In this study, Franklin et al. show that histone chaperones, like the transcription-coupled histone chaperone SPT6 and the replication-coupled chromatin assembly factor complex CAF-1, control cell differentiation and cell identity through distinct mechanisms. The work further highlights how histone chaperones and their roles in chromatin assembly are essential for the self-renewal of hematopoietic stem and progenitor cells and influence lineage choice.
Keywords: histone chaperones, transcription, DNA replication, transcription factors, stem cells, lineage choice
Abstract
The manipulation of DNA replication and transcription can be harnessed to control cell fate. Central to the regulation of these DNA-templated processes are histone chaperones, which in turn are emerging as cell fate regulators. Histone chaperones are a group of proteins with diverse functions that are primarily involved in escorting histones to assemble nucleosomes and maintain the chromatin landscape. Whether distinct histone chaperone pathways control cell fate and whether they function using related mechanisms remain unclear. To address this, we performed a screen to assess the requirement of diverse histone chaperones in the self-renewal of hematopoietic stem and progenitor cells. Remarkably, all candidates were required to maintain cell fate to differing extents, with no clear correlation with their specific histone partners or DNA-templated process. Among all the histone chaperones, the loss of the transcription-coupled histone chaperone SPT6 most strongly promoted differentiation, even more than the major replication-coupled chromatin assembly factor complex CAF-1. To directly compare how DNA replication- and transcription-coupled histone chaperones maintain stem cell self-renewal, we generated an isogenic dual-inducible system to perturb each pathway individually. We found that SPT6 and CAF-1 perturbations required cell division to induce differentiation but had distinct effects on cell cycle progression, chromatin accessibility, and lineage choice. CAF-1 depletion led to S-phase accumulation, increased heterochromatic accessibility (particularly at H3K27me3 sites), and aberrant multilineage gene expression. In contrast, SPT6 loss triggered cell cycle arrest, altered accessibility at promoter elements, and drove lineage-specific differentiation, which is in part influenced by AP-1 transcription factors. Thus, CAF-1 and SPT6 histone chaperones maintain cell fate through distinct mechanisms, highlighting how different chromatin assembly pathways can be leveraged to alter cell fate.
Histone chaperones are a diverse family of proteins that can bind specific combinations of histones and assemble nucleosomes to maintain the chromatin landscape during DNA replication, transcription, recombination, and/or repair (Burgess and Zhang 2013; Hammond et al. 2017; Grover et al. 2018; Franklin et al. 2021). For example, some histone chaperones bind exclusively to the replicative histone dimers and assemble nucleosomes throughout the genome during DNA replication, whereas others bind to histone variants and assemble nucleosomes at specific genomic loci independently of DNA replication (Mattiroli et al. 2015; Martire and Banaszynski 2020; Ray-Gallet and Almouzni 2021). Moreover, histone chaperones perform other complex functions, including in DNA and RNA metabolism and the recruitment of regulatory proteins to chromatin (Pardal et al. 2019; Franklin et al. 2021). Despite these general roles during various DNA transactions, accumulating evidence supports specific roles of histone chaperones in cell fate control (Filipescu et al. 2013; Cheloufi and Hochedlinger 2017; Franklin et al. 2021).
Our work and that of others have demonstrated a specific role for the chromatin assembly factor-1 (CAF-1) in safeguarding cell fate in various cellular contexts (Heyd et al. 2011; Cheloufi et al. 2015; Hatanaka et al. 2015; Ishiuchi et al. 2015; Cheloufi and Hochedlinger 2017; Volk et al. 2018; Cheng et al. 2019; Ng et al. 2019; Zhang et al. 2022). CAF-1 is a trimeric histone chaperone complex composed of CHAF1A, CHAF1B, and RBBP4 subunits (Smith and Stillman 1989) that is involved in the DNA replication-coupled deposition of newly synthesized replicative histone H3:H4 dimers. In addition to its role in chromatin assembly, CAF-1 is also involved in heterochromatin regulation (Quivy et al. 2008; Cheng et al. 2019). Although CAF-1 plays a general role in chromatin maintenance, studies in reprogramming, pluripotent stem cells, and hematopoietic cells have shown that its loss results in local chromatin changes that allow transcription factors to bind regulatory regions and promote the activation of fate genes (Cheloufi and Hochedlinger 2017; Franklin et al. 2021).
The role of histone chaperones in cell fate regulation goes beyond replication-coupled chromatin assembly. For example, SPT6 is a histone chaperone involved in transcription-coupled chromatin maintenance and RNA processing, regulating cell fate in different contexts (Bortvin and Winston 1996; Hartzog et al. 1998; DeGennaro et al. 2013; Jeronimo et al. 2019; Bobkov et al. 2020; Vos et al. 2020; Narain et al. 2021; Aoi et al. 2022; Miller and Winston 2023; Miller et al. 2023). Moreover, given its diverse functions, SPT6 has been implicated in both cell identity maintenance and cellular differentiation (Wang et al. 2013, 2017; Obara et al. 2020; Li et al. 2021; Vo et al. 2021; Miller et al. 2023). For example, the loss of SPT6 in mouse embryonic stem cells triggered their differentiation (Wang et al. 2017). In contrast, studies in epithelial and muscle cells point to the necessity of SPT6 in promoting cell differentiation (Wang et al. 2013; Li et al. 2021; Vo et al. 2021). Noticeably, this divergent behavior may depend on how SPT6 controls cell fate under self-renewal versus induced differentiation conditions.
Besides SPT6 and CAF-1, two major players in transcription- and replication-coupled nucleosome assembly, a number of other histone chaperones have been implicated in cell fate control, including those that associate with specific histone variants (Franklin et al. 2021). However, the implication of histone chaperones in cell fate regulation is based on studying individual factors in different cellular settings. Moreover, recent genome-wide screens do not typically recover histone chaperones as major hits and may be limited due to their pooled nature, coverage limitations, and sensitivity of the readout (Wang et al. 2021; Replogle et al. 2022; Do et al. 2024). To circumvent these challenges, we performed an arrayed screen to systematically assess the function of known histone chaperones in the maintenance of cell fate in the same cellular setting. Specifically, we used a well-established and robust ex vivo hematopoietic system that has been widely used to understand the mechanisms of normal differentiation and malignant dependencies (Wang et al. 2006; Sykes et al. 2016; Blanco et al. 2021; Franklin et al. 2022; Do et al. 2024). In this system, mouse bone marrow cells are conditionally immortalized at the granulocyte and macrophage progenitors (iGMPs) state through the expression of a transgene encoding the HOXA9 transcription factor fused to the estrogen receptor ligand binding domain (ER-HOXA9) (Schnabel et al. 2000; Wang et al. 2006; Sykes et al. 2016). iGMPs are maintained in their stem and progenitor state when cultured in estradiol (E2) under self-renewing culture conditions. Notably, E2 withdrawal induces their synchronous, homogeneous, and terminal differentiation into functional neutrophils. Here, we tested which histone chaperone pathways are implicated in the maintenance of their stem cell state.
The combination of this robust hematopoietic system and our focused screen approach revealed that inhibition of all histone chaperones under self-renewal conditions triggered some degree of iGMP differentiation, with SPT6 scoring as the top candidate. Given the dynamic relationship between transcription and replication (Chen et al. 2019; Gnan et al. 2020), and to understand how the manipulation of histone chaperone pathways coupled to these processes affect cell identity, we generated a dual-inducible iGMP system, where CAF-1 and SPT6 can be individually perturbed. Although inhibiting either pathway triggered iGMP differentiation, their mechanisms of action had distinct effects, ultimately leading to different lineage choices. Our study introduces a new paradigm to study histone chaperones and reveals how replication- and transcription-coupled processes function through different mechanisms to maintain the myeloid progenitor cell state.
Results
A histone chaperone screen reveals a major role for SPT6 in maintaining cell identity
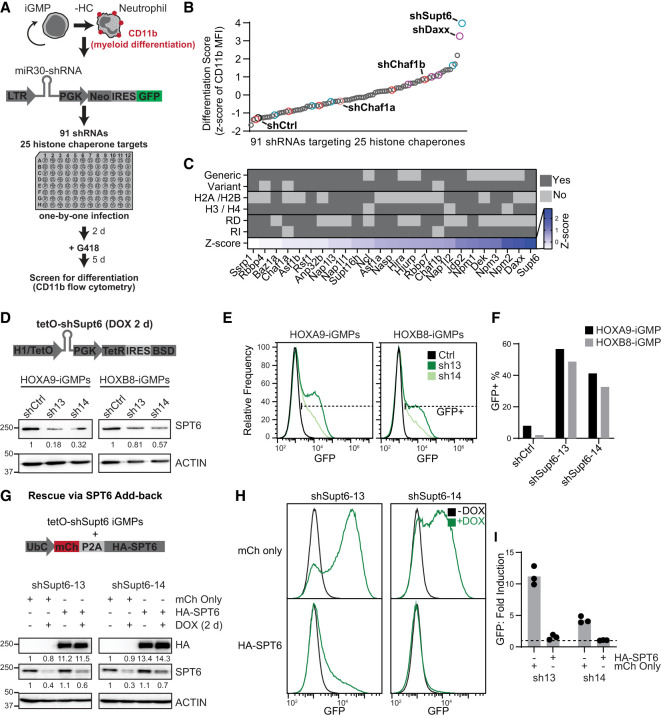
The iGMPs in our hematopoietic system can be maintained in culture as myeloid stem and progenitor cells while retaining the potential to be induced into functional macrophages or neutrophils, previously shown to recapitulate normal in vivo differentiation (Wang et al. 2006; Sykes et al. 2016). We reasoned that under self-renewing conditions, any histone chaperone candidate required for the maintenance of iGMPs will prevent their differentiation into mature myeloid cells. Therefore, under self-renewing conditions, we depleted histone chaperones and assessed differentiation using a pan-myeloid cell surface differentiation marker, CD11b (Fig. 1A). To determine the role of histone chaperones in maintaining iGMP cell identity, we designed a miR30-based shRNA library targeting 25 histone chaperones (De Koning et al. 2007; Das et al. 2010; Avvakumov et al. 2011), with each candidate defined as a protein that interacts with histones and exerts nucleosome assembly. We targeted each histone chaperone candidate with two to six shRNAs by individual retroviral transduction of iGMPs in parallel with a control Renilla luciferase shRNA (shCtrl) (Supplemental Table S1). iGMP differentiation was then quantified by measuring the expression of CD11b using flow cytometry. Targeting of all candidates induced differentiation to a greater degree than the control shRNA, suggesting a universal role of all histone chaperone pathways in the maintenance of the iGMP self-renewing state (Fig. 1B; Supplemental Fig. S1A,B).
Figure 1.
SPT6 and other histone chaperone pathways maintain iGMPs. (A) Schematic for the arrayed screen design targeting 25 different histone chaperones through individual retroviral infection with pLMN-miR30-shRNAs. Infected (GFP+) cells were analyzed for mean fluorescence intensity (MFI) of CD11b expression with flow cytometry. (HC) Histone chaperone. (B) Screen results showing the differentiation score of individual shRNAs in iGMPs. The differentiation score is based on the Z-score of CD11b geometric mean fluorescence intensity (MFI) (see the Materials and Methods; Supplemental Fig. S1A,B). Colored points represent shRNAs targeting Supt6 (blue), Daxx (purple), Chaf1b (red), Chaf1a (light red), and Renilla luciferase (shCtrl; black). Data represent the average Z-score of n = 3 experimental replicates. (C) Annotation table of targeted histone chaperones. Categories include histone chaperones known to interact with replicative histones, variant histones, H2A/H2B, or H3/H4 and histone chaperones known to be involved in replication-dependent (RD) and replication-independent (RI) pathways. Z-score represents the average of all shRNAs in B targeting the indicated histone chaperone. (D, top panel) Schematic of the tetO-shRNA (EZ-tet-pLKO-Blast; Addgene 85973) used for shCtrl and inducible depletion of SPT6. (Bottom panel) Western blot quantifying SPT6 with ACTIN as a loading control after 2 days of Dox induction. HOXA9-iGMPs and HOXB8-iGMPs transduced with two independent tetO-shRNAs targeting Supt6 (shSupt6-13/14) and a scrambled shRNA as control (shCtrl). (E) Histograms showing GFP expression quantified with flow cytometry in HOXA9-iGMPs and HOXB8-iGMPs after 2 day Dox induction of shCtrl or one of two independent Supt6 shRNAs (shSupt6-13/14). (F) Bar plots summarizing the percentage of GFP+ cells shown in F. Experiments represent one polyclonally transduced population per cell line and shRNA. Experiments with HOXA9-iGMPs from D–F were independently repeated three times with consistent results. (G, top panel) Schematic of rescue experiment performed by transducing tetO-shSupt6-HOXA9-iGMPs with pUltra expression vector containing the ubiquitin C promoter driving a polycistronic transcript. The HA-SPT6 vector encodes mCherry (mCh), P2A self-cleaving peptide, and HA fused to the SUPT6H N terminus (HA-SPT6; human SPT6 homolog). pUltra backbone with mCh still encoded was used as a control (mCh only). Cells treated with Dox for 2 days were analyzed with flow cytometry and Western blotting and compared with untreated iGMPs. (Bottom panel) Western blot quantifying HA and SPT6 expression with ACTIN as a loading control. Cells with exogenous HA-SPT6 or mCh only were measured before and after 48 h of Dox induction. (H)Histogram showing GFP expression quantified with flow cytometry. shSupt6-13 and shSupt6-14 HOXA9-iGMPs with mCh only or with HA-SPT6 are treated with Dox for 2 days. Representative of n = 3 clonal replicates. (I) Bar plot summarizing fold induction of GFP (= +Dox GFP MFI/−Dox GFP MFI) as shown in H. n = 3 clonal replicates for each shRNA and condition. The dotted line represents no fold induction (=1).
Although we did not identify a clear trend between the strength of differentiation and histone chaperone classifications, replication-independent histone chaperones scored among the top hits (Fig. 1C; Supplemental Fig. S1C). Interestingly, the top two candidates, SPT6 and DAXX, have distinct roles in regulating the chromatin landscape (Fig. 1C). DAXX deposits the histone H3.3 variant in heterochromatic regions (Drané et al. 2010; Lewis et al. 2010; He et al. 2015; Wasylishen et al. 2020), whereas SPT6 recycles histones H3:H4 during transcription, facilitating the passage of RNA Pol II through nucleosomes and maintaining centromeric regions (DeGennaro et al. 2013; Jeronimo et al. 2019; Bobkov et al. 2020; Miller et al. 2023). We validated that shRNAs targeting Supt6 and Daxx induced differentiation in two independently derived iGMP clones (Supplemental Fig. S1D). However, only SPT6 exhibited a positive correlation between the level of knockdown and the strength of iGMP differentiation (Supplemental Fig. S1E). Based on this correlation, the strong depletion of SPT6 protein, and the more profound differentiation effect upon SPT6 loss (Fig. 1B,C; Supplemental Fig. S1D–F), we focused subsequent analyses on SPT6.
Additional shRNAs were designed targeting distinct SPT6 mRNA sites using a colorless pLKO lentiviral shRNA vector that allowed us to take advantage of the pan-myeloid differentiation knock-in reporter transgene lysozyme-GFP (GFP) (Faust et al. 2000) built into our iGMP system (Supplemental Fig. S1G,H). Using two independent lysozyme-GFP iGMP clones immortalized with either ER-HOXA9 or ER-HOXB8 transgenes, we validated that SPT6 knockdown induced expression of both the GFP reporter and the CD11b surface marker (Supplemental Fig. S1I,J). Notably, constitutive depletion of SPT6 resulted in decreased viability of the iGMPs, hindering further analysis of our differentiation system (Supplemental Fig. S1K). To overcome this effect on cell viability, we generated a Dox-inducible Supt6 tetO-shRNA, permitting controlled SPT6 knockdown on demand in the iGMPs (Fig. 1D). Two independent Dox-inducible Supt6 shRNAs efficiently depleted SPT6 and consistently triggered differentiation in HOXA9 and HOXB8 iGMP clones (Fig. 1D–F; Supplemental Fig. S1G). Importantly, we ruled out potential off-target effects of these shRNAs by add-back of an RNAi-resistant SUPT6H (Supt6 human homolog) cDNA, which efficiently suppressed the differentiation phenotype triggered by Dox induction of both Supt6 shRNAs (Fig. 1G–I). Notably, overexpression of SPT6 alone did not affect the self-renewal state of iGMPs. Taken together, these results indicate that SPT6 is essential to maintain the self-renewing and undifferentiated state of myeloid progenitors.
