. Cell Rep. Med. 2024;5 doi: 10.1016/j.xcrm.2024.101416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ambrogio C., Martinengo C., Voena C., Tondat F., Riera L., di Celle P.F., Inghirami G., Chiarle R. NPM-ALK oncogenic tyrosine kinase controls T-cell identity by transcriptional regulation and epigenetic silencing in lymphoma cells. Cancer Res. 2009;69:8611–8619. doi: 10.1158/0008-5472.CAN-09-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vallois D., Dupuy A., Lemonnier F., Allen G., Missiaglia E., Fataccioli V., Ortonne N., Clavert A., Delarue R., Rousselet M.C., et al. RNA fusions involving CD28 are rare in peripheral T-cell lymphomas and concentrate mainly in those derived from follicular helper T cells. Haematologica. 2018;103:e360–e363. doi: 10.3324/haematol.2017.186767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou Y., Attygalle A.D., Chuang S.S., Diss T., Ye H., Liu H., Hamoudi R.A., Munson P., Bacon C.M., Dogan A., Du M.Q. Angioimmunoblastic T-cell lymphoma: histological progression associates with EBV and HHV6B viral load. Br. J. Haematol. 2007;138:44–53. doi: 10.1111/j.1365-2141.2007.06620.x. [DOI] [PubMed] [Google Scholar]
- 82.Chiba S., Sakata-Yanagimoto M. Advances in understanding of angioimmunoblastic T-cell lymphoma. Leukemia. 2020;34:2592–2606. doi: 10.1038/s41375-020-0990-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lwin T., Hazlehurst L.A., Li Z., Dessureault S., Sotomayor E., Moscinski L.C., Dalton W.S., Tao J. Bone marrow stromal cells prevent apoptosis of lymphoma cells by upregulation of anti-apoptotic proteins associated with activation of NF-kappaB (RelB/p52) in non-Hodgkin's lymphoma cells. Leukemia. 2007;21:1521–1531. doi: 10.1038/sj.leu.2404723. [DOI] [PubMed] [Google Scholar]
- 84.Blonska M., Agarwal N.K., Vega F. Shaping of the tumor microenvironment: Stromal cells and vessels. Semin. Cancer Biol. 2015;34:3–13. doi: 10.1016/j.semcancer.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mastini C., Campisi M., Patrucco E., Mura G., Ferreira A., Costa C., Ambrogio C., Germena G., Martinengo C., Peola S., et al. Targeting CCR7-PI3Kgamma overcomes resistance to tyrosine kinase inhibitors in ALK-rearranged lymphoma. Sci. Transl. Med. 2023;15 doi: 10.1126/scitranslmed.abo3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Niitsu N., Kohori M., Higashihara M., Bessho M. Phase II study of the irinotecan (CPT-11), mitoxantrone and dexamethasone regimen in elderly patients with relapsed or refractory peripheral T-cell lymphoma. Cancer Sci. 2007;98:109–112. doi: 10.1111/j.1349-7006.2006.00349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Saotome T., Takagi T., Sakai C., Kumagai K., Tamaru J. Combination chemotherapy with irinotecan and adriamycin for refractory and relapsed non-Hodgkin's lymphoma. Ann. Oncol. 2000;11:115–116. doi: 10.1023/a:1008368905546. [DOI] [PubMed] [Google Scholar]
- 88.Kang H.J., Kim W.S., Suh C., Park Y.H., Kim B.S., Yuh Y.J., Ryoo B.Y. Irinotecan plus cisplatin and dexamethasone (ICD) combination chemotherapy for patients with diffuse large B-cell lymphoma previously treated with Rituximab plus CHOP. Cancer Chemother. Pharmacol. 2008;62:299–304. doi: 10.1007/s00280-007-0607-4. [DOI] [PubMed] [Google Scholar]
- 89.Sharkey R.M., Govindan S.V., Cardillo T.M., Goldenberg D.M. Epratuzumab-SN-38: a new antibody-drug conjugate for the therapy of hematologic malignancies. Mol. Cancer Therapeut. 2012;11:224–234. doi: 10.1158/1535-7163.MCT-11-0632. [DOI] [PubMed] [Google Scholar]
- 90.Lemonnier F., Dupuis J., Sujobert P., Tournillhac O., Cheminant M., Sarkozy C., Pelletier L., Marçais A., Robe C., Fataccioli V., et al. Treatment with 5-azacytidine induces a sustained response in patients with angioimmunoblastic T-cell lymphoma. Blood. 2018;132:2305–2309. doi: 10.1182/blood-2018-04-840538. [DOI] [PubMed] [Google Scholar]
- 91.Ruan J., Moskowitz A., Mehta-Shah N., Sokol L., Chen Z., Kotlov N., Nos G., Sorokina M., Maksimov V., Sboner A., et al. Multicenter phase 2 study of oral azacitidine (CC-486) plus CHOP as initial treatment for PTCL. Blood. 2023;141:2194–2205. doi: 10.1182/blood.2022018254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cidado J., Boiko S., Proia T., Ferguson D., Criscione S.W., San Martin M., Pop-Damkov P., Su N., Roamio Franklin V.N., Sekhar Reddy Chilamakuri C., et al. AZD4573 Is a Highly Selective CDK9 Inhibitor That Suppresses MCL-1 and Induces Apoptosis in Hematologic Cancer Cells. Clin. Cancer Res. 2020;26:922–934. doi: 10.1158/1078-0432.CCR-19-1853. [DOI] [PubMed] [Google Scholar]
- 93.Hamlin P.A., Flinn I.W., Wagner-Johnston N., Burger J.A., Coffey G.P., Conley P.B., Michelson G., Leeds J.M., Der K., Kim Y., et al. Efficacy and safety of the dual SYK/JAK inhibitor cerdulatinib in patients with relapsed or refractory B-cell malignancies: Results of a phase I study. Am. J. Hematol. 2019;94:E90–E93. doi: 10.1002/ajh.25387. [DOI] [PubMed] [Google Scholar]
- 94.Shaw L.C., Poussin M., Rodriguez-Garcia A., Eggold J., Minutolo N.G., Wang J., Rook A.H., Schuster S.J., Powell D.J. TCRvbeta-CART therapy mediates high-precision targeting of malignant T-cell clones. Blood Adv. 2023;7:1885–1898. doi: 10.1182/bloodadvances.2022008798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu Y., Chen D., Lu Y., Dong S.C., Ma R., Tang W.Y., Wu J.Q., Feng J.F., Wu J.Z. A new immunotherapy strategy targeted CD30 in peripheral T-cell lymphomas: CAR-modified T-cell therapy based on CD30 mAb. Cancer Gene Ther. 2022;29:167–177. doi: 10.1038/s41417-021-00295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Patel R.P., Ghilardi G., Zhang Y., Chiang Y.H., Xie W., Guruprasad P., Kim K.H., Chun I., Angelos M.G., Pajarillo R., et al. CD5 deletion enhances the antitumor activity of adoptive T cell therapies. Sci. Immunol. 2024;9 doi: 10.1126/sciimmunol.adn6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Villacorta-Martin C., Craig A.J., Villanueva A. Divergent evolutionary trajectories in transplanted tumor models. Nat. Genet. 2017;49:1565–1566. doi: 10.1038/ng.3983. [DOI] [PubMed] [Google Scholar]
- 98.Chen J., Zhang Y., Petrus M.N., Xiao W., Nicolae A., Raffeld M., Pittaluga S., Bamford R.N., Nakagawa M., Ouyang S.T., et al. Cytokine receptor signaling is required for the survival of ALK- anaplastic large cell lymphoma, even in the presence of JAK1/STAT3 mutations. Proc. Natl. Acad. Sci. USA. 2017;114:3975–3980. doi: 10.1073/pnas.1700682114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ramsdale E., van Besien K., Smith S.M. Personalized treatment of lymphoma: promise and reality. Semin. Oncol. 2011;38:225–235. doi: 10.1053/j.seminoncol.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 100.Intlekofer A.M., Younes A. Precision therapy for lymphoma--current state and future directions. Nat. Rev. Clin. Oncol. 2014;11:585–596. doi: 10.1038/nrclinonc.2014.137. [DOI] [PubMed] [Google Scholar]