A novel dual-inducible system for perturbing replication- and transcription-coupled chromatin assembly
Perturbations of the DNA replication and transcription processes, not necessarily limited through manipulating nucleosome assembly, can have profound influence on cell fate. For example, a recent study demonstrated that replication stress mediated by nucleotide depletion triggers the differentiation of iGMPs (Do et al. 2024). Moreover, manipulation of transcription factors is known to alter cell fate trajectories (Takahashi and Yamanaka 2016). Our screen revealed that histone chaperones play a broad role in cell fate maintenance, with SPT6 having the most pronounced effect (Fig. 1). Among the candidates tested, SPT6 and CAF-1 stand out as major regulators of transcription- and replication-coupled chromatin maintenance. Consistently, upon closer examination of the scores for our 25 histone chaperone candidates in recent genome-wide CRISPR–Cas9 screens conducted in two independent human leukemic models as well as our iGMP system (Wang et al. 2021; Replogle et al. 2022; Do et al. 2024), we found that SPT6 and CAF-1 promote cellular differentiation across these distinct cellular contexts (Supplemental Fig. S2A). Given these observations, we reasoned that the manipulation of these two pathways individually can illuminate the mechanisms underlying the activity of each factor, which can inform potential strategies on how to control cell fate during these DNA-templated processes (Fig. 2A).
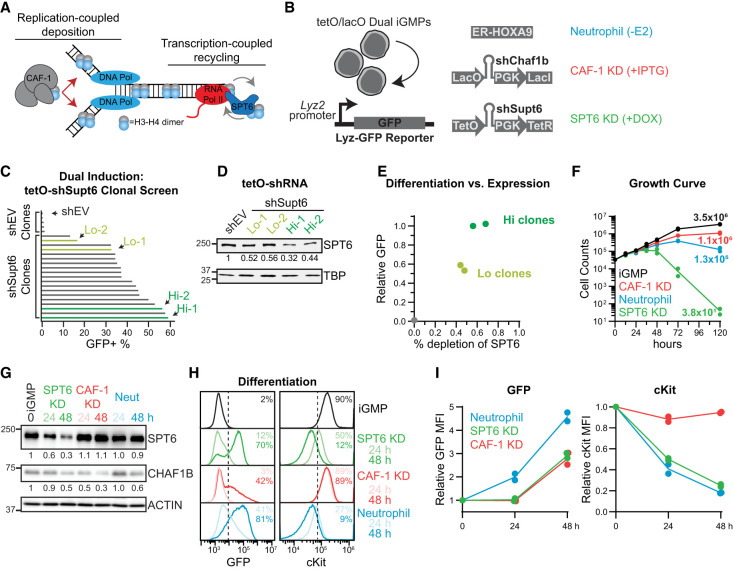
Figure 2.
Dually inducible depletion of SPT6 and CAF-1 perturbs replication- and transcription-coupled histone chaperone pathways. (A) CAF-1 and SPT6 are involved in replication- and transcription-coupled maintenance of chromatin, respectively. (B) Schematic of the dual-inducible iGMPs with corresponding genotypes and outcomes upon induction. (Lyz-GFP) Lysozyme-GFP. (C) Differentiation screen analysis of dual-inducible clones derived as seen in B. Bar plots show the percentage of GFP+ cells determined by flow cytometry after 5 days of continuous SPT6 KD (+Dox) treatment. TetO-pLKO backbone was used as a negative control (shEV). Green bars represent selected clones with high (Hi) or low (Lo) induction of GFP. n = 5 (shEV) and n = 19 (shSupt6) clonal replicates represented individually. (D) Western blot quantifying SPT6 with TBP as a loading control. Two clones with high (Hi) or low (Lo) GFP induction were compared with shEV cells. Cells were continuously treated with SPT6 KD for 48 h before quantification. (E) Differentiation versus expression. Differentiation signal (relative GFP) of selected Hi (dark green) and Lo (light green) clones. Relative GFP was normalized with the lower limit set to shEV and the upper limit set to Hi clones. Percent depletion of SPT6 was derived from the blot shown in D. For D and E, n = 2 clonal replicates for Hi and Lo populations, displayed individually. (F) Growth curve analysis performed by counting cells induced continuously with SPT6 KD, CAF-1 KD, or neutrophil differentiation for 0, 12, 24, 36, 48, 72, or 120 h. The Y-axis displays cell counts on a log10 scale. Lines connect the average counts of n = 2 clonal replicates per time point. The average number of cells recovered after 120 h is displayed next to the corresponding points. (G) Western blot analysis of SPT6 and CHAF1B after 24 and 48 h of SPT6 KD, CAF-1 KD, and neutrophil differentiation conditions compared with iGMPs. Western blot is representative of n = 2 clonal replicates with the average signal density normalized to the ACTIN loading control and displayed relative to iGMPs. (H) Flow cytometry quantification of GFP and cKit in iGMPs and after 24 or 48 h of SPT6, CAF-1, or neutrophil differentiation conditions. Dashed lines indicate the threshold for differentiation signal (GFP+ and cKit+). The mean percentages of GFP+ and cKit+ populations for n = 2 clonal replicates are displayed and colored according to condition and time point. (I) Line plots summarizing GFP and cKit expression in differentiated cells quantified by flow cytometry from H and displayed relative to iGMPs. The line connects the mean of n = 2 clonal replicates.
To generate an inducible iGMP system in which CAF-1 and SPT6 pathways could be perturbed in an isogenic cellular context, we built on our previously established system of IPTG-inducible CAF-1 depletion (Franklin et al. 2022). We transduced our validated Dox-inducible Supt6 tetO-shRNA into iGMPs containing the IPTG-inducible lacO-shRNA targeting the Chaf1b middle subunit of CAF-1 (Figs. 1D, 2B). This double IPTG- and Dox-inducible iGMP line allowed us to selectively perturb each pathway and compare the individual effects, referred to here as CAF-1 KD and SPT6 KD cell states (Fig. 2A,B).
Multiple double-inducible iGMP clones were derived that demonstrated pronounced differentiation upon Dox induction (Fig. 2C). Consistent with the 14 Supt6 shRNAs tested (Supplemental Fig. S1G), the differentiation strength of these clones correlated with the degree of SPT6 knockdown (Fig. 2C–E). Moreover, HOXA9 clones bearing the Dox-inducible Supt6 shRNA without the CAF-1 IPTG-inducible shRNA also exhibited a strong dose-dependent differentiation phenotype, measured by activation of the lysozyme-GFP reporter and loss of the stemness marker cKit (Supplemental Fig. S2B–G). Collectively, these multiple iGMP clones allowed us to selectively perturb SPT6 and CAF-1 pathways and compare their effects individually. Moreover, HOXA9 inactivation by E2 withdrawal within these dual-inducible clones allowed us to compare these conditions with normal differentiation (Wang et al. 2006; Sykes et al. 2016), referred to here as the neutrophil cell state (Fig. 2B).
Next, we analyzed viability, target protein depletion, and differentiation over time in two independently derived dual-inducible clones following SPT6 or CAF-1 depletion (Fig. 2F–I; Supplemental Fig. S3A–D). In both systems, target depletion was achieved as early as 24 h after Dox or IPTG treatment (Fig. 2G; Supplemental Fig. S3A,B). Consistent with the mixed-lineage state of CAF-1 KD iGMPs (Franklin et al. 2022), dual-inducible clones maintained stemness gene expression despite gaining GFP (Fig. 2H–I; Supplemental Fig. S3C,D). In contrast, SPT6 depletion exhibited a stronger phenotype resembling the neutrophil-induced condition but severely impairing cell growth after 48 h (Fig. 2F,H,I). Based on these findings, we focused subsequent analyses on the 24 and 48 h time points to investigate the mechanisms underlying the differentiated states resulting from SPT6 or CAF-1 depletion.
Distinct transcriptional and replicative effects induced by CAF-1 and SPT6 perturbation during the cell cycle
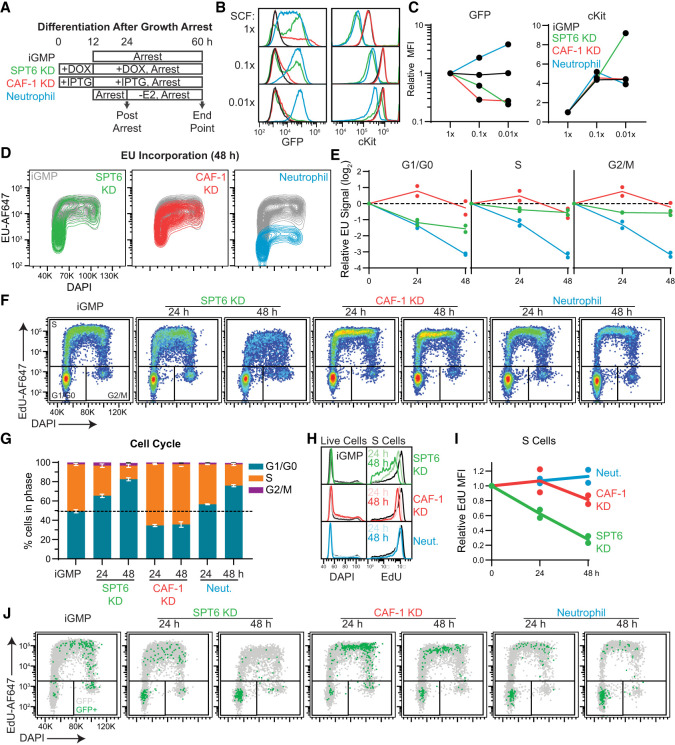
Progression through cell division provides a window of opportunity for cell fate changes, as demonstrated across various reprogramming and differentiation systems as well as during normal development and tissue homeostasis (Soufi and Dalton 2016; Liu et al. 2019; Martell et al. 2023). However, how histone chaperones influence cell fate decisions during the different phases of the cell cycle remains elusive. We previously reported that CAF-1 KD iGMPs couple differentiation with cell cycle progression, whereas iGMPs can differentiate into neutrophils during G1/G0 arrest (Franklin et al. 2022). Given the stronger differentiation phenotype induced by SPT6 depletion and the more abrupt effect on cell viability (Fig. 2), we addressed whether cell cycle progression in the SPT6 KD cells was also uncoupled from differentiation. SCF was omitted from the culture media to induce a G1/G0 arrest (Fig. 3A–C; Supplemental Fig. S4A,B), and differentiation was quantified in SPT6 KD, CAF-1 KD, and neutrophil cell states. As reported previously, HOXA9 inactivation exhibited a normal differentiation response concomitant with cell cycle arrest, whereas the differentiation of CAF-1 KD cells was dependent on cell cycle progression (Fig. 3B,C; Franklin et al. 2022; Do et al. 2024). Strikingly, cells arrested after SPT6 KD did not differentiate despite the phenotypic parallels with neutrophil differentiation (Fig. 3B,C). We complemented this result using a label retention assay (CellTrace) to track the differentiation dependency on cellular divisions. The number of cell divisions and differentiation of cells was tracked over 48 h using flow cytometry (Supplemental Fig. S4C,D). Remarkably, the SPT6 KD and CAF-1 KD cells responded similarly where cells undergoing the fewest number of divisions exhibited no differentiation. In contrast, neutrophils exhibited a higher degree of differentiation in populations with comparably fewer cell divisions (Supplemental Fig. S4C,D). Of note, faster-dividing cells exhibited better differentiation outcomes in all three conditions, reinforcing the idea that cell cycle progression promotes cell fate changes.
Figure 3.
Cell cycle dependencies upon CAF-1 and SPT6 perturbations. (A) iGMPs were arrested with 0.1× or 0.01× SCF (arrest), in parallel with normal 1× SCF treatment. For SPT6 KD and CAF-1 KD conditions, arrest was performed following 12 h of pretreatment with Dox and IPTG, respectively. For neutrophil differentiation conditions, cells were arrested for 12 h before inducing differentiation for 36 h. SCF concentrations were maintained throughout the 48 h (arrest). Samples were collected “postarrest” to confirm arrest (see Supplemental Fig. S4A,B) and at the “end point” as shown in the schematic. (B) Histograms showing cKit and GFP expression quantified by flow cytometry at the “end point” of each treatment, as shown in A. iGMP, SPT6 KD, CAF-1 KD, and neutrophil histograms are overlaid for each SCF condition. (C) Quantification of GFP and cKit expression levels shown in B for each treatment relative to 1× SCF. n = 2 clonal replicates. (D) Flow cytometry contour plot showing incorporation of 5-ethynyluridine (EU) versus total DNA DAPI staining in iGMPs and after 48 h of SPT6 KD, CAF-1 KD, and neutrophil differentiation. Untreated iGMPs are overlaid with each condition and colored accordingly. (E) Time-course analysis of EU incorporation relative to iGMPs for each condition as a function of cell cycle phase. n = 2 clonal replicates. Phases were gated based on DAPI signal (see also histograms in Supplemental Fig. S4E). (F) Cell cycle plots showing EdU incorporation and total DNA staining (DAPI) quantified with flow cytometry. G1/G0, S, and G2/M cell cycle phases are gated based on EdU and DAPI signal. Data were quantified in iGMPs and after 24 or 48 h of continuous treatment for each condition. Representative of n = 2 clonal replicates. The experiment was independently repeated three times with consistent results. (AF647) Alexa fluor-647. (G) The percentage of cells in each cell cycle phase as gated in F. The mean of n = 2 clonal replicates with error bars representing the range. (H) Histograms quantifying DNA content in the bulk population for each condition and EdU incorporation in S-phase cells from cells shown in F. Representative of n = 2 clonal replicates. (I) Geometric mean fluorescence intensity of EdU incorporation in S-phase cells as shown in H for n = 2 clonal replicates. (J) Cell cycle plots as shown in F, but with GFP− (gray) and GFP+ (green) cells displayed for each condition and time point. A reduced number of events is shown for clarity (see quantification of total populations in Supplemental Fig. S4F).
Given the distinct roles of SPT6 and CAF-1 in chromatin assembly (Fig. 2A), we assessed their impact on global transcription and DNA synthesis across the cell cycle. To this end, we measured EU and EdU incorporation using click chemistry as a function of DNA content, allowing us to track nascent RNA and DNA synthesis across different cell cycle phases.
Our analysis revealed striking differences, with SPT6 depletion specifically reducing RNA synthesis during G1/G0 (Fig. 3D,E; Supplemental Fig. S4E). Notably, SPT6-depleted cells gradually accumulated in G1/G0, mirroring neutrophil differentiation, whereas the remaining S-phase cells exhibited a sharp reduction in EdU incorporation, reflecting reduced replicative activity (Fig. 3F–I). In contrast, CAF-1-depleted cells accumulated in S phase, showing a slower and more moderate decline in EdU incorporation (Fig. 3F–I), consistent with the reduced DNA replication speed observed upon rapid CAF-1 degradation in human cells (Dreyer et al. 2024). Furthermore, analysis of lysozyme-GFP reporter expression as a function of DNA content revealed that upon SPT6 depletion, differentiated cells predominantly accumulated in G1/G0, whereas CAF-1 depletion led to their accumulation in S phase (Fig. 3J; Supplemental Fig. S4F).
Together, our data suggest that the differentiation induced by CAF-1 and SPT6 perturbations depends on cell cycle progression, with distinct effects on global DNA replication and transcription (Fig. 3; Supplemental Fig. S4). These effects may lead to a replication stress response, ultimately driving cellular differentiation, as recently demonstrated (Sykes et al. 2016; Do et al. 2024). To assess this, we examined the activity of the ataxia-telangiectasia mutated and Rad3-related (ATR) signaling pathway by probing the phosphorylation states of ATR targets Chk1 and RPA32, finding no response under either condition (Supplemental Fig. S5A). We then measured H2AX phosphorylation, which can respond to transcription-associated DNA damage independently of the ATR pathway (Mah et al. 2010; Barlow et al. 2013; Zeman and Cimprich 2014). Although CAF-1 depletion did not induce a response, SPT6 depletion led to a modest increase in H2AX phosphorylation compared with control cells treated with the DNA replication inhibitor hydroxyurea (Supplemental Fig. S5A). Moreover, immunofluorescence analysis of SPT6-depleted cells revealed pronounced punctate foci, suggesting local DNA damage sites (Supplemental Fig. S5B–D). Together, our results indicate that CAF-1 and SPT6 perturbations drive cellular differentiation in a cell cycle-dependent manner, with CAF-1 primarily affecting S phase, and SPT6 playing a more pronounced role in G1/G0.
Shared and unique transcriptional signatures of SPT6 and CAF-1 loss in iGMPs
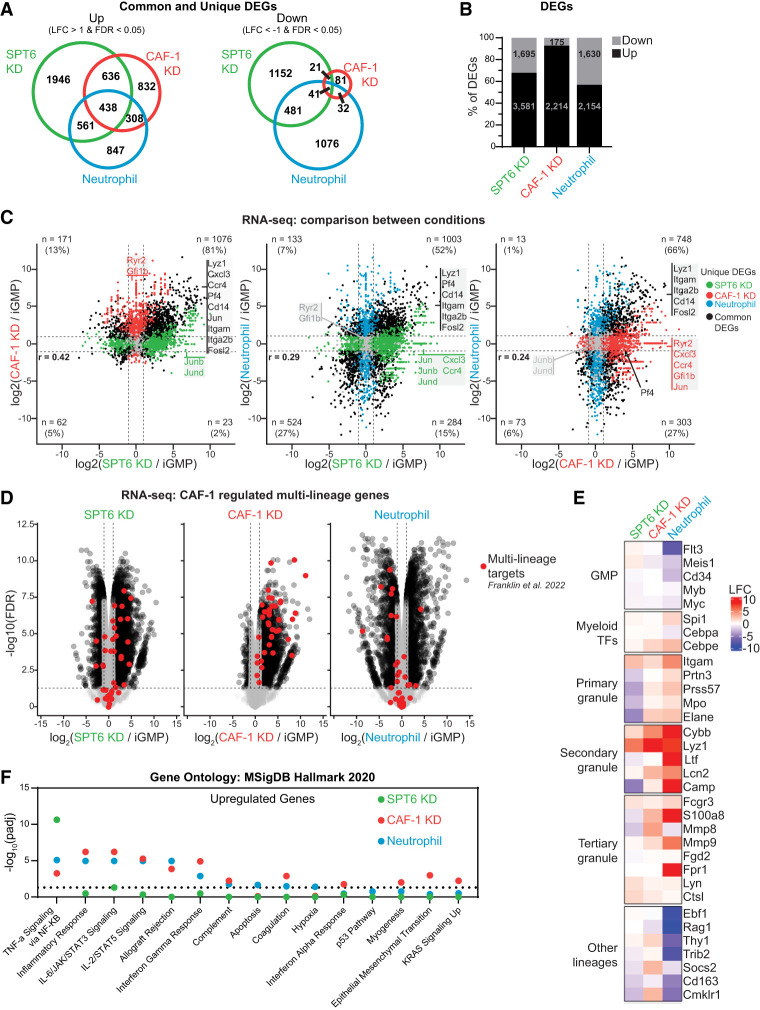
To determine the underlying transcriptional response of the cellular phenotypes observed upon SPT6 and CAF-1 depletion compared with normal neutrophil differentiation (Figs. 2, 3), we performed transcriptional profiling 48 h following induction of SPT6 KD, CAF-1 KD, or neutrophil differentiation. Global analysis of differentially expressed genes (DEGs) revealed that SPT6 KD cells manifested the highest number of transcriptional changes, with both SPT6 KD and CAF-1 KD cells demonstrating a bias toward transcriptional upregulation of DEGs (SPT6 ∼65% and CAF-1 ∼90%) (Fig. 4A,B). Pairwise comparisons upon depletion of either histone chaperone revealed that many CAF-1 KD upregulated genes are also upregulated in SPT6 KD iGMPs (Fig. 4C). In contrast, when compared with neutrophil differentiation, many CAF-1 KD or SPT6 KD upregulated genes are instead downregulated (Fig. 4C). Interestingly, when considering commonly downregulated genes between CAF-1 KD and SPT6 KD iGMPs (albeit below our twofold change cutoff), we found that replicative histone genes followed the same trend (Supplemental Fig. S6A). This is consistent with the fact that replicative histone genes are induced during S phase (Groth et al. 2007; Günesdogan et al. 2014; Marzluff and Koreski 2017; Mendiratta et al. 2019) and thus are sensitive to these perturbations and accompanying cell cycle defects and dependencies (Fig. 3; Supplemental Fig. S4). Additionally, we also detected the upregulation of several histone variant genes in all conditions (Supplemental Fig. S6A). This trend is particularly more pronounced in CAF-1 KD iGMPs for H3 variant genes H3f3b and H3f4, suggesting a compensatory mechanism to maintain chromatin, as reported previously (Ray-Gallet et al. 2011). Together, the common regulation of gene expression upon CAF-1 and SPT6 depletion supports their redundant role to promote cellular differentiation despite their independent molecular roles in DNA replication and transcription.
Figure 4.
Common and distinct transcriptional signatures upon SPT6 and CAF-1 loss in iGMPs. (A) Venn diagrams showing the overlap of upregulated or downregulated differentially expressed genes between SPT6 KD, CAF-1 KD, and neutrophils determined by RNA-seq after 48 h of treatment. Fold changes were calculated relative to iGMPs. (LFC) Log2 transformed fold change. (B) The percentages of total DEGs that are upregulated or downregulated within each condition. The number of DEGs is presented within the respective bars. (C) RNA-seq XY plots showing pairwise comparisons of gene log2 transformed fold changes (LFCs). For each plot, genes differentially expressed (FDR < 0.05 and |LFC| > 1) in both conditions are shown in black. Genes differentially expressed in only one of the displayed conditions (unique DEGs) are colored according to their respective conditions. Dotted lines indicate twofold change cutoffs. The number of common DEGs in each quadrant is given with the percentage of all common DEGs. Select genes are labeled following the same color scheme. LFCs represent n = 2 clonal replicates. Pearson's correlation coefficient (r) is provided with P-values < 2.2 × 10−16. (D) RNA-seq volcano plot. Log2 transformed fold changes (LFCs) of genes in each respective condition compared with iGMPs are displayed on the X-axis, and −log10 transformed false discovery rates (FDRs) are displayed on the Y-axis. Black circles represent individual gene quantifications from all differentially expressed genes (|LFC| > 1 and FDR < 0.05). Red dots represent CAF-1 silenced multilineage genes as described by Franklin et al. (2022). Nondifferentially expressed (nc/ns; |LFC| < 1 or FDR > 0.05) detected genes are displayed as gray dots. Data represent n = 2 clonal replicates. (E) Heat map showing log2 transformed fold changes (LFCs) for selected genes relative to iGMPs. SPT6 KD, CAF-1 KD, and neutrophil conditions are presented in columns, with genes presented in rows. Genes are grouped according to function as described by Do et al. (2024). LFC represents n = 2 clonal replicates. (F) EnrichR gene ontology analysis of upregulated DEGs (LFC > 1 and FDR < 0.05). Gene symbols of upregulated and downregulated genes (see Supplemental Fig. S4B) were used to find enriched gene sets from the MSigDB Hallmark 2020 data set. −log10 adjusted P-value (Padj) of enrichment scores are shown on the Y-axis. The names of individual gene sets are displayed on the X-axis. Dots indicate enrichment scores for SPT6 (green), CAF-1 (red), or HOXA9 (blue) KD DEGs. The dotted line represents Padj = 0.05.
To understand the significance of these transcriptional changes (Fig. 4A–C), we first focused on specific genes relevant to our differentiation system. To this end, we examined the behavior of neutrophil-specific genes and a previous hematopoietic multilineage gene set regulated by CAF-1 in our iGMPs and in primary mouse HSPCs (Fig. 4C–E; Franklin et al. 2022; Do et al. 2024). Consistent with our screen readout, we found commonly regulated myeloid transcription factors and neutrophil-specific genes in all three conditions (Fig. 4E). However, when considering the multilineage gene set, the gene expression changes support their common regulation by CAF-1 and SPT6 but not HOXA9, given that most of them are unchanged or downregulated during neutrophil differentiation (Fig. 4D,E). This finding, together with the observed bias toward transcriptional upregulation, suggests that both histone chaperone pathways restrict aberrant gene expression to maintain the identity of iGMPs (Fig. 4A–E).
We next examined transcriptional effects that are specific to SPT6, given its broad role in regulating gene transcription. Indeed, compared with CAF-1 KD cells and neutrophils, SPT6 KD iGMPs have a greater number of uniquely upregulated and downregulated genes (1946 and 1152, respectively) (Fig. 4A,B). Moreover, gene ontology analysis using the Hallmark gene sets revealed unique enrichment of the TNF-α signaling pathways in SPT6 KD-upregulated genes, whereas SPT6 KD-downregulated genes were enriched for cell cycle and Myc targets (Fig. 4F; Supplemental Fig. S6B). In contrast, CAF-1 KD- and neutrophil-upregulated genes revealed significant enrichment in distinct inflammatory responses such as interferon γ, with no specific enrichment of downregulated genes.
Although neutrophil-downregulated genes did not have any strongly enriched Hallmark gene sets, we confirmed that they were enriched for myeloid gene regulation using the transcription factor (TF) perturbation gene sets (Supplemental Fig. S6C). For example, in neutrophils, Myc-activated targets and CEBPA-repressed targets were downregulated. Furthermore, in neutrophil-upregulated genes, we noted myeloid TF networks being highly enriched, further confirming their myeloid commitment status.
Altogether, the common transcriptional responses in CAF-1 KD, SPT6 KD, and neutrophils cell states are consistent with the differentiation phenotype observed in all conditions (Fig. 4A–E). However, the unique transcriptional changes may explain differences in cell cycle dynamics and cell identities (Figs. 2, 3), which may be controlled by the divergent chromatin functions of SPT6 and CAF-1 (Figs. 1C, 2A).
SPT6 and CAF-1 safeguard distinct chromatin environments in iGMPs
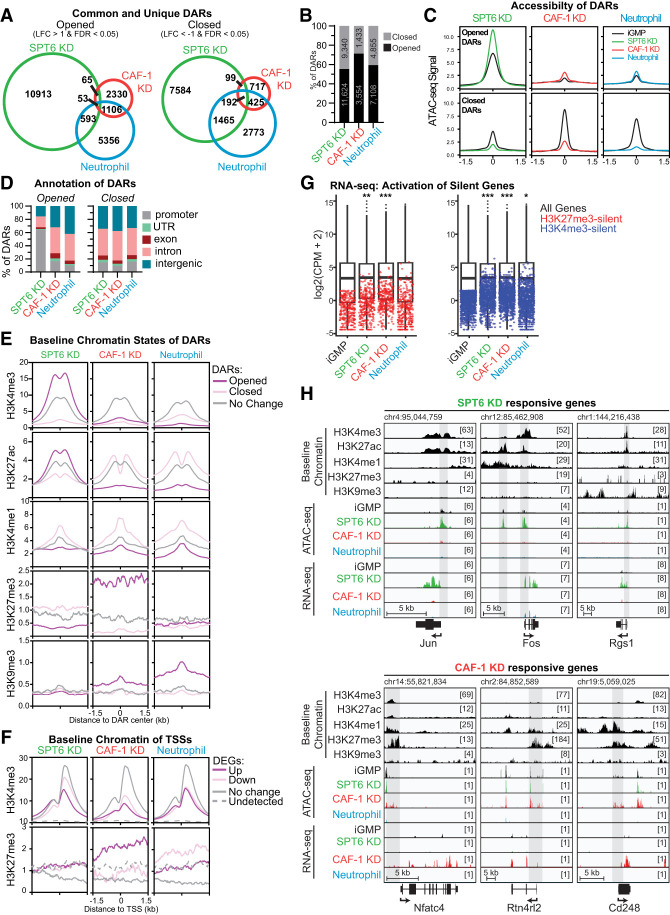
Given the common and distinct transcriptional signatures triggered by CAF-1 and SPT6 depletion (Fig. 4; Supplemental Fig. S6) and their known function in replication and transcription, we investigated how these perturbations may influence chromatin accessibility. To this end, we performed assay for transposase-accessible chromatin followed by sequencing (ATAC-seq) after 48 h in each condition. Differentially accessible regions (DARs) were computed globally by comparing SPT6, CAF-1, and neutrophil conditions with untreated iGMPs (Fig. 5A,B). The loss of CAF-1 triggered a relatively small number of DARs with a strong bias toward opening of chromatin (n = 3554 for open and n = 1433 for closed DARs). In contrast, SPT6 KD cells and neutrophil differentiation resulted in more accessibility changes, with SPT6 KD cells having the most sites (n = 11,624 for open and n = 9340 for closed DARs) (Fig. 5A,B).
Figure 5.
SPT6 and CAF-1 coordinate regulation of distinct chromatin environments. (A) Venn diagrams showing the overlap of opened (LFC > 1 and FDR < 0.05) or closed (LFC < −1 and FDR < 0.05) differentially accessible regions (DARs) between SPT6 KD, CAF-1 KD, and neutrophil differentiation. Opening and closing are quantified with ATAC-seq and relative to accessibility in iGMPs. (LFC) Log2 transformed fold change. (B) Stacked bar plot showing proportions of DARs that are opened or closed for each condition, with the number of DARs displayed inside the bar. (C) ATAC-seq metaplots showing mean counts per million over differentially accessible regions (DARs). Metaplots for each condition (colored lines) are overlaid on the corresponding ATAC-seq signal in iGMPs (black lines). The X-axis represents the center of each DAR as 0, with a ±1.5 kb genomic window. Metaplots represent n = 2 clonal replicates and quantify average counts per million over 10 bp bins. (D) Stacked bar plots showing proportions of opened and closed DARs annotated to genomic regions. Promoters are defined as within 1.5 kb upstream of or 500 bp downstream from annotated transcription start sites. UTRs are defined as DARs overlapping annotated 5′ or 3′ genic untranslated regions. Exons and introns are DARs that overlap annotated exonic or intronic regions, respectively. Intergenic regions are defined as all other DARs not overlapping one of the other genomic features. Annotations are presented in the order of priority and assigned with at least 1 bp overlap. (E) ChIP-seq (H3K27ac and H3K4me3) and CUT&RUN (H3K4me1, H3K27me3, and H3K9me3) metaplots showing mean signal in iGMPs (fold change enrichments over input or IgG, respectively). Mean signal is averaged in 10 bp bins over 3 kb windows centered on opened or closed DARs from each condition (|LFC| > 1 and FDR < 0.05) or on ATAC-seq peaks not differentially accessible (no change; |LFC| < 0 or FDR > 0.05). Metaplots represent n = 3 (H3K27ac, H3K4me1, H3K9me3, and H3K27me3) and n = 2 (H3K4me3) independent experiments. (F) H3K27me3 CUT&RUN and H3K4me3 ChIP-seq metaplots, as shown in E, showing mean signal in iGMPs. Mean signal is averaged in 10 bp bins over 3 kb windows centered on TSSs of differentially expressed genes (RNA-seq read counts with |LFC| > 1 and FDR < 0.05) in each condition or centered on randomly sampled TSSs from genes not differentially expressed (no change; |LFC| < 0 or FDR > 0.05) or from genes not detected (RNA-seq CPM = 0 in all conditions). (G) Box plots summarizing the gene-wise log2 transformed RNA-seq counts per million (CPM) of all detected genes (black box plots) in each condition. The median, second/third quartiles, and 1.5× interquartile range are represented as the center line, box bounds, and extended lines, respectively. Red and blue dots represent lowly expressed (<3 CPM in iGMPs) genes with H3K27me3-marked (red) or H3K4me3-marked (blue) promoters. Mean CPMs are represented for n = 2 clonal replicates. Pairwise Wilcoxon rank sum tests were performed with Holm's correction for multiple comparisons. (*) P = 0.018, (**) P = 0.00022, (***) P < 2.2 × 10−16. (H) Genomic snapshots of SPT6 KD- and CAF-1 KD-responsive genes from the IGV genome browser using the mm10 reference. Baseline chromatin tracks are displayed as base-pair resolution fold change over background (input for H3K27ac, H3K4me3 ChIP-seq, and IgG for H3K4me1, H3K27me3, and H3K9me3 CUT&RUN) in iGMPs. ATAC-seq tracks are base-pair resolution counts per million (CPM) in each indicated condition (iGMPs, SPT6 KD, CAF-1 KD, or neutrophils). RNA-seq tracks are displayed as base-pair resolution CPM in each indicated condition. Schematics for expressed genes are displayed below the snapshots, with UTRs, exons, and introns represented. Arrows indicate the direction of transcription. Bracketed numbers represent the scale maximum. Shaded boxes display regions of interest within the snapshot associated with accessibility changes and certain histone mark enrichments. A scale bar and starting coordinate are shown for each locus.
Remarkably, even though the three conditions resulted in cell differentiation, there was little overlap between the three conditions (n = 53 for open and n = 192 for closed DARs) (Fig. 5A). Moreover, the magnitude of opening and closing of these DARs differed, with SPT6 KD having the strongest effect on increased chromatin accessibility (Fig. 5C; Supplemental Fig. S7A). Consistent with the unique transcriptional response of SPT6 KD cells (Fig. 4A,B), a substantial number of open and closed DARs were also unique to SPT6 KD (Fig. 5A).