- 101.Sugio T., Miyawaki K., Kato K., Sasaki K., Yamada K., Iqbal J., Miyamoto T., Ohshima K., Maeda T., Miyoshi H., Akashi K. Microenvironmental immune cell signatures dictate clinical outcomes for PTCL-NOS. Blood Adv. 2018;2:2242–2252. doi: 10.1182/bloodadvances.2018018754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu T., Guo S., Ji Y., Zhu W. Role of cancer-educated mesenchymal stromal cells on tumor progression. Biomed. Pharmacother. 2023;166 doi: 10.1016/j.biopha.2023.115405. [DOI] [PubMed] [Google Scholar]
- 103.Le Naour A., Prat M., Thibault B., Mével R., Lemaitre L., Leray H., Joubert M.V., Coulson K., Golzio M., Lefevre L., et al. Tumor cells educate mesenchymal stromal cells to release chemoprotective and immunomodulatory factors. J. Mol. Cell Biol. 2020;12:202–215. doi: 10.1093/jmcb/mjz090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hill B.S., Sarnella A., D'Avino G., Zannetti A. Recruitment of stromal cells into tumour microenvironment promote the metastatic spread of breast cancer. Semin. Cancer Biol. 2020;60:202–213. doi: 10.1016/j.semcancer.2019.07.028. [DOI] [PubMed] [Google Scholar]
- 105.Zheng Z., Li P., Shen F., Shi Y., Shao C. Mesenchymal Stem/Stromal Cells in Cancer: from Initiation to Metastasis. Arch. Med. Res. 2022;53:785–793. doi: 10.1016/j.arcmed.2022.11.001. [DOI] [PubMed] [Google Scholar]
- 106.Valkenburg K.C., de Groot A.E., Pienta K.J. Targeting the tumour stroma to improve cancer therapy. Nat. Rev. Clin. Oncol. 2018;15:366–381. doi: 10.1038/s41571-018-0007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shojaei F., Ferrara N. Role of the microenvironment in tumor growth and in refractoriness/resistance to anti-angiogenic therapies. Drug Resist. Updates. 2008;11:219–230. doi: 10.1016/j.drup.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 108.Tarin D. Role of the host stroma in cancer and its therapeutic significance. Cancer Metastasis Rev. 2013;32:553–566. doi: 10.1007/s10555-013-9438-4. [DOI] [PubMed] [Google Scholar]
- 109.Siolas D., Hannon G.J. Patient-derived tumor xenografts: transforming clinical samples into mouse models. Cancer Res. 2013;73:5315–5319. doi: 10.1158/0008-5472.CAN-13-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kopetz S., Lemos R., Powis G. The promise of patient-derived xenografts: the best laid plans of mice and men. Clin. Cancer Res. 2012;18:5160–5162. doi: 10.1158/1078-0432.CCR-12-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu Y., Wu W., Cai C., Zhang H., Shen H., Han Y. Patient-derived xenograft models in cancer therapy: technologies and applications. Signal Transduct. Targeted Ther. 2023;8:160. doi: 10.1038/s41392-023-01419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vennin C., Chin V.T., Warren S.C., Lucas M.C., Herrmann D., Magenau A., Melenec P., Walters S.N., Del Monte-Nieto G., Conway J.R.W., et al. Transient tissue priming via ROCK inhibition uncouples pancreatic cancer progression, sensitivity to chemotherapy, and metastasis. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aai8504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Landis M.D., Lehmann B.D., Pietenpol J.A., Chang J.C. Patient-derived breast tumor xenografts facilitating personalized cancer therapy. Breast Cancer Res. 2013;15:201. doi: 10.1186/bcr3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Na Y.S., Ryu M.H., Park Y.S., Lee C.W., Lee J.K., Park Y., Park J.M., Ma J., Kang Y.K. Establishment of patient-derived xenografts from patients with gastrointestinal stromal tumors: analysis of clinicopathological characteristics related to engraftment