To understand whether changes were localized to specific genomic regions, we assessed the genomic annotations and chromatin features of open and closed DARs across the three conditions (Fig. 5D,E; Supplemental Fig. S7A). As expected, most DARs mapped to intronic and intergenic regions regardless of opening or closing, except for the opened DARs of SPT6 KD, where ∼65% mapped to gene promoters (Fig. 5D). This enrichment is approximately threefold higher than in neutrophils and CAF-1 KD, highlighting a potential specific role of SPT6 in regulating promoter elements that cannot be compensated for by other chromatin factors (Fig. 5D). This is unexpected because SPT6 participates in all steps of the transcriptional process, suggesting compensatory mechanisms outside promoter regions (DeGennaro et al. 2013; Gopalakrishnan and Winston 2021; Narain et al. 2021).
To examine whether the initial iGMP chromatin states can predict the accessibility changes, we used our previously described maps of active and repressive histone marks in iGMPs (Franklin et al. 2022) and compared their enrichment over the opened, closed, and unchanged chromatin regions in all three conditions (Fig. 5E; Supplemental Fig. S7A). Consistent with the finding of promoter sensitivity following SPT6 loss, we detected a stronger enrichment of H3K4me3 and H3K27ac at the opened DARs in SPT6 KD cells, suggesting that SPT6 maintains nucleosome density at active promoters. In contrast, we detected a strong enrichment of H3K27me3 and H3K9me3 at the opened DARs compared with the closed and unchanged DARs in CAF-1 KD cells. Notably, neutrophil differentiation did not show similar trends of histone mark enrichments, except for H3K9me3, suggesting unique roles for SPT6 and CAF-1 in the maintenance of promoters and H3K27me3 elements, respectively. The pairwise comparison of chromatin accessibility changes in all three conditions highlighted the unique roles of SPT6 and CAF-1 in maintaining nucleosomes at divergent chromatin environments, given the negligible overlap in DARs between these two conditions (Fig. 5A; Supplemental Fig. S7B).
This divergent effect of SPT6 KD and CAF-1 KD on chromatin regions may explain their distinct transcriptional signatures and subsequent differences in cell identity. Indeed, when we examined transcriptional changes of genes neighboring DARs, we found intriguing differences. Although the opened sites predominantly resulted in increased expression of neighboring genes in all three conditions, we found a regulatory decoupling at the closed DARs in SPT6 KD and CAF-1 KD conditions (Supplemental Fig. S7C,D). Here, a large proportion of these closed sites (∼55% for SPT6 KD and >90% for CAF-1 KD) had upregulated gene neighbors compared with neutrophil differentiation, where ∼30% of closed sites had upregulated gene neighbors (Supplemental Fig. S7C,D). Interestingly, we detected a stronger H3K4me1 enrichment in closed DARs in all conditions, suggesting a common mechanism for aberrant activation of neighboring genes at these closed sites (Fig. 5E; Supplemental Fig. S7A).
We next examined chromatin features over the promoters of DEGs, with a focus on H3K4me3 and H3K27me3 marks, given their profound enrichments in SPT6 KD and CAF-1 KD open DARs, respectively. H3K4me3 was enriched regardless of gene expression changes, whereas H3K27me3 was preferentially enriched at CAF-1 KD-upregulated DEGs (Fig. 5F). Consistent with this observation, low-expressed and nonexpressed genes (CPM < 1) with H3K27me3-marked promoters showed significant upregulation in CAF-1 KD compared with neutrophil differentiation and SPT6 KD cells, especially among the lowest-expressed genes (Fig. 5G). However, this unique CAF-1 KD sensitivity was not observed with H3K4me3-marked promoters (Fig. 5G). Together, these results suggest that H3K27me3-marked regions are specifically sensitive to the loss of CAF-1, resulting in increased gene expression in these regions. This may be due to loss of chromatin compaction or decoupling of repressive chromatin factors from H3K27me3 chromatin. We did not observe a similar trend with SPT6 KD, suggesting alternative mechanisms leading to transcriptional activation of SPT6-sensitive promoters.
Many of the opened promoters in SPT6 KD iGMPs involved signaling genes, including Rgs1, Jun, and Fos. This finding was consistent with the enrichment of the TNF-α–NF-κβ signaling pathway that we detected, without considering changes in chromatin accessibility (Figs. 4F, 5H; Supplemental Fig. S8A). Gene ontology analysis of the genes neighboring SPT6 KD open DARs revealed even stronger enrichment of the TNF-α–NF-κβ signaling pathway (Supplemental Figs. S7C,D, S8B). The enrichment in this subset of upregulated genes suggests an exquisite sensitivity of these promoters to SPT6 loss. In contrast, open sites in CAF-1 KD cells involved fate genes marked with H3K27me3, consistent with the mixed lineage and dysfunctional state of these cells (Fig. 5H; Supplemental Fig. S8A).
Taken together, our analysis of the chromatin state at differentially accessible regions revealed that promoters are sensitive to SPT6 loss, and heterochromatic regions (H3K27me3-enriched sites in particular) are sensitive to CAF-1 loss. Furthermore, these observations suggest that SPT6 and CAF-1 specifically maintain the chromatin landscape at distinct chromatin elements, and that the activity of histone chaperones at these sites is required to maintain cell fate.
Perturbation of SPT6 and CAF-1 leads to unique changes in transcription factor chromatin accessibility
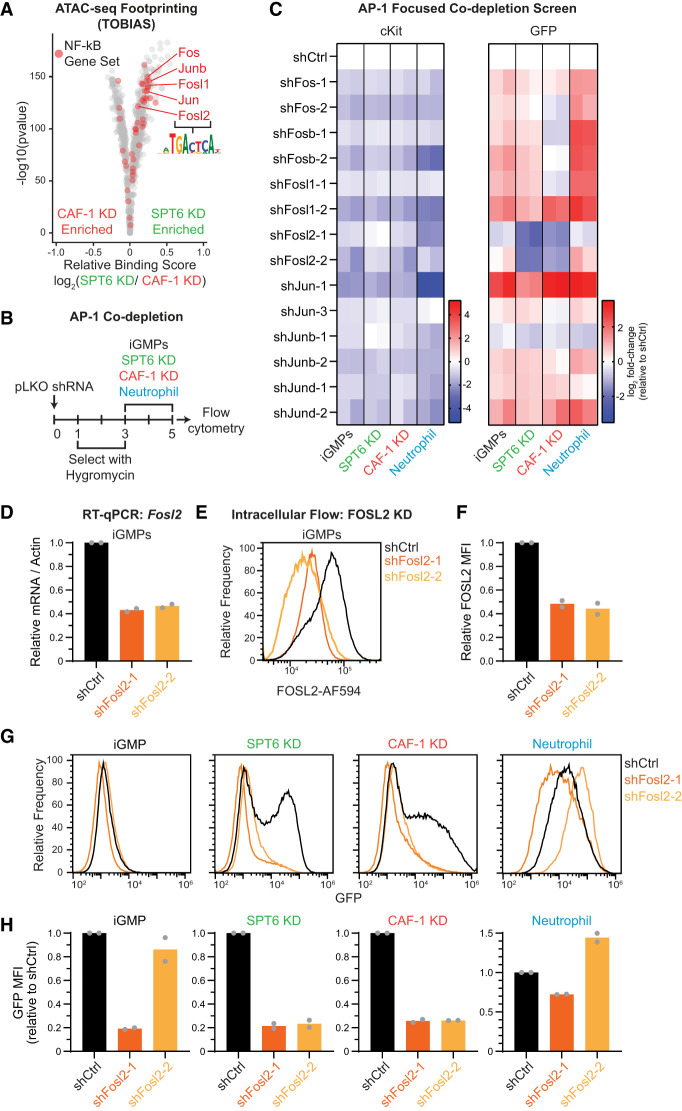
The transcriptional response at CAF-1- and SPT6-sensitive sites and subsequent cell fate change may be dependent on the availability of specific transcription factors (TFs) that can bind these regions and regulate target genes. The correlation between chromatin accessibility and transcription suggests that these TFs may act as activators (Supplemental Fig. S7C,D). To identify functionally relevant TF candidates, we used TOBIAS ATAC-seq footprinting to compute differential binding scores of TF binding sites (Bentsen et al. 2020). By comparing CAF-1 KD and SPT6 KD ATAC-seq maps, we noted that TF binding sites differed between the two conditions (Fig. 6A). Consistent with the enrichment of the TNF-α–NF-κB signaling pathway in SPT6 KD-upregulated genes, including the strong transcriptional activation of some of these factors in SPT6 KD cells (Figs. 4C, 5H; Supplemental Figs. S8, S9A), we found that binding sites for JUN/FOS family members were especially enriched in the footprint analysis (Fig. 6A). This class of proteins is known to form AP-1 TF complexes in various combinations and is encoded by early immediate genes that respond to various cellular processes, including proliferation, apoptosis, and differentiation (Liebermann et al. 1998).
Figure 6.
SPT6 uniquely maintains AP-1 (FOS/JUN family) binding sites. (A) TOBIAS ATAC-seq footprinting volcano plot showing the predicted changes in total transcription factor binding between SPT6 KD and CAF-1 KD. Red dots represent genes from the TNF-α signaling via NF-kB gene set (see Fig. 4F), with the FOS and JUN transcription factor family labeled. (B) Schematic of AP-1 TF miniscreen depleting FOS and JUN TFs in the context of SPT6 KD, CAF-1 KD, and neutrophil differentiation conditions. (C) Heat map of cKit+ and GFP+ population fold changes quantified by flow cytometry relative to scrambled shRNA (shCtrl) in each condition. Log2 fold changes for n = 2 clonal replicates are shown in individual columns. (D) Reverse transcription quantitative PCR (RT-qPCR) of Fosl2 mRNA for n = 2 clonal replicates in iGMPs transduced with shCtrl, shFosl2-1, or shFosl2-2, quantified 3 days after infection. (E) Histogram showing FOSL2 expression quantified by flow cytometry. iGMPs were transduced with two independent shRNAs targeting Fosl2 and compared with shCtrl cells transduced with scrambled shRNA. Depletion was quantified 3 days after infection. Histograms are representative of n = 2 clonal replicates. (F) Bar plot summarizing FOSL2 expression with mean fluorescence intensity (MFI) shown in E relative to the shCtrl. The mean of n = 2 clonal replicates. (G) Histograms showing GFP expression quantified with flow cytometry 5 days after transducing nontargeting shRNA (shCtrl) or Fosl2 shRNAs, including 2 days of inducing with SPT6 KD, CAF-1 KD, and neutrophil differentiation. Representative of n = 2 clonal replicates. (H) Bar plots summarizing GFP mean fluorescence intensity (MFI; from samples in G) relative to shCtrl.
To functionally determine whether JUN/FOS family members are driving the differentiation phenotype in SPT6 KD cells, we performed a focused AP-1 TF shRNA screen (Fig. 6B,C). iGMPs with individual constitutive AP-1 TF knockdown were assessed for effects on cellular differentiation induced by SPT6 KD, CAF-1 KD, and neutrophil conditions. Unexpectedly, the depletion of the majority of these TFs in conjunction with inducing differentiation in all three conditions further enhanced the phenotype. However, we found that the depletion of these TFs alone prior to the induction of the three conditions was sufficient to induce differentiation, arguing that they are required for the maintenance of iGMPs (Fig. 6C). This initial differentiation state triggered by the AP-1 TF knockdown alone in iGMPs may then sensitize the response after induction of SPT6 KD, CAF-1 KD, and neutrophil differentiation. Notably, FOSL2 was the only candidate whose depletion showed a consistent rescue of the differentiation induced in SPT6 KD and CAF-1 KD states (Fig. 6C–H). However, similar to the other AP-1 TF candidates, its depletion also reduced cKit signal (Fig. 6C). These results suggest complex functions of the AP-1 TFs in maintaining cell fate and inducing differentiation. This complexity may depend on the gene expression of various AP-1 TF combinations during different cell states and their dosage. Consistent with this idea, examining previously published results by ENCODE and Tabula Muris projects (Schaum et al. 2018; Luo et al. 2020), we found differential expression of AP-1 TF across different hematopoietic lineages, with FOSL2 in particular being enriched among the myeloid differentiated cells (Supplemental Fig. S9B,C).
Taken together, our TF motif predictions and AP-1 TF loss-of-function analyses suggest that although these TFs are commonly required to promote differentiation in the context of CAF-1 and SPT6 manipulations, their activity in SPT6 KD cells may depend on the accessibility of the SPT6-responsive sites (Figs. 5, 6A).
CAF-1 and SPT6 perturbations trigger alternative differentiation paths
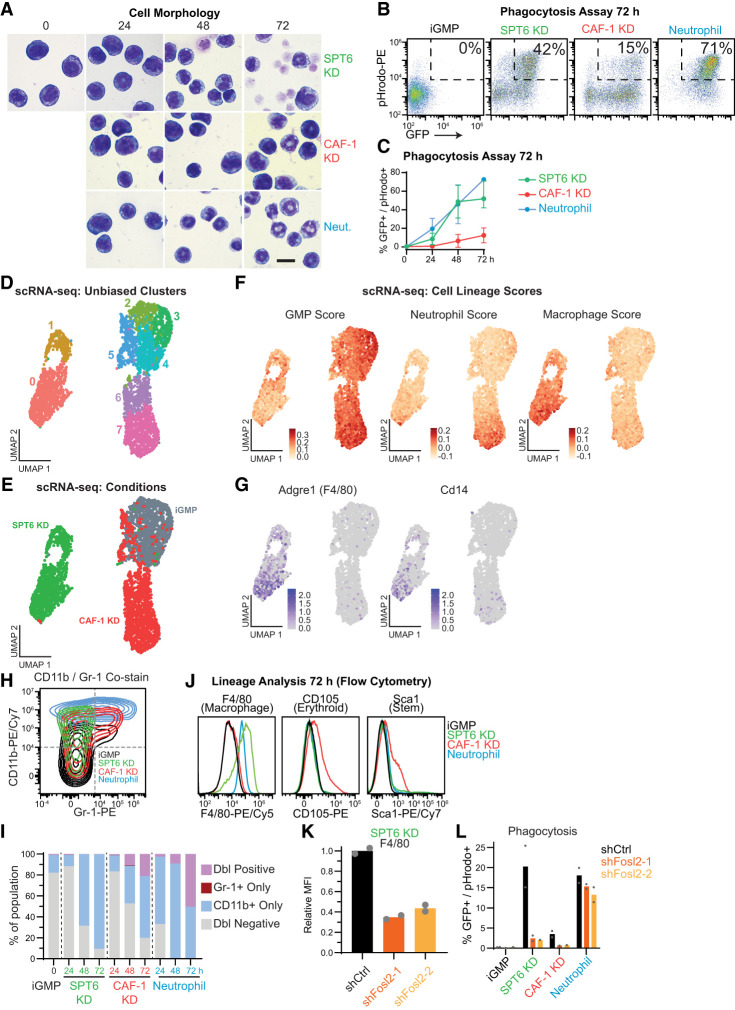
Given the distinct chromatin and transcriptional changes between the CAF-1- and SPT6-depleted iGMPs (Figs. 4–6), we wondered whether the respective differentiation states mimic the cellular morphology and phagocytic ability of neutrophils. Interestingly, even with extended time of either perturbation, their morphology was different from that of neutrophils (Fig. 7A). Moreover, consistent with the previously reported partial differentiation of CAF-1 KD iGMPs into a mixed-lineage state (Franklin et al. 2022), CAF-1 KD dual-inducible iGMPs were dysfunctional in engulfing bacterial particles (Fig. 7B,C). In contrast, SPT6 KD iGMPs gained a significant phagocytotic function despite being morphologically different from neutrophils (Fig. 7B,C).
Figure 7.