success. Sci. Rep. 2020;10:7996. doi: 10.1038/s41598-020-64552-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Brehm M.A., Kenney L.L., Wiles M.V., Low B.E., Tisch R.M., Burzenski L., Mueller C., Greiner D.L., Shultz L.D. Lack of acute xenogeneic graft- versus-host disease, but retention of T-cell function following engraftment of human peripheral blood mononuclear cells in NSG mice deficient in MHC class I and II expression. FASEB J. 2019;33:3137–3151. doi: 10.1096/fj.201800636R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bondarenko G., Ugolkov A., Rohan S., Kulesza P., Dubrovskyi O., Gursel D., Mathews J., O'Halloran T.V., Wei J.J., Mazar A.P. Patient-Derived Tumor Xenografts Are Susceptible to Formation of Human Lymphocytic Tumors. Neoplasia. 2015;17:735–741. doi: 10.1016/j.neo.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Butler K.A., Hou X., Becker M.A., Zanfagnin V., Enderica-Gonzalez S., Visscher D., Kalli K.R., Tienchaianada P., Haluska P., Weroha S.J. Prevention of Human Lymphoproliferative Tumor Formation in Ovarian Cancer Patient-Derived Xenografts. Neoplasia. 2017;19:628–636. doi: 10.1016/j.neo.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Leiting J.L., Hernandez M.C., Yang L., Bergquist J.R., Ivanics T., Graham R.P., Truty M.J. Rituximab Decreases Lymphoproliferative Tumor Formation in Hepatopancreaticobiliary and Gastrointestinal Cancer Patient-Derived Xenografts. Sci. Rep. 2019;9:5901. doi: 10.1038/s41598-019-42470-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bayda N., Tilloy V., Chaunavel A., Bahri R., Halabi M.A., Feuillard J., Jaccard A., Ranger-Rogez S. Comprehensive Epstein-Barr Virus Transcriptome by RNA-Sequencing in Angioimmunoblastic T Cell Lymphoma (AITL) and Other Lymphomas. Cancers. 2021;13 doi: 10.3390/cancers13040610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fujisawa M., Nguyen T.B., Abe Y., Suehara Y., Fukumoto K., Suma S., Makishima K., Kaneko C., Nguyen Y.T.M., Usuki K., et al. Clonal germinal center B cells function as a niche for T-cell lymphoma. Blood. 2022;140:1937–1950. doi: 10.1182/blood.2022015451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sueyoshi K., Komura D., Katoh H., Yamamoto A., Onoyama T., Chijiwa T., Isagawa T., Tanaka M., Suemizu H., Nakamura M., et al. Multi-tumor analysis of cancer-stroma interactomes of patient-derived xenografts unveils the unique homeostatic process in renal cell carcinomas. iScience. 2021;24 doi: 10.1016/j.isci.2021.103322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cappelli L.V., Fiore D., Phillip J.M., Yoffe L., Di Giacomo F., Chiu W., Hu Y., Kayembe C., Ginsberg M., Consolino L., et al. Endothelial cell-leukemia interactions remodel drug responses, uncovering T-ALL vulnerabilities. Blood. 2023;141:503–518. doi: 10.1182/blood.2022015414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fiore D., Cappelli L.V., Zumbo P., Phillips J.M., Liu Z., Cheng S., Yoffe L., Ghione P., Di Maggio F., Dogan A., et al. A Novel JAK1 Mutant Breast Implant-Associated Anaplastic Large Cell Lymphoma Patient-Derived Xenograft Fostering Pre-Clinical Discoveries. Cancers. 2020;12 doi: 10.3390/cancers12061603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.RC T. A language and environment for statistical computing. MSOR connections. 2014;1 [Google Scholar]
- 125.Chang, W., Cheng, J., and Allaire, J. (2023). Web Application Framework.
- 126.Wickham, H., Averick, M., and Bryan, J. (2019). Welcome to the Tidyverse. vignettes/paperRmd.