SPT6 and CAF-1 perturbations induce divergent differentiation paths. (A) Morphology of iGMPs after 0, 24, 48, or 72 h of SPT6 KD, CAF-1 KD, or neutrophil differentiation. Magnification, 1000×. Scale bar, 10 µm. Experiment was repeated independently in two clones with similar results. (B) Phagocytosis flow cytometric assay. Density plots of iGMPs and 72 h after induction for each condition, showing GFP signal versus fluorescently labeled bacterial particles (pHrodo). (C) Phagocytosis time-course analysis reflecting the percentage of GFP+/pHrodo+ cells (B shows a representative example of gating) for iGMPs and after 24, 48, or 72 h of SPT6 KD, CAF-1 KD, and neutrophil differentiation. The line connects the mean of two clonal replicates with the range displayed. (D) UMAP analysis of single-cell transcriptomes in iGMPs (n = 1419), 48 h of SPT6 KD (n = 1220), and 48 h of CAF-1 KD (n = 1425). Single cells are colored according to unsupervised cluster assignment (see the Materials and Methods). (E) UMAP analysis as in D with cells colored according to treatment (iGMPs, SPT6 KD, or CAF-1 KD). (F) UMAP feature plots representing lineage scores summarizing average signal over background for gene sets enriched in primary granulocyte–macrophage progenitors (GMPs), macrophages, or neutrophils (see the Materials and Methods; Supplemental Fig. S10B,C). (G) UMAP feature plot displaying the log-normalized expression of select macrophage-specific marker genes Adgre1 and Cd14. (H) Representative flow cytometry contour plot representing CD11b (pan-myeloid marker) and Gr-1 (neutrophil-specific marker) expression in iGMPs or after 72 h of SPT6 KD, CAF-1 KD, or neutrophil differentiation. Contours are colored according to each condition. (I) Time-course analysis of percent populations based on CD11b versus Gr1 expression. Representative gating as shown in H. (J) Representative flow cytometry analysis measuring the expression of F4/80 (macrophage-specific), CD105 (erythroid-specific), or Sca1 (stemness marker) in iGMPs or 72 h after SPT6 KD, CAF-1 KD, or neutrophil differentiation. (K) Bar plot showing F4/80 expression after 48 h of SPT6 KD in cells transduced with shCtrl, shFosl2-1, or shFosl2-2 (as shown in Fig. 6B). n = 2 clonal replicates. (L) Phagocytosis flow cytometric assay quantifying the percentage of phagocytic differentiated cells (GFP+/pHrodo+) in iGMPs and after 48 h of SPT6 KD, CAF-1 KD, and neutrophil differentiation in cells transduced with shCtrl, shFosl2-1, or shFosl2-2 (as shown in Fig. 6B). Mean of n = 2 clonal replicates.
These observations prompted us to further investigate the identities that CAF-1- and SPT6-depleted iGMPs differentiate into. To this end, we conducted transcriptome analysis at single-cell resolution (scRNA-seq) to capture the differentiation granularity upon CAF-1 or SPT6 depletion. Using uniform manifold approximation and projection (UMAP) analysis (Becht et al. 2019), we identified divergent clusters distinguishing CAF-1 KD and SPT6 KD populations (Fig. 7D,E). Remarkably, SPT6 KD cells exhibited a more distinct differentiation trajectory compared with CAF-1 KD cells, supporting the more robust differentiation response measured in our screen using a pan-myeloid differentiation marker as a readout (Figs. 1, 2, 7D,E). Interestingly, unsupervised clustering of variable genes revealed distinct single-cell expression signatures, with SPT6 KD cells upregulating macrophage-related genes such as Adgre1 and Laptm5, whereas CAF-1 KD cells predominantly activated neutrophil-associated genes, including Elane, Ly6c2, Lcn2, and Mmp8 (Supplemental Fig. S10A).
Given the robust differentiation of SPT6-depleted cells into functionally phagocytotic cells and iGMPs’ potential to differentiate into macrophages or neutrophils, we wondered whether cells were adopting a different fate. To determine differentiation paths in either condition, we computed GMP, neutrophil, and macrophage lineage scores using publicly available data sets from primary cells (Supplemental Material; Supplemental Fig. S10B,C; Choi et al. 2019). Consistent with the induction of a mixed-lineage state, we found that CAF-1 KD cells exhibited high GMP and neutrophil lineage scores (Fig. 7F). Interestingly, the SPT6 KD iGMPs display a distinctive, higher macrophage lineage score (Fig. 7F). Moreover, macrophage markers such as Adgre1 and Cd14 as well as the phagocytosis gene signature are enriched specifically in SPT6-depleted populations (Fig. 7G; Supplemental Fig. S10F). Notably, pairwise comparisons of macrophage versus neutrophil lineage scores confirmed a macrophage-like bias in SPT6-depleted cells, as evidenced by both scRNA-seq and bulk RNA-seq analyses (Supplemental Fig. S10D,E).
To further validate the macrophage transcriptional signature of SPT6 KD cells, we assessed the expression of cell surface markers common and specific to neutrophils and macrophages. In line with our RNA-seq analyses, neutrophils and CAF-1 KD iGMPs showed an increased proportion of double-positive CD11b/Gr-1 population, indicating a normal bias toward neutrophil lineage as they gain the neutrophil-specific marker Gr-1. In contrast, SPT6-depleted cells only induced the common myeloid marker CD11b (Fig. 7H,I). Assessment of additional hematopoietic surface markers confirmed that SPT6 perturbation directs cells toward a macrophage lineage, as judged by expression of the macrophage-specific marker F4/80 (ADGRE1) (Fig. 7J; Supplemental Fig. S10G). In contrast, CAF-1 KD cells uniquely induced the Sca1 stem cell marker and CD105 erythroid marker, consistent with a mixed-lineage state. Taken together, these results highlight the divergent differentiation pathways with perturbations of CAF-1 and SPT6.
Finally, we investigated whether the macrophage-like differentiation bias in SPT6 depletion is dependent on the activity of AP-1 TFs. We first analyzed previously published FOSL2 ChIP-seq data sets from primary macrophages (Abe et al. 2024) over our opened and closed chromatin sites in all three conditions (Fig. 5) and found that FOSL2 binding is enriched over opened elements compared with closed sites, a trend specific to SPT6-sensitive sites (Supplemental Fig. S10H). Consistent with these observations, codepletion of SPT6 and FOSL2 results in a reduction of the F4/80 macrophage marker and a strong suppression of phagocytosis (Fig. 7K–L; Supplemental Fig. S10I,J). Together, our results suggest that in response to SPT6 depletion, the AP-1 TF activity pushes cells toward a macrophage-like differentiation trajectory.
Discussion
We performed a histone chaperone-focused screen, revealing their universal role in maintaining cellular identity, with SPT6 identified as the top-scoring candidate (Fig. 1). Using CAF-1 and SPT6 as models for replication- and transcription-coupled histone chaperones (Fig. 2), we demonstrated how these processes preserve cellular identity by regulating specific chromatin sites. We found that heterochromatic regions are particularly sensitive to CAF-1 depletion, whereas active promoter elements are sensitive to SPT6 depletion (Fig. 5).
Integrating RNA-seq, ATAC-seq, and histone modification profiles, we linked these chromatin-sensitive sites to the activation of H3K27me3 silenced genes in CAF-1-deficient cells and to AP-1 transcription factor activity in SPT6-deficient cells (Figs. 5, 6). Although both perturbations rely on cell cycle progression to drive differentiation, they have distinct effects on global DNA replication and transcription: SPT6 depletion disrupts transcription primarily in G1/G0, whereas CAF-1 depletion impairs S-phase progression (Fig. 3).
Interestingly, SPT6 depletion biases cells toward a macrophage-like state, whereas CAF-1 depletion promotes differentiation toward a neutrophil-like state, albeit with a mixed-lineage signature (Fig. 7). Based on these findings, we propose a model in which, in self-renewing stem and progenitor cells, CAF-1 maintains H3K27me3-repressed genes during S phase, whereas SPT6 sustains active promoters during G1/G0. Inhibiting CAF-1 results in the promiscuous activation of various transcription factors, driving a multilineage state, whereas inhibiting SPT6 activates AP-1 transcription factors, promoting a specific lineage choice (Supplemental Fig. S11).
SPT6 in different cellular contexts and effects on chromatin and transcription
To our knowledge, this is the first study pointing to the role of SPT6 in the maintenance of hematopoietic stem and progenitor identity. Our findings complement evidence linking SPT6 to cell fate control in other systems (Wang et al. 2013, 2017; Obara et al. 2020; Li et al. 2021). Compared with CAF-1, which broadly safeguards cellular identity (Cheloufi et al. 2015; Cheloufi and Hochedlinger 2017; Volk et al. 2018; Cheng et al. 2019; Franklin et al. 2022), SPT6's role is more dynamic. Unlike its prodifferentiation role in myogenesis and epidermal formation, SPT6 depletion in embryonic stem cells and hematopoietic progenitors induces differentiation, underscoring the importance of cellular context. Here, as in ESCs, master regulatory programs maintain the progenitor state and suppress differentiation genes. SPT6 depletion disrupts this balance, leading to differentiation.
It will be interesting to compare chromatin sensitivity sites and gene expression changes upon SPT6 depletion across these different cellular contexts. This may reveal specific genic features leading to transcriptional activation or repression. We show here that SPT6 depletion had the most profound effect on transcriptional output (Figs. 3, 4). At genic regions, this could occur through SPT6 regulation of promoter pause release, transcription elongation, histone marking, or post-transcriptional processing, given its complex protein domains. Several reports show a role for mammalian SPT6 in transcriptional initiation and pause–release (Endoh et al. 2004; Li et al. 2021; Narain et al. 2021). Interestingly, however, a recent study in DLD-1 cells counterintuitively found that SPT6 depletion can induce RNA Pol II pause–release (Aoi et al. 2022). This supports SPT6 as a dynamic gatekeeper of transcription fidelity more than simply a proelongation factor. Although further research is necessary to reconcile these results with SPT6's known role in promoting elongation, it is tempting to speculate that SPT6 may dynamically regulate pausing or elongation depending on the specific regulatory factors at promoters (Aoi et al. 2022). In addition to its role in transcriptional regulation, SPT6 has also been involved in post-transcriptional gene regulation such as RNA stability, splicing, and export (Yoh et al. 2007, 2008; Dronamraju et al. 2018; Oqani et al. 2019). Notably, EU labeling of nascent RNA reveals a global transcriptional effect (Fig. 3), suggesting that SPT6 influences ribosomal RNA transcription (Engel et al. 2015). Future studies will reveal how these diverse roles of SPT6 are involved in the regulation of cell identity genes in various cellular contexts.
Coordination between histone chaperone pathways coupled to DNA replication and transcription during cell cycle
Our results support the role of SPT6 and CAF-1 in the maintenance of iGMPs via different mechanisms. Their loss triggers iGMP differentiation while exerting divergent effects on chromatin accessibility, activity of transcription factors, and transcriptional response, ultimately leading to alternate lineage choice. Specifically, SPT6 affects active chromatin at promoter regions, whereas CAF-1 affects heterochromatic regions. Thus, despite the separable roles of SPT6 and CAF-1 in transcription- and replication-dependent chromatin assembly and their specific requirement at distinct chromatin elements that we uncovered here, the two pathways are in place to maintain stem cell identity. Notably, the maintenance of chromatin at these regions by SPT6 and CAF-1 does not seem to be compensated for by other histone chaperone pathways. Our cell cycle analysis suggests that CAF-1 maintains cell fate primarily through its action during S phase, whereas SPT6 maintains cell fate through its activity during G1/G0 phases. However, we cannot rule out additional roles for each histone chaperone throughout the cell cycle. For example, SPT6 has been recently linked to function during S phase in yeast cells and in Drosophila embryos (Narain et al. 2021; Miller and Winston 2023; Miller et al. 2023; Kemp et al. 2024). It will be interesting to investigate how each chaperone maintains chromatin as a function of cell cycle phases and how they interact with replication and transcription machineries. Such roles are undoubtedly related to the necessary coordination between DNA replication and transcription during the cell cycle.
Materials and methods
Cell culture
Immortalized granulocyte–macrophage progenitors (iGMPs) (Wang et al. 2006; Sykes et al. 2016) with different combinations of transgenes, generated in this study, were cultured as described previously, with the respective constitutive or induced transgene expression (Supplemental Material; Franklin et al. 2022).
Histone chaperone shRNA screen
miR-30 shRNA libraries were a gift from Chris Vakoc. Sixty-thousand iGMPs per well in a 96 well plate were infected with 200 µL of viral supernatant (see “Virus Production and Infection” in the Supplemental Material). Two days after infection, cells were selected with G418 antibiotic for 5 days and then analyzed by flow cytometry after staining for CD11b. After gating shRNA-positive cells, the geometric means of CD11b signal (PE/Cyanine7 channel) were collected, and Z-score was normalized based on the mean and standard deviations of all samples.
SPT6 rescue
HOXA9-iGMP clones containing tetO-shSupt6 shRNA13 or shRNA14 (three independent clones for each shRNA) were transduced with pULTRA-HA-SPT6 or pULTRA-hot (mCherry-only) lentiviral vectors (see “Plasmid Cloning”; Fig. 1G). pULTRA-HA-SPT6 was transduced at low efficiency, so mCherry+ cells were purified by fluorescence-activated cell sorting (FACS). Purified HA-SPT6 iGMPs were then induced with Dox, and GFP fold induction was compared with mCherry-only transduced iGMPs.
Flow cytometry
Cell surface markers were live-cell-stained with 1:200 antibody dilutions in 100 µL of FACS buffer (PBS with 5% FBS, 1 mM EDTA) for 20 min in the dark at 4°C or on ice. After incubation, cells were washed twice with excess FACS buffer and then resuspended in FACS buffer for flow cytometry. Intracellular FOSL2 was stained by fixing cells with 4% formaldehyde for 15 min at room temperature and incubating them with 1:100 FOSL2 antibody (Millipore Sigma MABS1261) in a saponin-based permeabilization buffer for 2 h at 4°C. Cells were washed twice and incubated with antirat AF594 antibody (A-11007) 1:500 in saponin-based permeabilization buffer for 1 h in the dark at 4°C. The mouse antibodies used—CD11b-PE/Cyanine7 (101216), cKit-PE/Cyanine5 (105809), Gr-1-PE (108407), F4/80-PE/Cyanine5 (123112), CD105-PE (120407), and Sca1-PE/Cyanine7 (108113)—were purchased from BioLegend. Flow cytometry data were acquired with NovoExpress (v1.5) on NovoCyte 2100Y/Quanteon cytometers at University of California Riverside. Flow data were analyzed with FlowJo (v9 and v10). Flow cytometry analysis included gating cells by forward versus side scatter to exclude debris and dead cells followed by gating height versus area scatter to isolate live, single-cell events. Further gates were then applied as needed for parameters of interest based on negative and positive controls.
Growth curve
Cells were seeded at low density (3000 cells/mL) on the same day for each condition and then counted at subsequent time points with a hemacytometer. SPT6 KD, CAF-1 KD, and neutrophil differentiation treatments were as described in “Cell Culture and Transgene Expression” in the Supplemental Material.
Cell cycle (EdU) and global transcription (EU) analysis
For 5-ethynyl uridine (EU) incorporation assay, iGMPs untreated or after 24 and 48 h of SPT6 KD, CAF-1 KD, and neutrophil differentiation were labeled with 1 mM EU for 1 h at 37°C.
For cell cycle analysis (EdU), iGMPs untreated or after 24 and 48 h of SPT6 KD, CAF-1 KD, and neutrophil differentiation were labeled with 10 µM 5-ethynyl-2′-deoxyuridine (EdU) for 1 h at 37°C (Click-iT Plus EdU kit, Thermo C10646).
After EU or EdU labeling, cells were collected at 1 million cells/mL, fixed with 4% formaldehyde, and permeabilized with a saponin-based wash. EU and EdU were labeled in a reaction cocktail with an Alexa fluor 647 picolyl azide for 30 min at room temperature (Click-iT Plus EdU). After labeling, total DNA content was stained with 250 ng/mL DAPI or 1× FxCycle violet (also referred to as DAPI) either overnight at 4°C or for 1 h at room temperature. Cells were then analyzed by flow cytometry in FACS buffer (PBS with 5% FBS, 1 mM EDTA). After gating out debris, dead cells, and doublets, the gates for cell cycle phases were made based on DNA content versus EdU signal. For EU-labeled samples, G1/G2, S, and G2/M populations were gated based on DAPI signal only, gating on the classical G1 and G2 peaks with intermediate cells as S phase.
Growth arrest
For uninduced cells, growth arrest was achieved by incubating cells in media with depleted SCF (0.1× or 0.01× SCF) and compared with self-renewal culture conditions (1× SCF). SPT6 KD and CAF-1 KD cells were induced for 12 h before withdrawing SCF to achieve depletion at the time of arrest. For neutrophil differentiation, because HOXA9 translocation happens within hours (Sykes et al. 2016; Blanco et al. 2021; Franklin et al. 2022; Do et al. 2024), cells were induced after 12 h of arrest. DNA content was analyzed by DAPI staining and flow cytometry after 12 h of arrest (postarrest) and by flow cytometry for GFP after 48 h of arrested treatment (end point).
CellTrace assay
CellTrace (Thermo Fisher C34564) was used according to the manufacturer's recommendation. Uninduced iGMPs were collected in PBS and stained with 1:1000 CellTrace reagent at 1 million cells/mL. Cells were incubated for 20 min at 37°C during staining. Complete cell culture media was added to stained cells at 5× the sample's volume to quench extra staining reagent and incubated for 5 min at 37°C. Cells were pelleted and resuspended in complete media. SPT6 KD, CAF-1 KD, and neutrophil conditions were induced after staining, and cells were collected after 48 h to analyze CellTrace signal by flow cytometry.