- 127.Huber W., Carey V.J., Gentleman R., Anders S., Carlson M., Carvalho B.S., Bravo H.C., Davis S., Gatto L., Girke T., et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cacciapuoti M.T., Cappelli L.V., Fiore D., Toruno P., Kayembe C., Tam W., Inghirami G. In Vivo and Ex Vivo Patient-Derived Tumor Xenograft Models of Lymphoma for Drug Discovery. Curr. Protoc. 2021;1 doi: 10.1002/cpz1.96. [DOI] [PubMed] [Google Scholar]
- 129.Sarachakov A., Varlamova A., Svekolkin V., Polyakova M., Valencia I., Unkenholz C., Pannellini T., Galkin I., Ovcharov P., Tabakov D., et al. Spatial mapping of human hematopoiesis at single-cell resolution reveals aging-associated topographic remodeling. Blood. 2023;142:2282–2295. doi: 10.1182/blood.2023021280. [DOI] [PubMed] [Google Scholar]
- 130.van Dongen J.J.M., Langerak A.W., Brüggemann M., Evans P.A.S., Hummel M., Lavender F.L., Delabesse E., Davi F., Schuuring E., García-Sanz R., et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 131.Leek J.T., Storey J.D. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–1735. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Reynisson B., Barra C., Kaabinejadian S., Hildebrand W.H., Peters B., Nielsen M. Improved Prediction of MHC II Antigen Presentation through Integration and Motif Deconvolution of Mass Spectrometry MHC Eluted Ligand Data. J. Proteome Res. 2020;19:2304–2315. doi: 10.1021/acs.jproteome.9b00874. [DOI] [PubMed] [Google Scholar]
- 134.Greenbaum J., Sidney J., Chung J., Brander C., Peters B., Sette A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63:325–335. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.McGinnis C.S., Murrow L.M., Gartner Z.J. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 2019;8:329–337. doi: 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R.P., Wolters P.J., Abate A.R., et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mabbott N.A., Baillie J.K., Brown H., Freeman T.C., Hume D.A. An expression atlas of human primary cells: inference of gene function from coexpression networks. BMC Genom. 2013;14:632. doi: 10.1186/1471-2164-14-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 doi: 10.48550/arXiv.1303.3997. Preprint at. [DOI] [Google Scholar]
- 141.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Trifonov V., Pasqualucci L., Tiacci E., Falini B., Rabadan R. SAVI: a statistical algorithm for variant frequency identification. BMC Syst. Biol. 2013;7 doi: 10.1186/1752-0509-7-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gehring J.S., Fischer B., Lawrence M., Huber W. SomaticSignatures: inferring mutational signatures from single-nucleotide variants. Bioinformatics. 2015;31:3673–3675. doi: 10.1093/bioinformatics/btv408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hamilton N., Ferry A. Ternary Diagrams Using ggplot2. J. Stat. Software. 2018;87:1–17. [Google Scholar]
- 145.Talevich E., Shain A.H., Botton T., Bastian B.C. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput. Biol. 2016;12 doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mermel C.H., Schumacher S.E., Hill B., Meyerson M.L., Beroukhim R., Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Leshchiner I., Livitz D., Gainor J., Rosebrock D., Spiro O., Martinez A., Mroz E., Lin J.J., Stewart C., Kinm J., et al. Comprehensive analysis of tumour initiation, spatial and temporal progression under multiple lines of treatment. bioRxiv. 2018 doi: 10.1101/508127. Preprint at. [DOI] [Google Scholar]
- 148.Miller C.A., McMichael J., Dang H.X., Maher C.A., Ding L., Ley T.J., Mardis E.R., Wilson R.K. Visualizing tumor evolution with the fishplot package for R. BMC Genom. 2016;17:880. doi: 10.1186/s12864-016-3195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bolotin D.A., Poslavsky S., Davydov A.N., Frenkel F.E., Fanchi L., Zolotareva O.I., Hemmers S., Putintseva E.V., Obraztsova A.S., Shugay M., et al. Antigen receptor repertoire profiling from RNA-seq data. Nat. Biotechnol. 2017;35:908–911. doi: 10.1038/nbt.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Goncharov M., Bagaev D., Shcherbinin D., Zvyagin I., Bolotin D., Thomas P.G., Minervina A.A., Pogorelyy M.V., Ladell K., McLaren J.E., et al. VDJdb in the pandemic era: a compendium of T cell receptors specific for SARS-CoV-2. Nat. Methods. 2022;19:1017–1019. doi: 10.1038/s41592-022-01578-0. [DOI] [PubMed] [Google Scholar]
- 151.Chen Q., Meng Z., Su R. WERFE: A Gene Selection Algorithm Based on Recursive Feature Elimination and Ensemble Strategy. Front. Bioeng. Biotechnol. 2020;8:496. doi: 10.3389/fbioe.2020.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Melamed R.D., Wang J., Iavarone A., Rabadan R. An information theoretic method to identify combinations of genomic alterations that promote glioblastoma. J. Mol. Cell Biol. 2015;7:203–213. doi: 10.1093/jmcb/mjv026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Schubert M., Klinger B., Klünemann M., Sieber A., Uhlitz F., Sauer S., Garnett M.J., Blüthgen N., Saez-Rodriguez J. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun. 2018;9:20. doi: 10.1038/s41467-017-02391-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Redmond D., Poran A., Elemento O. Single-cell TCRseq: paired recovery of entire T-cell alpha and beta chain transcripts in T-cell receptors from single-cell RNAseq. Genome Med. 2016;8:80. doi: 10.1186/s13073-016-0335-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 157.Zerbino D.R., Achuthan P., Akanni W., Amode M.R., Barrell D., Bhai J., Billis K., Cummins C., Gall A., Girón C.G., et al. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A: PDX and PDX-Dlines list, B: patient clinical annotation, C: primary-PDX identity, D: all implanted samples source
A: WES, B: bulk RNA-seq, C: scRNA-seq, D: DeepSeq, E: TCR_data, F: BCR_data, G: EBV status, H: TCR_EBV, I: EBV match TCR, J: MHC12_AITL
A: Fusions detected in the TCL PDX cohort
A: Copy-number aberrations estimated from WES data in the TCL PDX cohort
A: NY-AI-AM, B: NY-AI-CL, C: NY-AT-FC, D: TO-ALCL-BELLI, E: COH1, F: TO-ALCL-DN03, G: IL114A, H: IL-2, I: IL6, J: IL17, K: IL19, L: IL33, M: IL34, N: IL35, O: IL36, P: IL38B, Q: IL40, R: IL41, S: IL49, T: IL54, U: IL56, V: IL65, W: IL69, X: IL76, Y: IL78, Z: IL79, AA: IL81, AB: IL86, AC: IL88, AD: IL89, AE: IL92, AF: IL94, AG: IL98, AH: IL104, AI: IL107, AJ: IL122A, AK: IL123, AL: IL126A, AM: IL126B, AN: IL129A, AO: IL135A, AP: IL138A, AQ: TO-ALCL-MARC, AR: TO-ALCL-MARI, AS: MT05, AT: NY-NK-CD, AU: NY-PTCL-RV, AV: NY-ALCL-SG, AW: NY-NK-VP
A: clonal evolution analysis reporting cancer cell fraction estimates and mutation clonality
A: datasets utilized to generate microenvironment signatures, B: TME subtype of 70 primary PTCL
A: list of 433 compounds utilized in the high-throughput (HTS) drug screening
Data Availability Statement
-
•
Raw sequencing data have been deposited to the Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra) and are publicly available. Accession numbers are listed in the key resources table (WES: PRJNA1198080; RNA-seq: PRJNA1214670; scRNA-seq: PRJNA1218277). Also, this paper analyzes existing, publicly available data, whose accession numbers are listed in the key resources table.
-
•
Codes used in this study are deposited at https://github.com/marchionniLab/ing-2023 (https://doi.org/10.5281/zenodo.14847458) and https://github.com/Mew233/DeepPTCL (https://doi.org/10.5281/zenodo.14845644) and listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.