Phagocytosis assay
Uninduced, SPT6 KD, CAF-1 KD, or neutrophil differentiated iGMPs were resuspended in media along with fluorescently labeled, heat-killed Escherichia coli bioparticles (Thermo Fisher P35361). Bioparticles were resuspended in live-cell imaging buffer (Thermo Fisher A14291DJ) at 1 mg/mL. Bioparticles were diluted 1:10 in fresh, complete cell culture media with 1 million cells/mL. Cells with bioparticles were incubated for 2 h at 37°C, collected, and analyzed for flow cytometry.
Microscopy
iGMPs before or after 24, 48, and 72 h of SPT6 KD, CAF-1 KD, and neutrophil differentiation were spun onto glass slides (30,000–40,000 cells each) with a Cytospin at 600 rpm for 5 min at room temperature. For morphology, cells were fixed with methanol and sequentially stained with eosin Y and methylene blue/azure A (modified Wright-Giemsa stain and Richard-Allan Scientific three-step stain). Images were acquired using a Nikon Eclipse 80i upright microscope.
For immunofluorescence, iGMP, 24 h, and 48 h conditions were prepared in parallel with cells treated for 4 h with 5 mM hydroxyurea (HU+) or vehicle (HU−). Cells were fixed with 4% formaldehyde for 10 min at room temperature., permeabilized with 0.1% Triton-X in phosphate-buffered saline (PBST), and saturated with 10% FBS in PBST. Cells were washed three times with PBST and incubated overnight at 4°C with primary antibody targeting γH2AX (1:100; Cell Signaling Technology 9718S) in 10% FBS-PBST. Cells were washed three times and incubated with antirabbit secondary antibody (1:1000; Invitrogen A21245) in 10% FBS-PBST for 1 h at room temperature. Coverslips were mounted with VectaShield Plus DAPI mounting media (Vector Laboratories H-2000), and images were acquired on a Leica DMi8 microscope with Leica application suite X (v3.3). Images were analyzed with ImageJ to quantify fluorescent signal and number of foci per cell. Briefly, single-channel images of DAPI were loaded and reduced to 8 bits, thresholded, and watershed. Nuclei were defined as regions of interest with Analyze Particles, and these regions were then used to analyze the γH2AX channel. Here, images were reduced to 8 bits, smoothened with Gaussian blur, and then quantified with Measure for each region of interest (nuclei). For focus counts, images were then analyzed with Find Maxima, and points were counted per nucleus.
Whole-cell lysates
Cells were collected in 1× Lamelli buffer (pure water with 200 mM Tris-HCl at pH 6.8, 1% [w/v] SDS, 250 µg/mL bromophenol blue, 10% [w/v] glycerol, 1% β-mercaptoethanol) at 10 million cells/mL to create whole-cell lysates. Whole-cell lysates were processed by centrifugation at >15,000g for 2 min followed by DNA shearing with sonication using a Branson probe sonifier at 15%–30% amplitude, depending on lysate volume, for 10 sec. If lysates foamed, sonication ended abruptly, and processing proceeded. Alternatively, aspiration with a 27 gauge needle was used to shear and solubilize DNA instead of sonication. After shearing, samples were boiled for 3 min at 100°C in a heat block and then placed for 5 min on ice. Lysates were centrifuged at maximum speed for 30 sec. If lysates were still too viscous, sonication and boiling were repeated. Lysates were then analyzed or stored indefinitely at −80°C.
SDS-PAGE and Western blot analysis
Whole-cell lysates were size-separated on 6%–15% SDS–polyacrylamide gels in running buffer (pure water with 3 mg/mL Tris, 14.4 mg/mL glycine, 1% [w/v] SDS) and transferred to a nitrocellulose membrane (Bio-Rad 1620097) for 1–2 h at 120–145 V and 4°C in transfer buffer (pure water with 3 mg/mL Tris, 14.4 mg/mL glycine). Membranes were blocked for 1 h in Tris-buffered saline (TBS; pure water with 8 mg/mL NaCl, 2.4 mg/mL Tris at pH adjusted to 7.6 with HCl) with 0.05% Tween-20 (TBST) and 5% (w/v) dry skim milk, washed three times in TBST for 5 min, and incubated overnight at 4°C with primary antibody diluted 1:500–1:2000 in TBST with 3% BSA. Blots were washed three times for 5 min in TBST, incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies, and diluted 1:2000 in TBST with 5% (w/v) dry skim milk for 1 h at room temperature, and blots were washed three times for 5 min in TBST. β-Actin was detected with HRP-conjugated primary antibody (Millipore Sigma A3854), diluted 1:20,000 in TBST with 5% (w/v) dry skim milk, and incubated with the membrane for 15–30 min at room temperature. HRP was detected by Western Lightning Plus (PerkinElmer NEL103E001EA) or SuperSignal West Pico Plus (Thermo Fisher 34578) chemiluminescent substrate. Chemiluminescent signal was detected by exposing membranes to X-ray film in a dark room, followed by film development and image scanning, or detected with a Bio-Rad ChemiDoc imaging system. β-Actin (ACTIN) and TATA-binding protein (TBP) were used as loading controls.
The following antibodies were used: anti-CHAF1B (Santa Cruz Biotechnology sc-393662), anti-SPT6 (Bethyl A300-801A), anti-pRPA32-S4/8 (Bethyl A300-245A), anti-HA (Cell Signaling Technologies 3724), anti-γH2AX-S139 (Cell Signaling Technologies 9718S), anti-H2AX (Cell Signaling Technologies 2595S), anti-pChk1-S345 (Cell Signaling Technologies 2348S), anti-Chk1 (Cell Signaling Technologies 2360S), anti-pRPA32-S33 (Cell Signaling Technologies 10148), anti-RPA32 (Cell Signaling Technologies 2208S), anti-TBP (Abcam ab818), anti-β-Actin (Millipore Sigma A3854), antimouse IgG (Millipore Sigma AP124P), antirabbit IgG (Millipore Sigma AP307P), and antirat IgG (Millipore Sigma A9037).
Plasmid cloning
For inducible CAF-1 depletion, Chaf1b targeting shRNAs were cloned into the pLKO-TRC912 1X LacI IPTG-inducible vector (Broad Institute) harboring a puromycin-selectable marker described previously (Franklin et al. 2022). Inducible Supt6 targeting shRNAs were cloned into the EZ-tet-PLKO-Blast plasmid (Addgene 85973) via restriction digestion and ligation. shRNAs targeting Jun, Junb, Jund, Fos, Fosb, Fosl1, and Fosl2 were cloned into the pLKO.1-blast plasmid (Addgene 26655) with restriction digestion and ligation. Briefly, 21-mer shRNA sequences targeting the respective gene mRNAs were retrieved from the GPP web portal (Broad Institute). Sense and antisense oligos were designed with these 21-mer sequences, and phosphorylation and annealing reactions were set up with 10 µM sense oligo, 10 µM antisense oligo, 1× T4 DNA Ligation buffer (NEB B0202S), and 500 U/mL T4 polynucleotide kinase (NEB M0201S) and filled with pure water to 10 µL each. Oligos were phosphorylated by incubating reactions in a thermocycler for 30 min at 37°C and then denatured and annealed by incubating them for 5 min at 95°C and gradually decreasing (−5°C/min) temperature to 25°C.
Plasmids were digested for 4 h at 37°C in 35 µL reactions containing 5 µg of plasmid, 1.5 µL of each restriction enzyme needed, appropriate 1× buffer, and pure water for the balance volume. If restriction enzymes could be heat-deactivated, they were incubated for 20 min at 65°C/80°C, depending on enzyme requirements. Otherwise, reactions were bound to a silica column, washed twice with an ethanol-based wash, and eluted in 35 µL of pure water using a PCR purification kit (Thermo Fisher K310002). After heat inactivation or purification, digested plasmid was dephosphorylated by adding 140 U/mL Quick CIP (recombinant calf intestinal alkaline phosphatase; NEB M0525) and, if samples were purified, 1× appropriate buffer.
Ligation reactions were then set up with ∼5 nmol/µL digested plasmid, 1:200 annealed oligos, 1× T4 DNA ligase buffer, and 20,000 U/mL T4 DNA ligase (NEB M0525) and filled with pure water. Ligation reactions were incubated overnight at 16°C. Reactions excluding annealed oligos were used to test plasmid self-ligation efficiency.
Ligated plasmids were transformed into NEB stable competent E. coli by incubating 100 µL of competent cell stock with 23 µL of the ligation reaction for 10 min on ice followed by heat shock for 30 sec at 42°C and then incubation for 5 min on ice. The full volume of transformed bacteria was then spread evenly over LB agar containing antibiotic. Bacteria were incubated overnight at 37°C, and single colonies were picked and inoculated into 5 mL of LB broth. Liquid bacteria broth was then incubated overnight at 37°C. Cultures were collected via maximum-speed centrifugation, and plasmids were isolated from bacterial cell pellets. If needed, bacterial cultures were stored for up to 5 days at 4°C, and bacteria cell pellets were stored indefinitely at −20°C.
Plasmid isolation was performed using the GeneJET plasmid minipreparation kit (Thermo Fisher K0502) by resuspending bacterial cell pellets in resuspension solution with RNase A, lysing the cells with lysis solution, neutralizing the lysis with neutralization solution, and centrifuging at maximum speed for 15 min to pellet cell debris and chromosomal DNA. Supernatants were then transferred to GeneJET columns, and plasmids were bound to the silica membrane. Plasmids were washed twice with ethanol-based wash solution and then dried by centrifugation at maximum speed for 3 min. Plasmids were eluted in 50 µL of pure water. Plasmids were sent for Sanger sequencing at Azenta (formerly known as GeneWiz) to confirm successful insertion of shRNA sequences. A list of the shRNA targeting sequences used is in Supplemental Table S1.
The HA-SPT6 expression plasmid was cloned using NEB HiFi DNA assembly (E2621). The pULTRA-hot (Addgene 24130) plasmid was digested with EcoRI and BamHI and purified with PureLink Quick PCR purification kit (K310002). The SPT6 CDS was amplified from an SPT6 expression vector (provided as a gift by Lucas Farnung), and the fragment was gel-purified with PureLink Quick gel extraction kit (K210012; PCR primers are listed in Supplemental Table S1). The digested plasmid and purified fragment were assembled with a synthesized oligo (Supplemental Table S1), resulting in an HA tag added on the N terminus of human SPT6 inserted downstream from the mCherry-P2A sequence in pULTRA-hot (see Fig. 1G). pULTRA-hot without modification was used as a control.
Bulk RNA-seq data analysis
See the Supplemental Material for RNA-seq library generation. For bulk RNA-seq data analysis, we trimmed our PE150 reads with fastp (Chen 2023) (v0.23.2) using default settings. We then aligned reads to the mouse mm10 reference genome (Gencode M25 primary assembly) with STAR (v2.7) (Dobin et al. 2013), setting “‐‐outFilterMultimapNmax 20 ‐‐alignSJoverhangMin 8 ‐‐alignSJDBoverhangMin 1 ‐‐outFilterMismatchNmax 999 ‐‐outFilterMismatchNoverReadLmax 0.04 ‐‐alignIntronMin 20 ‐‐alignIntronMax 1000000 ‐‐alignMatesGapMax 1000000 ‐‐sjdbScore 1” according to ENCODE standards (Hitz et al. 2023). Read counts for individual genes were quantified with subread featureCounts (v2.0.3; setting “-B -O -p ‐‐countReadPairs -s 2 -C -g gene_id” for stranded, paired-end counting excluding multimapping reads) using the Gencode M25 primary assembly annotation file. Read counts were used for differential expression analysis with edgeR (v3.14) (Robinson et al. 2010). Briefly, analysis included filtering genes with very low counts, TMM normalization of library sizes, dispersion estimates, and general linear model QLF testing. Adjusted P-values were corrected with the Benjamini–Hochberg method. Differentially expressed genes (DEGs) were defined by at least twofold change with an FDR of <0.05. Gene set enrichment was performed by submitting gene symbol lists to the Enrichr (Kuleshov et al. 2016) web browser interface and extracting q-values from the enriched gene sets from the MSigDB Hallmark 2020 and TF perturbations followed by expression libraries. Data were then plotted with Prism. DEG expression data were plotted in R using ggplot2 and pheatmap.
For analysis of macrophage and neutrophil scores, genes enriched in primary GMPs, neutrophils, and macrophages were extracted from the Haemopedia RNA-seq database (Choi et al. 2019) through the Haemosphere web portal (https://www.haemosphere.org, v4.9.5). GMP, macrophage, and neutrophil gene sets were derived by extracting gene symbols from the High Expression Search function for “GMP,” “Mac,” and “NeutBM” cell types, respectively, which identified genes enriched in those cell types compared with all other cell types in the data set (Haemopedia mouse RNA-seq data set). Using these gene sets, scores were calculated by taking the average RPKM of each gene set for each condition and normalizing it to the average expression of genes in all three gene sets.
To determine expression of Fos and Jun family transcription factors in mouse bone marrow, we obtained raw paired-end reads from ENCODE's rRNA-depleted, total RNA-seq project (ENCSR654KWB) (Luo et al. 2020). Transcript abundance estimates were quantified by pseudoaligning the paired reads to a custom transcriptome containing the Gencode VM26 transcriptome (mm39) and mouse consensus transposable elements (RepBase 25.05) with kallisto quant (kb_python 0.27.3 [kallisto|bustools]). Abundance estimates were further analyzed with sleuth (v0.30.1) in R. A heat map of z-scaled TPMs normalized across cell types was generated in R with pheatmap (v.1.0.12).
10x Chromium single-cell RNA-seq data analysis
See the Supplemental Material for scRNA-seq library generation. Reads from iGMPs, SPT6 KD, and CAF-1 KD single cells were pseudoaligned to a custom transcriptome using kb count from kallisto | bustools (kb_python 0.27.3) (Melsted et al. 2021). The transcriptome was generated by combining the mm39 Gencode VM26 reference genome (primary assembly with comprehensive gene annotation) with consensus mouse transposable elements (RepBase 25.05) and with an eGFP coding sequence and indexing with kallisto. Downstream analysis was performed in R (v4.3) with Seurat (v5.1) (Hao et al. 2024). Initial quality control was performed using DropletUtils (v1.20.0) to remove background noise (empty droplets or ambient RNA) for each sample. Further quality control excluded cells with <1700 genes (potential dead/dying cells), >6500 genes (potential doublets), >40,000 UMIs (potential doublets), or mitochondrial gene percentage of >10%. After quality control, we analyzed 1419 cells for iGMPs, 1425 cells for CAF-1 KD, and 1220 cells for SPT6 KD. Raw reads for Tabula Muris bone marrow (Schaum et al. 2018) were obtained from GSE109774 and pseudoaligned using the same custom transcriptome but without eGFP coding sequences and filtered for quality control using DropletUtils (v1.20.0) to remove background noise, and cells with <1000 genes, >7000 genes, >60,000 UMIs, or mitochondrial gene percentage of >3% were removed. Following the Seurat pipeline, raw counts for each data set were log-normalized based on total reads per cell and scaled according to all detected genes. Then, principal component analysis (PCA) was performed for linear dimension reduction. Next, nearest-neighbor graph construction, clustering, and nonlinear dimension reduction were performed using the first 14 dimensions for iGMP scRNA-seq data and the first 30 dimensions for Tabula Muris bone marrow scRNA-seq data to capture the majority of the variation within each data set. For the Tabula Muris bone marrow scRNA-seq data set, the cell type for each cluster was annotated using scType (Ianevski et al. 2022). Specifically, clusters annotated as neutrophil, macrophage, classical monocytes, intermediate monocytes, erythroid-like, and erythroid precursor cells were subset for downstream analysis. Cell identity scores were generated using gene sets enriched in primary GMPs, neutrophils, or macrophages from Haemosphere (Choi et al. 2019), generated as described in “Bulk RNA-Seq Data Analysis.” These gene sets were validated for scRNA-seq by scoring the Tabula Muris data set (Supplemental Fig. S10B,C). To evaluate the iGMP, SPT6 KD, CAF-1 KD, and Tabula Muris data sets with GMP, neutrophil, and macrophage lineage scoring, AddModuleScore from Seurat was used to calculate the average expression level based on the enriched genes adjusted for average expression of randomly selected control genes for each cell. The following packages were used to generate plots: Seurat (v5.1), ComplexHeatMap (v2.16.0), and ggplot2 (v3.5.1).
For Fosl2 expression in bone marrow scRNA-seq, expression data were downloaded from the Tabula Muris (Schaum et al. 2018) web portal using FACS method and marrow tissue filters with the Fosl2 gene symbol (accessed June 2024).
ATAC-seq data analysis
See the Supplemental Material for ATAC-seq library generation. Reads were processed with the ENCODE Uniform Processing Pipeline (Hitz et al. 2023) to produce quality control data, read alignments, and signal tracks. For this pipeline, reads were trimmed with cutadapt and aligned against the mm10 reference genome (mm10 no alt analysis set ENCODE) using bowtie2 (v0.7.17) (Li and Durbin 2010). The pipeline filtered out reads mapping to the mitochondrial genome or to multiple locations and duplicate reads based on read start position. The pipeline then shifted reads to account for Tn5 insertions and called peaks with the filtered, shifted reads using MACS2, where individual reads were centered on the cut sites. MACS2 was also used for signal track generation; however, we modified the pipeline slightly to generate base-pair resolution counts per million signal files instead of the default fold change over background. For differential accessibility analysis, a unified set of peaks was created by merging the peaks from all conditions. For each sample, MACS3 callpeak was used on the Tn5 shifted reads, with individual reads centered on the cut site, using an FDR threshold of 0.01 (“‐‐nomodel ‐‐shift ‐‐extsize 150”). For each condition, peaks common between replicates were kept (bedtools intersect defaults) and then all peaks were merged into a single file (bedtools merge defaults), creating a unified peak set. ATAC-seq read counts were then calculated over these unified peaks with featureCounts (“-B -p ‐‐countReadPairs -s 0 -C”). Unified peaks were annotated with the closest gene using the R package ChIPseeker annotatePeak function, defining promoters as peaks within 1500 bp upstream of or 500 bp downstream from annotated TSSs. For annotations, the R libraries org.Mm.eg.db and TxDb.Mmusculus.UCSC.mm10.knownGene were loaded and used. Briefly, analysis included filtering genes with very low counts, TMM normalization of library sizes, dispersion estimates, and general linear model QLF testing. Adjusted P-values were corrected with the Benjamini–Hochberg method.
Differentially accessible regions (DARs) were defined by at least twofold change with an FDR of <0.05. ATAC-seq quantifications were plotted in R with ggplot2 and pheatmap. ATAC-seq footprinting was performed using the TOBIAS pipeline (Bentsen et al. 2020) with default settings. For TOBIAS, the filtered, deduplicated alignments were used as input files with all motifs downloaded from the JASPAR database (vertebrate nonredundant).
ChIP-seq data analysis
See the Supplemental Material for ChIP-seq library generation. FOSL2 ChIP-seq from bone marrow-derived macrophages was downloaded from GSE247941 (Abe et al. 2024). Paired-end (PE) reads for H3K4me3, H3K27ac, and FOSL2 ChIP-seq were processed with the ENCODE Uniform Processing Pipeline (Hitz et al. 2023) to produce quality control data, read alignments, and signal tracks. Fastq files for n = 2 (H3K4me3, FOSL2, and inputs) or n = 3 (H3K27ac and inputs) replicates were used with the pipeline. For this pipeline, reads were mapped to mm10 (mm10 no alt analysis set ENCODE; bowtie2, v2.2.9), duplicates and reads mapping to multiple locations were removed, and peaks were called with MACS2 (v2.2.6) using all default parameters for paired-end sequencing. The pipeline produced base-pair resolution signal over background fold change signal files for pooled replicates with MACS2. These signal files were used with deepTools computeMatrix (reference point or scale regions with “-missingDataAsZero ‐‐binSize 10”; other parameters were adjusted as presented in the figures) and plotHeatmap (default parameters) to generate heat maps and meta-analysis profiles.
Significance testing for FOSL2 binding over ATAC-seq DARs was performed by first summarizing mean signal over each set of DARs with deepTools multiBigwigSummary (default settings) using log2 fold enrichment signal tracks.
CUT&RUN/CUT&Tag data analysis
CUT&RUN and CUT&Tag assays were prepared as described previously (Skene et al. 2018; Kaya-Okur et al. 2019; Franklin et al. 2022). See the Supplemental Material for details. Paired-end (PE) fastqs were processed with the ENCODE Uniform Processing Pipeline (Hitz et al. 2023) to produce quality control data, read alignments, and signal tracks. Fastq files for n = 3 replicates (H3K4me1, H3K27me3, H3K9me3, and IgG) were used with the pipeline. For this pipeline, reads were mapped to mm10 (mm10 no alt analysis set ENCODE; bowtie2, v2.2.9), duplicates were removed, multimapping and discordant reads were excluded, and peaks were called with MACS2 using all default parameters for paired-end sequencing. The pipeline produced base-pair resolution signal over background fold change signal files for pooled replicates with MACS2. These were used with deepTools computeMatrix (reference point or scale regions with “-missingDataAsZero ‐‐binSize 10”; other parameters were adjusted as presented in the figures) and plotHeatmap (default parameters) to generate heat maps and meta-analysis profiles.
H3K27me3-marked regions (related to Fig. 5) were determined by quantifying CUT&RUN reads over 15 kb bins genome-wide with deepTools bamCoverage. Using reads per kilobase, the top 5% of regions was taken as H3K27me3-marked regions. H3K27me3 silenced genes were genes with an annotated TSS within these regions and filtered for genes with RNA-seq CPM <1 in iGMPs. H3K27me3 CUT&RUN results were confirmed with independent CUT&Tag assays.
Sequencing quality control
Quality control of ATAC-seq, RNA-seq, ChIP-seq, and CUT&RUN reads included analyzing reads for off-species enrichment, GC bias, and library complexity. All libraries were confirmed to lack contamination of off-target model species, E. coli (except for CUT&RUN E. coli spiked libraries), and mycoplasma using fastScreen with standard parameters. In cases where GC bias was present, all batched libraries were confirmed to have similar GC bias. All batched libraries were confirmed to have similar library complexity.
Data availability
The RNA-seq, ATAC-seq, and scRNA-seq data generated in this study have been made available in the Gene Expression Omnibus (GEO) under superseries GSE288727. ChIP-seq and CUT&RUN data that have been previously published in the GEO database are publicly available under superseries accession numbers GSE158229 and GSE247945. Genome assemblies used in this study (GRCm38/mm10) are publicly available and were downloaded from Gencode (M25 primary assembly) for RNA sequencing analysis or from ENCODE (mm10 no alt analysis set ENCODE) for DNA sequencing analysis. Gencode annotations (M25 primary assembly annotations) were downloaded for use with bulk RNA sequencing analysis. Gencode annotations and genome assembly for GRCm39/mm39 (M26 reference chromosome annotations and M26 primary assembly) were downloaded for use with single-cell RNA sequencing analysis.
Code availability
No custom programs were used in this analysis. Published pipelines are referenced and were installed according to the instructions. Default parameters were used, and consequential decisions in types of files supplied, databases, or sources are noted here. Tools and packages used for analyzing signal files, read counts, or other custom analysis are specified here, and consequential nondefault parameters are given. Inconsequential, nondefault parameters such as output file name, number of processors used, or plot colors are excluded. In addition, plot-specific, nondefault parameters that are provided in the figures, such as genomic window distance around TSSs in the metaplots, are not provided here.
Supplemental Material
Acknowledgments
We thank Melanie Oakes at University of California Irvine for support with sequencing, and Rachel Behar at the University of California Riverside Stem Cell Core and Mary Hamer at the University of California Riverside School of Medicine Core. We are grateful to members of the Cheloufi, Murn, Ninova, Sykes, and Volk laboratories for discussions and critical reading of the manuscript. We thank Bill Marzluff for guidance, Mikhail Trostnikov for technical help with image analysis, Lucas Farnung for providing the SPT6 expression plasmid, and Johannes Zuber for help with retroviral packaging reagents. We thank the Life Science Editors Foundation (LSEF) team and Francesca Mattiroli for critical reading and helpful suggestions. S.C., R.F., J.F., M.C., and B.Z. were supported by U.S. National Institutes of Health (NIH) grant R35 GM151004, the City of Hope/University of California Riverside Biomedical Research Initiative (CUBRI) and University of California Cancer Research Coordinating Committee (CRCC-C21CR2175) seed grants. R.F. and M.C. are supported by the TRANSCEND training grant from the California Institute of Regenerative Medicine (award EDUC4-12752). S.C. was supported by the LSEF Justice, Equity, Diversity, and Inclusion (JEDI) award. D.B.S. was supported by the American Society of Hematology, the St. Baldrick's Foundation, and the Children's Leukemia Research Association. J.M. was supported by NIH grant GM144693. M.N. and K.W. are supported by NIH grants R00HD099316 and R35GM151016. J.-S.R. was supported by the National Research Foundation of Korea (NRF; RS-2024-00338254) and the Brain Korea 21 (BK21) FOUR Program, awarded to Yonsei University. B.T.D. was supported by F30HL156404 from the National Heart, Lung, and Blood Institute. M.G.V.H. was supported by the Ludwig Center at Massachusetts of Institute of Technology (MIT), the MIT Center for Precision Cancer Medicine, and the National Cancer Institute (R35CA242379 and P30CA1405141). This work was made possible in part through access to the Genomics High-Throughput Facility Shared Resource of the Cancer Center Support Grant (P30CA-062203) at the University of California Irvine, and NIH shared instrumentation grants 1S10RR025496-01, 1S10OD010794-01, and 1S10OD021718-01.
Author contributions: S.C. conceived the study. S.C., R.F., S.P., J.F., M.C., and B.Z. performed and analyzed the experiments. R.F. and B.Z. performed the bioinformatics analysis. D.B.S. provided the iGMP lines and guidance. C.R.V. and J.-S.R. provided the histone chaperone shRNA library. K.W., M.N., and J.M. contributed key reagents and provided guidance. B.T.D. and M.G.V.H. contributed key data for replication stress response and genome-wide screen analysis and provided guidance. S.C., R.F., and B.Z. wrote the manuscript with input from all coauthors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.352316.124.
Freely available online through the Genes & Development Open Access option.
Competing interest statement
C.R.V. has received consulting fees from Flare Therapeutics, Roivant Sciences, and C4 Therapeutics; has served on the advisory boards of KSQ Therapeutics, Syros Pharmaceuticals, and Treeline Biosciences; has received research funding from Boehringer-Ingelheim and Treeline Biosciences; and owns stock in Treeline Biosciences. M.G.V.H. is a scientific advisor for Agios Pharmaceuticals, iTeos Therapeutics, Sage Therapeutics, Pretzel Therapeutics, Lime Therapeutics, Faeth Therapeutics, Droia Ventures, MPM Capital, and Auron Therapeutics. The othter authors declare no competing interests.
References
- Abe Y, Kofman ER, Ouyang Z, Cruz-Becerra G, Spann NJ, Seidman JS, Troutman TD, Stender JD, Taylor H, Fan W, et al. 2024. A TLR4/TRAF6-dependent signaling pathway mediates NCoR coactivator complex formation for inflammatory gene activation. Proc Natl Acad Sci 121: e2316104121. 10.1073/pnas.2316104121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoi Y, Shah AP, Ganesan S, Soliman SHA, Cho B-K, Goo YA, Kelleher NL, Shilatifard A. 2022. SPT6 functions in transcriptional pause/release via PAF1C recruitment. Mol Cell 82: 3412–3423.e5. 10.1016/j.molcel.2022.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avvakumov N, Nourani A, Côté J. 2011. Histone chaperones: modulators of chromatin marks. Mol Cell 41: 502–514. 10.1016/j.molcel.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Barlow JH, Faryabi RB, Callén E, Wong N, Malhowski A, Chen HT, Gutierrez-Cruz G, Sun H-W, McKinnon P, Wright G, et al. 2013. Identification of early replicating fragile sites that contribute to genome instability. Cell 152: 620–632. 10.1016/j.cell.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, Newell EW. 2019. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 37: 38–44. 10.1038/nbt.4314 [DOI] [PubMed] [Google Scholar]
- Bentsen M, Goymann P, Schultheis H, Klee K, Petrova A, Wiegandt R, Fust A, Preussner J, Kuenne C, Braun T, et al. 2020. ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation. Nat Commun 11: 4267. 10.1038/s41467-020-18035-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco MA, Sykes DB, Gu L, Wu M, Petroni R, Karnik R, Wawer M, Rico J, Li H, Jacobus WD, et al. 2021. Chromatin-state barriers enforce an irreversible mammalian cell fate decision. Cell Rep 37: 109967. 10.1016/j.celrep.2021.109967 [DOI] [PubMed] [Google Scholar]
- Bobkov GOM, Huang A, van den Berg SJW, Mitra S, Anselm E, Lazou V, Schunter S, Feederle R, Imhof A, Lusser A, et al. 2020. Spt6 is a maintenance factor for centromeric CENP-A. Nat Commun 11: 2919. 10.1038/s41467-020-16695-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortvin A, Winston F. 1996. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272: 1473–1476. 10.1126/science.272.5267.1473 [DOI] [PubMed] [Google Scholar]
- Burgess RJ, Zhang Z. 2013. Histone chaperones in nucleosome assembly and human disease. Nat Struct Mol Biol 20: 14–22. 10.1038/nsmb.2461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Hochedlinger K. 2017. Emerging roles of the histone chaperone CAF-1 in cellular plasticity. Curr Opin Genet Dev 46: 83–94. 10.1016/j.gde.2017.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Elling U, Hopfgartner B, Jung YL, Murn J, Ninova M, Hubmann M, Badeaux AI, Euong Ang C, Tenen D, et al. 2015. The histone chaperone CAF-1 safeguards somatic cell identity. Nature 528: 218–224. 10.1038/nature15749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. 2023. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. iMeta 2: e107. 10.1002/imt2.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-H, Keegan S, Kahli M, Tonzi P, Fenyö D, Huang TT, Smith DJ. 2019. Transcription shapes DNA replication initiation and termination in human cells. Nat Struct Mol Biol 26: 67–77. 10.1038/s41594-018-0171-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Zhang X, Wang Y, Gan H, Xu X, Lv X, Hua X, Que J, Ordog T, Zhang Z. 2019. Chromatin assembly factor 1 (CAF-1) facilitates the establishment of facultative heterochromatin during pluripotency exit. Nucleic Acids Res 47: 11114–11131. 10.1093/nar/gkz858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Baldwin TM, Wong M, Bolden JE, Fairfax KA, Lucas EC, Cole R, Biben C, Morgan C, Ramsay KA, et al. 2019. Haemopedia RNA-seq: a database of gene expression during haematopoiesis in mice and humans. Nucleic Acids Res 47: D780–D785. 10.1093/nar/gky1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das C, Tyler JK, Churchill MEA. 2010. The histone shuffle: histone chaperones in an energetic dance. Trends Biochem Sci 35: 476–489. 10.1016/j.tibs.2010.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGennaro CM, Alver BH, Marguerat S, Stepanova E, Davis CP, Bähler J, Park PJ, Winston F. 2013. Spt6 regulates intragenic and antisense transcription, nucleosome positioning, and histone modifications genome-wide in fission yeast. Mol Cell Biol 33: 4779–4792. 10.1128/MCB.01068-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koning L, Corpet A, Haber JE, Almouzni G. 2007. Histone chaperones: an escort network regulating histone traffic. Nat Struct Mol Biol 14: 997–1007. 10.1038/nsmb1318 [DOI] [PubMed] [Google Scholar]
- Do BT, Hsu PP, Vermeulen SY, Wang Z, Hirz T, Abbott KL, Aziz N, Replogle JM, Bjelosevic S, Paolino J, et al. 2024. Nucleotide depletion promotes cell fate transitions by inducing DNA replication stress. Dev Cell 59: 2203–2221.e15. 10.1016/j.devcel.2024.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drané P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. 2010. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev 24: 1253–1265. 10.1101/gad.566910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer J, Ricci G, van den Berg J, Bhardwaj V, Funk J, Armstrong C, van Batenburg V, Sine C, VanInsberghe MA, Tjeerdsma RB, et al. 2024. Acute multi-level response to defective de novo chromatin assembly in S-phase. Mol Cell 84: 4711–4728.e10. 10.1016/j.molcel.2024.10.023 [DOI] [PubMed] [Google Scholar]
- Dronamraju R, Hepperla AJ, Shibata Y, Adams AT, Magnuson T, Davis IJ, Strahl BD. 2018. Spt6 association with RNA polymerase II directs mRNA turnover during transcription. Mol Cell 70: 1054–1066.e4. 10.1016/j.molcel.2018.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh M, Zhu W, Hasegawa J, Watanabe H, Kim D-K, Aida M, Inukai N, Narita T, Yamada T, Furuya A, et al. 2004. Human Spt6 stimulates transcription elongation by RNA polymerase II in vitro. Mol Cell Biol 24: 3324–3336. 10.1128/MCB.24.8.3324-3336.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel KL, French SL, Viktorovskaya OV, Beyer AL, Schneider DA. 2015. Spt6 is essential for rRNA synthesis by RNA polymerase I. Mol Cell Biol 35: 2321–2331. 10.1128/MCB.01499-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust N, Varas F, Kelly LM, Heck S, Graf T. 2000. Insertion of enhanced green fluorescent protein into the lysozyme gene creates mice with green fluorescent granulocytes and macrophages. Blood 96: 719–726. 10.1182/blood.V96.2.719.014k29_719_726 [DOI] [PubMed] [Google Scholar]
- Filipescu D, Szenker E, Almouzni G. 2013. Developmental roles of histone H3 variants and their chaperones. Trends Genet 29: 630–640. 10.1016/j.tig.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Franklin R, Murn J, Cheloufi S. 2021. Cell fate decisions in the wake of histone H3 deposition. Front Cell Dev Biol 9: 654915. 10.3389/fcell.2021.654915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin R, Guo Y, He S, Chen M, Ji F, Zhou X, Frankhouser D, Do BT, Chiem C, Jang M, et al. 2022. Regulation of chromatin accessibility by the histone chaperone CAF-1 sustains lineage fidelity. Nat Commun 13: 2350. 10.1038/s41467-022-29730-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnan S, Liu Y, Spagnuolo M, Chen C-L. 2020. The impact of transcription-mediated replication stress on genome instability and human disease. Genome Instab Dis 1: 207–234. 10.1007/s42764-020-00021-y [DOI] [Google Scholar]
- Gopalakrishnan R, Winston F. 2021. The histone chaperone Spt6 is required for normal recruitment of the capping enzyme Abd1 to transcribed regions. J Biol Chem 297: 101205. 10.1016/j.jbc.2021.101205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A, Corpet A, Cook AJL, Roche D, Bartek J, Lukas J, Almouzni G. 2007. Regulation of replication fork progression through histone supply and demand. Science 318: 1928–1931. 10.1126/science.1148992 [DOI] [PubMed] [Google Scholar]
- Grover P, Asa JS, Campos EI. 2018. H3–H4 histone chaperone pathways. Annu Rev Genet 52: 109–130. 10.1146/annurev-genet-120417-031547 [DOI] [PubMed] [Google Scholar]
- Günesdogan U, Jäckle H, Herzig A. 2014. Histone supply regulates S phase timing and cell cycle progression. eLife 3: e02443. 10.7554/eLife.02443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond CM, Strømme CB, Huang H, Patel DJ, Groth A. 2017. Histone chaperone networks shaping chromatin function. Nat Rev Mol Cell Biol 18: 141–158. 10.1038/nrm.2016.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Stuart T, Kowalski MH, Choudhary S, Hoffman P, Hartman A, Srivastava A, Molla G, Madad S, Fernandez-Granda C, et al. 2024. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat Biotechnol 42: 293–304. 10.1038/s41587-023-01767-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog GA, Wada T, Handa H, Winston F. 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev 12: 357–369. 10.1101/gad.12.3.357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka Y, Inoue K, Oikawa M, Kamimura S, Ogonuki N, Kodama EN, Ohkawa Y, Tsukada Y, Ogura A. 2015. Histone chaperone CAF-1 mediates repressive histone modifications to protect preimplantation mouse embryos from endogenous retrotransposons. Proc Natl Acad Sci 112: 14641–14646. 10.1073/pnas.1512775112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Kim H, Huang R, Lu W, Tang M, Shi F, Yang D, Zhang X, Huang J, Liu D, et al. 2015. The Daxx/Atrx complex protects tandem repetitive elements during DNA hypomethylation by promoting H3K9 trimethylation. Cell Stem Cell 17: 273–286. 10.1016/j.stem.2015.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyd F, Chen R, Afshar K, Saba I, Lazure C, Fiolka K, Möröy T. 2011. The p150 subunit of the histone chaperone Caf-1 interacts with the transcriptional repressor Gfi1. Biochim Biophys Acta 1809: 255–261. 10.1016/j.bbagrm.2011.04.009 [DOI] [PubMed] [Google Scholar]
- Hitz BC, Lee J-W, Jolanki O, Kagda MS, Graham K, Sud P, Gabdank I, Strattan JS, Sloan CA, Dreszer T, et al. 2023. The ENCODE uniform analysis pipelines. bioRxiv 10.1101/2023.04.04.535623 [DOI] [Google Scholar]
- Ianevski A, Giri AK, Aittokallio T. 2022. Fully-automated and ultra-fast cell-type identification using specific marker combinations from single-cell transcriptomic data. Nat Commun 13: 1246. 10.1038/s41467-022-28803-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiuchi T, Enriquez-Gasca R, Mizutani E, Bošković A, Ziegler-Birling C, Rodriguez-Terrones D, Wakayama T, Vaquerizas JM, Torres-Padilla M-E. 2015. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat Struct Mol Biol 22: 662–671. 10.1038/nsmb.3066 [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Poitras C, Robert F. 2019. Histone recycling by FACT and Spt6 during transcription prevents the scrambling of histone modifications. Cell Rep 28: 1206–1218.e8. 10.1016/j.celrep.2019.06.097 [DOI] [PubMed] [Google Scholar]
- Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, Ahmad K, Henikoff S. 2019. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 10: 1930. 10.1038/s41467-019-09982-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp JP, Geisler MS, Hoover M, Cho C-Y, O'Farrell PH, Marzluff WF, Duronio RJ. 2024. Cell cycle-regulated transcriptional pausing of Drosophila replication-dependent histone genes. bioRxiv 10.1101/2024.12.16.628706 [DOI] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44: W90–W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Elsaesser SJ, Noh K-M, Stadler SC, Allis CD. 2010. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci 107: 14075–14080. 10.1073/pnas.1008850107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26: 589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Xu X, Tiwari M, Chen Y, Fuller M, Bansal V, Tamayo P, Das S, Ghosh P, Sen GL. 2021. SPT6 promotes epidermal differentiation and blockade of an intestinal-like phenotype through control of transcriptional elongation. Nat Commun 12: 784. 10.1038/s41467-021-21067-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebermann DA, Gregory B, Hoffman B. 1998. AP-1 (Fos/Jun) transcription factors in hematopoietic differentiation and apoptosis. Int J Oncol 12: 685–1385. 10.3892/ijo.12.3.685 [DOI] [PubMed] [Google Scholar]
- Liu L, Michowski W, Kolodziejczyk A, Sicinski P. 2019. The cell cycle in stem cell proliferation, pluripotency and differentiation. Nat Cell Biol 21: 1060–1067. 10.1038/s41556-019-0384-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Hitz BC, Gabdank I, Hilton JA, Kagda MS, Lam B, Myers Z, Sud P, Jou J, Lin K, et al. 2020. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res 48: D882–D889. 10.1093/nar/gkz1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah L-J, El-Osta A, Karagiannis TC. 2010. γH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia 24: 679–686. 10.1038/leu.2010.6 [DOI] [PubMed] [Google Scholar]
- Martell DJ, Merens HE, Caulier A, Fiorini C, Ulirsch JC, Ietswaart R, Choquet K, Graziadei G, Brancaleoni V, Cappellini MD, et al. 2023. RNA polymerase II pausing temporally coordinates cell cycle progression and erythroid differentiation. Dev Cell 58: 2112–2127.e4. 10.1016/j.devcel.2023.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martire S, Banaszynski LA. 2020. The roles of histone variants in fine-tuning chromatin organization and function. Nat Rev Mol Cell Biol 21: 522–541. 10.1038/s41580-020-0262-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Koreski KP. 2017. Birth and death of histone mRNAs. Trends Genet 33: 745–759. 10.1016/j.tig.2017.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiroli F, D'Arcy S, Luger K. 2015. The right place at the right time: chaperoning core histone variants. EMBO Rep 16: 1454–1466. 10.15252/embr.201540840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melsted P, Booeshaghi AS, Liu L, Gao F, Lu L, Min K(, da Veiga Beltrame E, Hjörleifsson KE, Gehring J, Pachter L. 2021. Modular, efficient and constant-memory single-cell RNA-seq preprocessing. Nat Biotechnol 39: 813–818. 10.1038/s41587-021-00870-2 [DOI] [PubMed] [Google Scholar]
- Mendiratta S, Gatto A, Almouzni G. 2019. Histone supply: multitiered regulation ensures chromatin dynamics throughout the cell cycle. J Cell Biol 218: 39–54. 10.1083/jcb.201807179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CLW, Winston F. 2023. The conserved histone chaperone Spt6 is strongly required for DNA replication and genome stability. Cell Rep 42: 112264. 10.1016/j.celrep.2023.112264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CLW, Warner JL, Winston F. 2023. Insights into Spt6: a histone chaperone that functions in transcription, DNA replication, and genome stability. Trends Genet 39: 858–872. 10.1016/j.tig.2023.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narain A, Bhandare P, Adhikari B, Backes S, Eilers M, Dölken L, Schlosser A, Erhard F, Baluapuri A, Wolf E. 2021. Targeted protein degradation reveals a direct role of SPT6 in RNAPII elongation and termination. Mol Cell 81: 3110–3127.e14. 10.1016/j.molcel.2021.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng C, Aichinger M, Nguyen T, Au C, Najar T, Wu L, Mesa KR, Liao W, Quivy J-P, Hubert B, et al. 2019. The histone chaperone CAF-1 cooperates with the DNA methyltransferases to maintain Cd4 silencing in cytotoxic T cells. Genes Dev 33: 669–683. 10.1101/gad.322024.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara EAA, Aguilar-Morante D, Rasmussen RD, Frias A, Vitting-Serup K, Lim YC, Elbæk KJ, Pedersen H, Vardouli L, Jensen KE, et al. 2020. SPT6-driven error-free DNA repair safeguards genomic stability of glioblastoma cancer stem-like cells. Nat Commun 11: 4709. 10.1038/s41467-020-18549-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oqani RK, Lin T, Lee JE, Kang JW, Shin HY, Il Jin D. 2019. Iws1 and Spt6 regulate trimethylation of histone H3 on lysine 36 through Akt signaling and are essential for mouse embryonic genome activation. Sci Rep 9: 3831. 10.1038/s41598-019-40358-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardal AJ, Fernandes-Duarte F, Bowman AJ. 2019. The histone chaperoning pathway: from ribosome to nucleosome. Essays Biochem 63: 29–43. 10.1042/EBC20180055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivy J-P, Gérard A, Cook AJL, Roche D, Almouzni G. 2008. The HP1-p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nat Struct Mol Biol 15: 972–979. 10.1038/nsmb.1470 [DOI] [PubMed] [Google Scholar]
- Ray-Gallet D, Almouzni G. 2021. The histone H3 family and its deposition pathways. Adv Exp Med Biol 1283: 17–42. 10.1007/978-981-15-8104-5_2 [DOI] [PubMed] [Google Scholar]
- Ray-Gallet D, Woolfe A, Vassias I, Pellentz C, Lacoste N, Puri A, Schultz DC, Pchelintsev NA, Adams PD, Jansen LET, et al. 2011. Dynamics of histone H3 deposition in vivo reveal a nucleosome Gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol Cell 44: 928–941. 10.1016/j.molcel.2011.12.006 [DOI] [PubMed] [Google Scholar]
- Replogle JM, Saunders RA, Pogson AN, Hussmann JA, Lenail A, Guna A, Mascibroda L, Wagner EJ, Adelman K, Lithwick-Yanai G, et al. 2022. Mapping information-rich genotype–phenotype landscapes with genome-scale Perturb-seq. Cell 185: 2559–2575.e28. 10.1016/j.cell.2022.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaum N, Karkanias J, Neff NF, May AP, Quake SR, Wyss-Coray T, Darmanis S, Batson J, Botvinnik O, Chen MB, et al. 2018. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562: 367–372. 10.1038/s41586-018-0590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel CA, Jacobs Y, Cleary ML. 2000. HoxA9-mediated immortalization of myeloid progenitors requires functional interactions with TALE cofactors Pbx and Meis. Oncogene 19: 608–616. 10.1038/sj.onc.1203371 [DOI] [PubMed] [Google Scholar]
- Skene PJ, Henikoff JG, Henikoff S. 2018. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc 13: 1006–1019. 10.1038/nprot.2018.015 [DOI] [PubMed] [Google Scholar]
- Smith S, Stillman B. 1989. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 58: 15–25. 10.1016/0092-8674(89)90398-X [DOI] [PubMed] [Google Scholar]
- Soufi A, Dalton S. 2016. Cycling through developmental decisions: how cell cycle dynamics control pluripotency, differentiation and reprogramming. Development 143: 4301–4311. 10.1242/dev.142075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes DB, Kfoury YS, Mercier FE, Wawer MJ, Law JM, Haynes MK, Lewis TA, Schajnovitz A, Jain E, Lee D, et al. 2016. Inhibition of dihydroorotate dehydrogenase overcomes differentiation blockade in acute myeloid leukemia. Cell 167: 171–186.e15. 10.1016/j.cell.2016.08.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2016. A decade of transcription factor-mediated reprogramming to pluripotency. Nat Rev Mol Cell Biol 17: 183–193. 10.1038/nrm.2016.8 [DOI] [PubMed] [Google Scholar]
- Vo DT, Fuller MR, Tindle C, Anandachar MS, Das S, Sahoo D, Ghosh P. 2021. SPT6 loss permits the transdifferentiation of keratinocytes into an intestinal fate that resembles Barrett's metaplasia. iScience 24: 103121. 10.1016/j.isci.2021.103121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk A, Liang K, Suraneni P, Li X, Zhao J, Bulic M, Marshall S, Pulakanti K, Malinge S, Taub J, et al. 2018. A CHAF1B-dependent molecular switch in hematopoiesis and leukemia pathogenesis. Cancer Cell 34: 707–723.e7. 10.1016/j.ccell.2018.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos SM, Farnung L, Linden A, Urlaub H, Cramer P. 2020. Structure of complete Pol II–DSIF–PAF–SPT6 transcription complex reveals RTF1 allosteric activation. Nat Struct Mol Biol 27: 668–677. 10.1038/s41594-020-0437-1 [DOI] [PubMed] [Google Scholar]
- Wang GG, Calvo KR, Pasillas MP, Sykes DB, Häcker H, Kamps MP. 2006. Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nat Methods 3: 287–293. 10.1038/nmeth865 [DOI] [PubMed] [Google Scholar]
- Wang AH, Zare H, Mousavi K, Wang C, Moravec CE, Sirotkin HI, Ge K, Gutierrez-Cruz G, Sartorelli V. 2013. The histone chaperone Spt6 coordinates histone H3K27 demethylation and myogenesis. EMBO J 32: 1075–1086. 10.1038/emboj.2013.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AH, Juan AH, Ko KD, Tsai P-F, Zare H, Dell'Orso S, Sartorelli V. 2017. The elongation factor Spt6 maintains ESC pluripotency by controlling super-enhancers and counteracting polycomb proteins. Mol Cell 68: 398–413.e6. 10.1016/j.molcel.2017.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Zhou H, Nadorp B, Cayanan G, Chen X, Yeaton AH, Nomikou S, Witkowski MT, Narang S, Kloetgen A, et al. 2021. Surface antigen-guided CRISPR screens identify regulators of myeloid leukemia differentiation. Cell Stem Cell 28: 718–731.e6. 10.1016/j.stem.2020.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylishen AR, Sun C, Moyer SM, Qi Y, Chau GP, Aryal NK, McAllister F, Kim MP, Barton MC, Estrella JS, et al. 2020. Daxx maintains endogenous retroviral silencing and restricts cellular plasticity in vivo. Sci Adv 6: eaba8415. 10.1126/sciadv.aba8415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoh SM, Cho H, Pickle L, Evans RM, Jones KA. 2007. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev 21: 160–174. 10.1101/gad.1503107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoh SM, Lucas JS, Jones KA. 2008. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev 22: 3422–3434. 10.1101/gad.1720008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman MK, Cimprich KA. 2014. Causes and consequences of replication stress. Nat Cell Biol 16: 2–9. 10.1038/ncb2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang Y, Qiao P, Wang X, Yu R, Sun H, Xing X, Zhang Y, Su J. 2022. CHAF1b, chromatin assembly factor-1 subunit b, is essential for mouse preimplantation embryos. Int J Biol Macromol 195: 547–557. 10.1016/j.ijbiomac.2021.11.181 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq, ATAC-seq, and scRNA-seq data generated in this study have been made available in the Gene Expression Omnibus (GEO) under superseries GSE288727. ChIP-seq and CUT&RUN data that have been previously published in the GEO database are publicly available under superseries accession numbers GSE158229 and GSE247945. Genome assemblies used in this study (GRCm38/mm10) are publicly available and were downloaded from Gencode (M25 primary assembly) for RNA sequencing analysis or from ENCODE (mm10 no alt analysis set ENCODE) for DNA sequencing analysis. Gencode annotations (M25 primary assembly annotations) were downloaded for use with bulk RNA sequencing analysis. Gencode annotations and genome assembly for GRCm39/mm39 (M26 reference chromosome annotations and M26 primary assembly) were downloaded for use with single-cell RNA sequencing analysis.